Abstract
Objectives
The goals of this study were to determine the role of OCT3 in the pharmacologic action of metformin and to identify and functionally characterize genetic variants of OCT3 with respect to the uptake of metformin and monoamines.
Methods
For the pharmacologic studies, we evaluated metformin-induced activation of AMPK, a molecular target of metformin. We used quantitative PCR and immunostaining to localize the transporter and isotopic uptake studies in cells transfected with OCT3 and its nonsynonymous genetic variants for functional analyses.
Results
Quantitative PCR and immunostaining showed that OCT3 was expressed high on the plasma membrane of skeletal muscle and liver, target tissues for metformin action. Both the OCT inhibitor, cimetidine, and OCT3-specific shRNA significantly reduced the activating effect of metformin on AMPK. To identify genetic variants in OCT3, we used recent data from the 1000 Genomes Project and the Pharmacogenomics of Membrane Transporters project. Six novel missense variants were identified. In functional assays, using various monoamines and metformin, 3 variants, T44M (c.131C>T), T400I (c.1199C>T) and V423F (c.1267G>T), showed altered substrate specificity. Notably, in cells expressing T400I and V423F, the uptakes of metformin and catecholamines were significantly reduced but the uptakes of metformin, MPP+ and histamine by T44M were significantly increased more than 50%. Structural modeling suggested that these two variants may be located in the pore-lining (T400) or proximal (V423) membrane-spanning helixes.
Conclusion
Our study suggests that OCT3 plays a role in the therapeutic action of metformin and that genetic variants of OCT3 may modulate metformin and catecholamine action.
Keywords: Organic cation transporters, monoamines, metformin, pharmacogenomics, muscle cells
Introduction
Organic cation transporters (OCTs) participate in the disposition of a variety of cationic substances including endogenous amines and xenobiotics in tissues such as liver, kidney, and placenta (1–6). In addition to this general detoxification function, the OCT family has also been proposed to represent Uptake2, a catecholamine removal system found in peripheral tissues with sympathetic innervations (4). Although OCT1 (SLC22A1) and OCT2 (SLC22A2) seem to be restricted mainly to excretory organs such as the liver and kidney, OCT3 (SLC22A3) shows a much broader tissue distribution, including skeletal muscle, heart, brain and placenta. A significant contribution in the handling of organic cations at the periphery was confirmed recently in mice in which the Orct3/Slc22a3 gene was disrupted. These mice showed impaired Uptake2 activity in brain, heart and in embryos (3, 7). A recent study indicates that OCT3 is critical in the physiological compensation of serotonin transport in the brain of knockout mice of the serotonin transporter (8). Several genome-wide association studies have linked OCT3 to the risk loci for coronary artery disease and prostate cancer (9–11).
Metformin is first line therapy for the treatment of Type 2 Diabetes (T2D) and is among the most widely prescribed drugs in the U.S.(12). Previous studies suggest that organic cation transporters, OCT1 and OCT2, play a critical role in the disposition and response to metformin and that genetic variants of these transporters are associated with variation in pharmacokinetics and anti-diabetic activity of the drug (6, 13–15). MATE1 has also been shown to play a role in metformin disposition (14, 16, 17). To date, however, there have been few studies of the role of OCT3 in metformin uptake and activity (18). Further, there have been no reports of the effects of genetic variants of OCT3 on metformin uptake, disposition or pharmacologic action. However, functionally significant genetic variants of OCT3 in the coding and proximal promoter have been identified (19, 20) and associated with human disease. In particular, in a case-control study of 84 Caucasian children and adolescents with obsessive-compulsive disorder (OCD), a non-synonymous variant of OCT3, M370I, which had a 40% decline of transport capacity for norepinephrine and a genetic variant in the promoter region g.−81G>delGA of OCT3, which showed increased luciferase activity, were associated with OCD(19).
In this study, we first determined that OCT3 modulated the pharmacologic action of metformin on its target, AMPK, and showed that the transporter was highly expressed in the skeletal muscle, one of the major sites of metformin action. Following these studies, we comprehensively examined the coding region of OCT3 using data from the 1000 Genomes Project and direct sequencing of a large DNA sample (n=247) from ethnically diverse healthy volunteers to identify all OCT3 nonsynonymous (missense) variants. We then assessed the functional significance of these nonsynonymous variants by investigating their transport function, expression, and subcellular localization and analyzed the effects of the variants on OCT3 structure-function relationships by structural modeling. We discovered several natural missense variants with significant functional effects. In particular, the uptake of metformin and catecholamine were modulated by several of the variants. Based on its tissue expression pattern, our studies suggest that OCT3 may play a role in skeletal muscle as one of the determinants of the pharmacologic effect of metformin (21).
Methods
Chemicals
All radiolabeled chemicals were purchased from American Radiolabeled Chemicals Inc. (St Louis, MI) or PerkinElmer (Boston, MA). The specific activity of each radiolabeled chemical was: [3H] MPP+ (1-methyl-4-phenylpyridinium) (85.5 mCi/mmol), [3H] Histamine (14.2 mCi/mmol), [3H] Norepinephrine (44.7 mCi/mmol), [3H] Serotonin (20.3 mCi/mmol), [3H] Dopamine (32.7 mCi/mmol), [3H] Epinephrine (80.0 mCi/mmol), [3H] Choline (76.1 mCi/mmol), [3H] Tyramine (50 mCi/mmol), [14C] TEA (Tetraethylammonium) (50 mCi/mmol) and [14C] metformin (26 mCi/mmol). HEK293 Flp-In Cells, Lipofectamine 2000 and pcDNA5/FRT expression vector were purchased from Invitrogen (Carlsbad, CA). BCA Protein Assay Kit was purchased from Pierce Thermo Fisher (Rockford, IL). Unlabeled chemicals were purchased from Sigma (St Louis, MI). Cell culture supplies were purchased from the Cell Culture Facility (UCSF, CA). All other chemicals were of reagent grade and commercially available.
Determination of mRNA expression level in tissues and cells
Total RNA from various human tissues or cell lines’ was purchased from Clontech (Palo Alto, CA) or ScienCell Research Labortories (Carlsbad, CA) and reverse-transcribed PCR was used Superscript III Reverse Transcriptase (Invitrogen) according to the manufacturer's protocols. The resulting cDNA was used as template for quantitative real-time PCR using Taqman Gene Expression Assays for human transporter OCT1 (assay ID: Hs00427550_m1), OCT2 (assay ID: Hs00533907_m1), OCT3 (assay ID: Hs01009568_m1), and human glyceraldehyde-3-phosphate dehydrogenase (assay ID: Hs99999905_m1). Quantitative real-time PCR was carried out in 96-well reaction plates in a volume of 10 µL using the Taqman Fast Universal Master Mix (Applied Biosystems). Reactions were run on the Applied Biosystems 7500 Fast Real-Time PCR System with the following profile: 95°C for 20 s followed by 40 cycles of 95°C for 3 s and 60°C for 30 s. The relative expression of each mRNA was calculated by the comparative method (ΔΔCt method). Firstly, the ΔCt values for each samples were obtained by subtracting the Ct value of glyceraldehyde-3-phosphate dehydrogenase mRNA from the Ct value of the target mRNA. Then, the ΔΔCt values for each sample were obtained by subtracting the highest ΔCt value obtained for the sample (which in this case is sample “OCT2 in smooth muscle”) from the ΔCt value of each sample. The mRNA expression reported as relative difference was calculated using the arithmetic formula 2−ΔΔCt (ABI Prism 7700 Sequence Detection system User Bulletin No. 2, P/N 4303859). For validation of shRNA inhibition of OCT3 expression in human primary muscle cells by electrophoresis, specific RT-PCR primers (forward: 5’-CAGCAGACAGGTATGGCAGG-3’ and reverse: 5’-GGAAGATCACAAACACAGGGAAGT -3’) were used. β-actin was used as control.
Metformin treatment in skeletal muscle cells
Human primary skeletal muscle cells from Lonza (Walkersville, MD) were cultured in the F-10 medium, supplemented with penicillin (100 U/ml), streptomycin (100 µg/ml), 20% fetal bovine serum (Invitrogen) and 2.5 ng/ml bFGF (Sigma). Cells were grown in collagen-coated 10 cm plates. Before treatment, cells at 70% confluency, were adapted with serum free medium with bFGF for 24 hours (22). For the cimetidine inhibition study, cells were treated for 2 hours as follows in reduced serum medium Opti-MEM (Invitrogen): Control without any treatment but solvent DMSO (10 µL), metformin only (1.5 mM), cimetidine only (1 mM) and metformin (1.5 mM) plus cimetidine (1 mM). For the OCT3 specific shRNA inhibition study, the HuSH 29mer shRNA Constructs against SLC22A3 in pGFP-V-RS vector (Catalog No. TG301651) was purchased from Origene (Rockville, MD). Cells were transfected 24 hours with tranfection mixture following the manufacture’s manual (lipofectamine 2000 with empty pGFP-V-RS (4µg) or pGFP-V-RS-shRNA (4µg) in the 6-well plate respectively). Cells were treated for 6 hours as follows in Opti-MEM medium: Control with empty pGFP-V-RS, metformin only (1.5 mM) plus empty pGFP-V-RS, shRNA only (4µg) and metformin (1.5 mM) plus shRNA. After 2 hours treatment, cells were rinsed 3 times in cold PBS and then harvested by a cell harvester (BD Bioscience) in 2 ml cold PBS. Cold PBS was removed after centrifuging at 500 × g for 5 minutes. Cell pellets were subject to the western blotting assay.
Western blotting and immunostaining
Cultured cells or human liver tissue (Asterand, Detroit, MI) were lysed using a tissue homogenizer at 4°C for 20 minutes in the RIPA buffer (Sigma) with the protease inhibitors dissolved from complete protease inhibitor cocktail tablet (Roche Applied Science). After centrifugation for 20 minutes at 16,100 g at 4°C, the supernatants were removed for determination of protein content and separated on 10% SDS-PAGE gels. 40 micrograms of proteins from the supernatant were separated and transferred to PVDF membrane. The membranes were blocked overnight at 4°C with Tris-buffered saline with 0.05% Tween 20 and 5% nonfat milk. Immunoblotting was performed following standard procedures, and the signals were detected by chemiluminescence reagents (GE healthcare, NJ). Primary antibodies were directed against: AMPKα (1:1000), AMPKα phosphorylated at Thr172 (1:500), GFP (1:2000), β-actin (1:2000) and Na+/K+ ATPase (1:1000) (Cell Signaling Technology, Danvers, MA), and hOCT3 (1:1000) (GenWay, San Diego, CA). For the quantification of western blot bands, the ImageJ method was used which was downloaded from http://rsb.info.nih.gov/ij/index.html. The quantification method followed the manual of the software. Each band from anti-hOCT3 was also normalized to its loading control anti-Na+/K+ ATPase. For the immunostaining of tissue section, paraffin-embedded normal human tissue slides was purchased from IMGENEX (San Diego, CA). Following the supplier’s protocol, the slides was placed in a rack, and the following washes were performed: 1. Xylene: 2 × 3 min; 2. Xylene 1:1 with 100% ethanol: 3 min; 3. 100% ethanol: 2 × 3 min; 4. 95% ethanol: 3 min; 5. 70 % ethanol: 3 min; 6. 50 % ethanol: 3 min; 7. Running cold tap water to rinse. Prior to antibody application, sections were steamed in 0.01M citric acid buffer (pH 6.0) for 15 min then cooled in 1X PBS for 15 min at room temperature. Sections were blocked in 10% lamb serum and 0.5% triton-X100 for 45 min then incubated in rabbit anti-OCT3 (1:100) (GenWay) for 1 hour or overnight at 4 °C. Following antibody incubation, sections were washed in PBS then incubated for 1 hour in Alexafluor 488 goat anti-rabbit (1:500) (Invitrogen) then placed on coverslips. Immunofluorescent images were captured using a Retiga CCD-cooled camera and associated QCapture Pro software (QImaging Surrey, BC Canada).
Identification of OCT3 genetic variants
OCT3 variants were identified by direct sequencing of genomic DNA as described previously from DNA samples collected from an ethnically diverse population of 247 individuals (100 African Americans, 100 European Americans, 30 Asian-Americans, 10 Mexican Americans and 7 Pacific Islanders). The study protocol was reviewed and approved by the Committee on the Human Research at UCSF. The reference cDNA sequence of OCT3 was obtained from GenBank (http://www.ncbi.nlm.nih.gov, accession number NM_021977.2). Primers were designed manually to span the exons and 50–200 bp of flanking intronic sequence per exon. The primer sequences can be found at http://www.pharmgkb.org. The sequencing data of 1000 Genomes Project were obtained from http://pharmacogenetics.ucsf.edu/ which combined the sequencing data from the Pharmacogenetics of Membrane Transporters Database and the 1000 Genomes Project. 1000 genome data is from the 2009_02 release of Pilot 1 data for three ethic groups (CEU, Caucasians in Utah; JPT/CHB, Japanese in Tokyo and Chinese in Beijing; YRI, Yoruba in Ibadan in Nigeria) which is based on human build 36.3.
Construction of genetic variants of OCT3
Human OCT3 cDNA (GenBank accession number NM_021977) was cloned from human prostate cDNA library and then subcloned into the mammalian expression vector pcDNA5/FRT or modified GFP-tagged pcDNA5/FRT at C-terminus to obtain OCT3 reference (wild-type) or GFP-OCT3, which corresponds to the highest-frequency amino-acid sequence in all ethnic groups. The reference sequence clone was used as the template for mutagenesis. Variant cDNA clones were constructed by site-directed mutagenesis of the reference clone using Pfu Turbo DNA polymerase (Invitrogen). Sequences of variant cDNA clones were confirmed by direct sequencing, and the full cDNA of each variant was sequenced to verify that only the intended mutation was introduced.
Transport studies
HEK-293 Flip-in cells were cultured in Dulbecco's modified Eagle's medium (DME H-21, UCSF Cell Culture Facility), supplemented with penicillin (100 U/ml), streptomycin (100 µg/ml), and 10% fetal bovine serum. For all of the reference and variants of OCT3, detailed functional studies were performed using stably transfected Flp-In-293 cells generated according to the manufacturer's protocol (Invitrogen).
All substrates were labeled with 3H except TEA and metformin, which were labeled with 14C. 14C labeled metformin and TEA had a concentration at 35 µM. Time course studies were performed to verify the linear range uptake of each substrate. Later a 1-minute uptake was performed with each compound except metformin for which the uptake was determined at 5 minutes. Data are shown as mean ± SD from 3 repeated experiments and each experiment was performed in triplicate. Results are expressed as the percent of activity of the OCT3-reference. Transport of radiolabeled substrates was tested in at least three separate experiments for all variants. Kinetics studies were performed with varying concentrations of unlabeled substrates added to the uptake buffer. Initial rates of uptake (V), expressed as pmol/min/mg protein, were fit to the equation: V=Vmax*[S]/(Km+[S])+Kdiff*S where Kdiff represents a diffusion constant for non-OCT3-mediated uptake, [S] is the substrate concentration, and Vmax and Km are the Michaelis-Menten kinetic parameters.
Green fluorescent protein fusion constructs
To visualize subcellular localization of OCT3 variants, variant cDNA constructs were subcloned in frame with green fluorescent protein (GFP) at the C terminus of the pcDNA5/FRT expression vector (23). For the colocalization study, the Image-iT™ LIVE Plasma Membrane and Nuclear Labeling Kit (Invitrogen) was used with red-fluorescent Alexa Fluor® 594 wheat germ agglutinin (WGA) to specifically label the plasma membrane. Cells, plated on poly-D-lysine-coated glass coverslips (BD Biosciences, San Diego, CA), were visualized using a Retiga CCD-cooled camera and associated QCapture Pro software (QImaging Surrey, BC Canada).
Biotinylation of the cell surface
The biotinylation of the HEK cell surface proteins was performed with the Pierce Cell Surface Protein Isolation Kit (Pierce). Briefly, each of the HEK cells from 10 cm2 culture plates was biotinylated with 10 mL sulfo-NHS-SS-biotin solution in PBS. Plates were placed in a rocking platform or orbital shaker and gently agitated for 30 minutes at 4°C. Reactions were stopped by adding quench solution. The cells were scraped from the plate bottom in 10 mL of PBS containing 490 mM oxidized glutathione. The cells were pelleted and lysed for 30 min on ice with 1 mL of RIPA lysis buffer with protease inhibitor cocktails (Sigma). The cell lysate was cleared by centrifugation (16,100g, 10 min). 500 µl clarified cell lysate was added to NeutrAvidin Agarose slurry. Unbound proteins were removed by washing buffers. Captured proteins were eluted from streptavidin sepharose by incubation with 400 µl SDS-PAGE sample buffer with 50 mM DTT for 60 minutes on an end-over-end rotator at room temperature. Samples were eluted by centrifuging for 2 minutes at 1,000 × g. The eluted samples were subject to immunoblotting assays.
Structural modeling
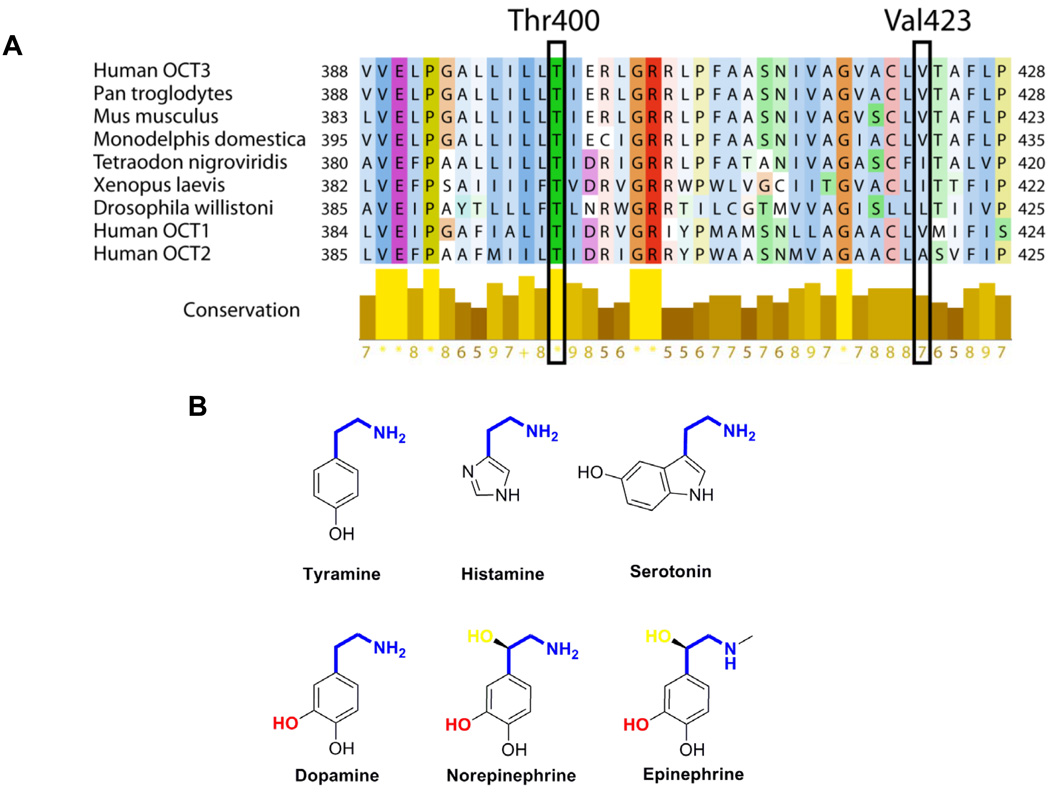
We created two different models for OCT3 (Fig. S3, Supplemental Digital Content 1, http://links.lww.com/FPC/A210). The coordinates of the first model (Fig. S3A and B, Supplemental Digital Content 1, http://links.lww.com/FPC/A210) were downloaded from MODBASE (24), a database of annotated comparative protein structure models. Briefly, models in MODBASE were calculated by MODPIPE, an automated system for comparative protein structure modeling (24) relying on various modules of MODELLER (25). Each model in MODBASE is predicted to have the correct fold based on a composite score calculated from the coverage of the modeled sequence, sequence identity with the template, the fraction of gaps in the alignment, the compactness of the model, and several statistical potential scores (24). This OCT3 model (Fig S3A and B, Supplemental Digital Content 1, http://links.lww.com/FPC/A210) was based on the crystal structure of the glycerol-3-phosphate transporter from E. coli (GlpT, PDB identifier 1PW4) (26). While the sequence similarity between the template structure and OCT3 is not high (i.e., 18% sequence identity), we verified the fold of the model using a variety of additional fold assignment methods such as Phyre (27); this method ranked 1PW4 as their top hit. Furthermore, they agreed on the alignment in the region that included Thr400. The model was assessed by Z-DOPE, which is a normalized atomic distance-dependent statistical potential based on known protein structures (28). The Z-DOPE score of the model was −1.08, where a score lower than −1 indicates a “reliable” model (ie, 80% of its Cα atoms are within 3.5 Å of their correct positions). The second OCT3 model (Fig. S3C–D, Supplemental Digital Content 1, http://links.lww.com/FPC/A210) was based on the crystal structure of lactose permease (LacY) from E. coli (PDB identifier 2CFQ) (29, 30). 100 models were generated based on a comprehensive alignment between LacY and OCTs from various organisms (30), using the standard ‘automodel’ routine of MODELLER (25). The models were then assessed using Z-DOPE, where the score of the highest-ranked model was 0.59. While such a score generally does not indicate a reliable model, this model is based on a reliable alignment that was verified experimentally for the rOCT1 model (30); in addition, our scoring scheme was developed for globular proteins and is less reliable for assessing membrane proteins. For the multiple sequence alignment (Fig. 6A), sequences of representative OCT3 homologs were downloaded from NCBI in June 2009. Their multiple sequence alignment was obtained by MUSCLE (31), and visualized using Jalview. The aligned residues were colored based on their type (using the “Clustlx” color scheme in Jalview) and their level of conservation (the more conserved the residue in a given position, the stronger the color).
Figure 6. Structural analysis of OCT3 genetic variants, T400 and V423F.
A. Multiple sequence alignments of OCT3 in various species and with its orthologs, human OCT1 and OCT2. The proteins in each species are known as OCT3 or predicted organic cation transporters. T400 and V423F are highlighted in the frame. The color scheme in the protein sequence is based on the amino acid residues and the score under the yellow bar indicates their conservation rate at their position with conserved residues labeled as *. B. Chemical structures of endogenous monoamine substrates of OCT3 are illustrated here. The common shared moiety, ethylamine is shown as blue and the unique hydroxyl group in the ethylamine of norepinephrine and epinephrine is colored as yellow. The catecholamine specific hydroxyl group in the phenyl ring is indicated as red.
Statistical analysis
Data are expressed as mean ± the standard deviation (Mean ± SD). For statistical analysis, multiple comparisons were analyzed using one-way analysis of variance followed by Dunnett’s two-tailed test. OCT3 reference was the basis for comparison unless stated otherwise. The data were analyzed using GraphPad Prism 4.0 (GraphPad Software Inc.). A P value less than 0.05 was considered statistically significant.
Results
OCT3 tissue expression pattern and immunostaining
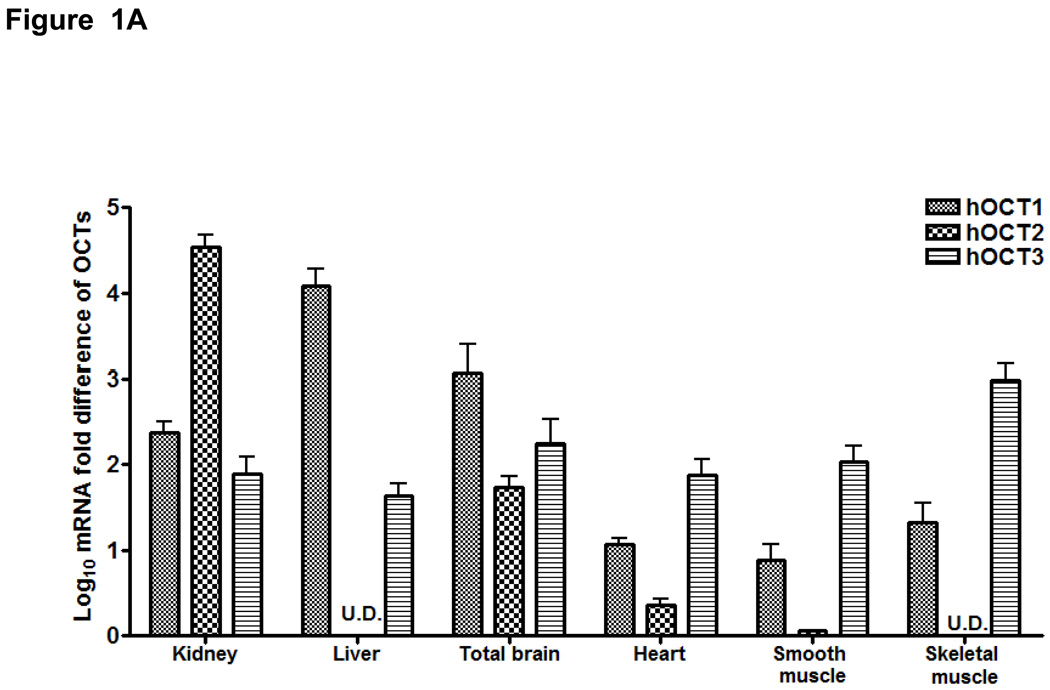
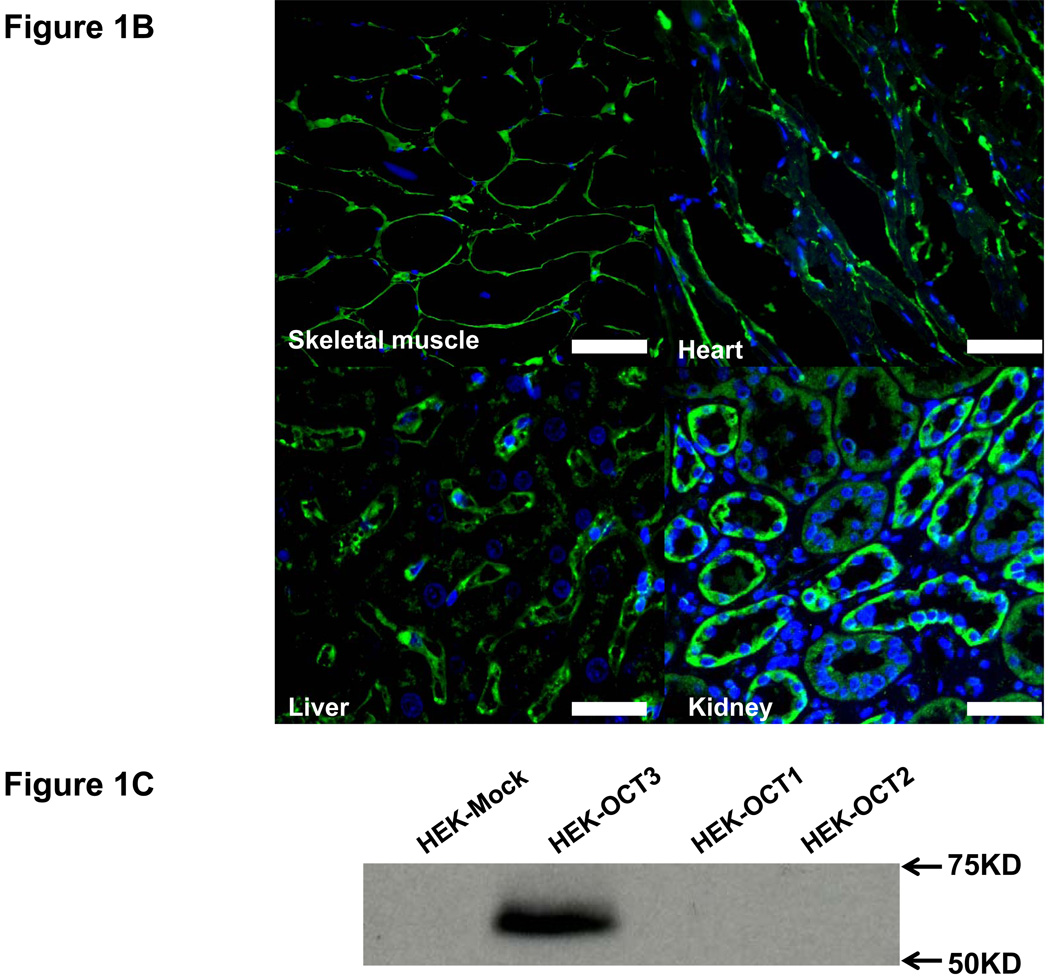
We screened 44 tissues for the expression level of OCT3 mRNA transcripts (Fig. 1S, Supplemental Digital Content 2, http://links.lww.com/FPC/A211). Among them, prostate had the highest expression level consistent with a previous report (32). Other major OCT3 expression sites were cardiac myocytes, skeletal muscle, placenta, uterus, liver, adrenal gland, salivary gland and appendix. The rest of the tissues showed modest or low expression compared with the above sites. Notably, cardiac myocytes was a new tissue type identified with high OCT3 expression. To elucidate the biological and pharmacological role of OCT3 relative to its paralogs, we compared the expression level of hOCT1-3 in various tissues using qRT-PCR (Fig. 1A). As expected, OCT1 was highly expressed in the liver whereas OCT2 was mainly expressed in the kidney and slightly detectable or undetectable in the skeletal muscle, liver and smooth muscle. In contrast, OCT3 was much higher than OCT1/2 in muscle tissues, especially in skeletal muscle. Immunostaining showed that OCT3 expression was strong at the plasma membrane of skeletal muscle and heart (Fig. 1B). In kidney and liver, immunostaining indicated the presence of OCT3 in proximal tubules and on the sinusoidal membrane of the hepatocytes, respectively. Western blotting also showed that OCT3 antibody was very specific without cross activity with OCT1/2 (Fig. 1C); i.e., a single band around 60 kD was detected only in HEK-OCT3 but not in HEK-OCT1/2. These data suggest that OCT3 plays a role in the disposition of endogenous monoamines and drugs in liver, kidney, skeletal muscle and heart.
Figure 1. Tissue distribution of OCT3 and its paralogs.
A. mRNA expression levels of OCT1, OCT2 and OCT3 in various tissues. OCT1-3 mRNA expression was normalized to hGAPDH expression for each tissue. Data are shown as mean ± SD from a single experiment with triplicate wells and plotted as log. U.D. (undetermined). B. Immunofluorescence of human skeletal muscle, heart, kidney and liver sections probed with rabbit anti-OCT3 at 1:100 and detected with 2nd antibody Alexa Fluor® 488 (green). Nuclei were stained with DAPI (blue). Scale bar: 50 µm. C. OCT3 antibody specificity tested in HEK-Mock, HEK-OCT1-3.
Effect of metformin on AMPK in skeletal muscle cells
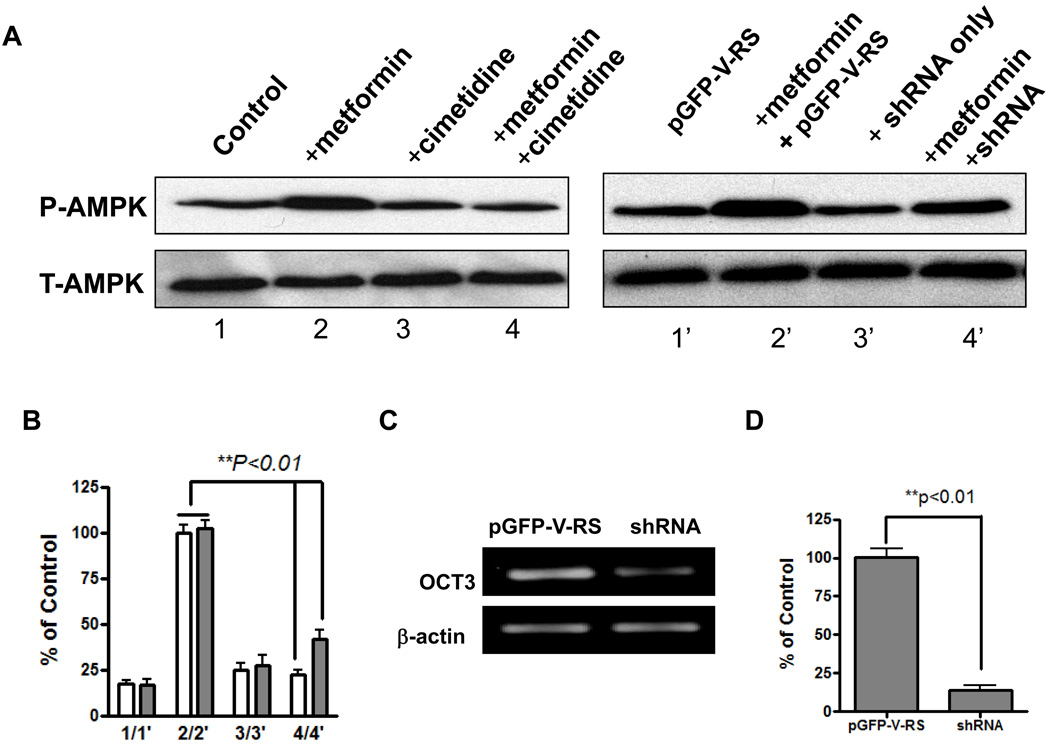
Metformin activates AMPK in the liver and skeletal muscle, which results in lower plasma glucose levels and increased rates of synthesis of glycogen (21, 33, 34). To explore the role of OCT3 in the action of metformin in the skeletal muscle, we performed an AMPK activation study with metformin (Fig. 2). Adult primary human muscle cells grown in culture were treated with metformin in the presence or absence of the OCT inhibitor cimetidine, and the phosphorylation of AMPK α2 Thr172 was measured. Without the OCT inhibitor, cimetidine, metformin significantly increased the phosphorylation of AMPK α2 Thr172 as compared with control cells without treatment. When metformin was applied along with cimetidine, AMPK phosphorylation level was similar to the control cells (without treatment) (Fig.2A and B). Because cimetidine is a general inhibitor of OCTs, OCT3 specific shRNA was applied to silence the specific expression of OCT3 in the muscle cells. The co-application of metformin and OCT3 shRNA significantly reduced AMPK phosphorylation compared with metformin alone. OCT3 shRNA, however, did not reduce AMPK activation to the same extend as cimetidine, implying that other transporters (e.g.,OCT1) may also play a role in the muscle cell uptake of metformin (Fig. 2A and B). The reduction on AMPK phosphorylation by cimetidine and OCT3 specific shRNA were about 80% and 60%, respectively. The knock-down of OCT3 expression by shRNA in human primary muscle cells was validated by RT-PCR (Fig. 2C and 2D). Both the electrophoresis and TaqMan quantitative RT-PCR indicated that the mRNA of OCT3 was substantially silenced by its shRNA. There was only 18% remaining in the shRNA treated sample compared with the one treated with empty vector (Fig.2D).
Figure 2. Metformin activation of AMPK in skeletal muscle cells.
A. Western blot assay of AMPK activation mediated by OCT3 in human primary skeletal muscle cells. Metformin (1.5 mM) stimulated AMPK α2 (T-AMPK) Thr172 phosphorylation (P-AMPK) in human adult primary skeletal muscle cell and that the OCT inhibitor, cimetidine (1 mM) and shRNA, inhibited its action on AMPK phosphorylation. pGFP-V-RS: empty vector without shRNA. B, Densitometry of immunoblotting assays. The bar chart represents the intensity of each band (P-AMPK) normalized to the total AMPK based on 3 independent experiments. C, Validation of shRNA knock-down of mRNA expression level of OCT3 in human primary skeletal muscle cells by RT-PCR. D. TaqMan quantitative PCR analysis of OCT3 mRNA expression level in shRNA or empty vector treated human primary skeletal muscle cells.
Identification of novel OCT3 variants
In the 247 DNA samples analyzed by direct sequencing, 13 nucleotide substitutions in the survey region of the OCT3 gene, which included the proximal promoter region, the coding region and 50–200 bp of flanking intronic sequence for each exon, were identified. Only 2 of these variants had been previously reported. The nucleotide positions and allele frequencies of the variants are shown in Table 1. Of the 13 variants identified, 7 occurred in the coding region, and 6 were found in the noncoding regions of the sequence interrogated in our screen. Variants in intronic regions mainly occurred in the African-American sample at low allele frequencies. Only IVS1 (+85) and IVS5 (+12) were detected at higher frequencies, i.e., 14.2% and 10%, respectively. Of the 4 synonymous variants identified (Table 1), 2 were highly polymorphic: R120R (c.360T>C) which was found in all ethnic groups at varying allele frequencies; and A411A (c.1233G>A) with an allele frequency of 45 % in the Asian sample, and lower allele frequencies in other ethnic groups. A411A (c.1233G>A) might serve as a cryptic splice acceptor site (19, 20). The 3 nonsynonymous variants included 3 amino-acid substitutions that occurred at very low allele frequencies (Table 1). T44M (c.131C>T) was found as singleton in African and Caucasian American samples but at presumably higher frequencies in Pacific Islanders (14.5%) though the sample set was much smaller (n = 7 samples or 14 chromosomes). A116S (c.346G>T) had an allele frequency of 1.7% in the African American sample but was not present in other ethnic groups. T400I (c.1199C>T) was a singleton in the European American sample. For the nsSNPs from 1000 Genomes Project (Table 2), A116S (c.346G>T) was only found in the YRI sample at 5.4% allele frequency which is higher than the direct sequencing result (1.7%). L186F (c.558G>T) had allele frequencies of 7.6% in Chinese and Japanese (CHB/JPT) DNA samples and 4.5% in African (YRI) samples. V388M (c.1162G>A) was a singleton in the Caucasian (CEU) group and L423F (c.1267G>T) had an allele frequency at 6.8% only in samples from Chinese and Japanese (CHB/JPT).
Table 1.
Genetic variants of OCT3 (SLC22A3) identified in 247 DNA samples from the Pharmacogenomics of Membrane Transporters Project
| dbSNP ID numbers |
Genomic position |
CDS position |
Nucleotide substitution |
Amino acid substitution |
Allele Frequency | ||||
|---|---|---|---|---|---|---|---|---|---|
| AA (n=200) |
EA (n=200) |
AS (n=60) |
ME (n=20) |
PA (n=14) |
|||||
| Coding variants | |||||||||
| rs68187715 | 160689572 | 131 | C>T | T44M | 0.006 | 0.006 | 0 | 0 | 0.143 |
| rs8187716 | 160689693 | 252 | C>A | P84P | 0.011 | 0 | 0 | 0 | 0 |
| rs8187717 | 160689787 | 346 | G>T | A116S | 0.017 | 0 | 0 | 0 | 0 |
| rs668871 | 160689801 | 360 | T>C | R120R | 0.417 | 0.494 | 0.617 | 0.5 | 0.571 |
| rs8187725 | 160778144 | 1199 | C>T | T400I | 0 | 0.005 | 0 | 0 | 0 |
| rs2292334 | 160778178 | 1233 | G>A | A411A | 0.126 | 0.364 | 0.446 | 0.35 | 0.5 |
| rs8187722 | 160784748 | 1494 | A>G | L498L | 0.025 | 0 | 0 | 0 | 0 |
| Intronic variants | |||||||||
| rs8187718 | 160689955 | IVS1(+85) | C>T | None | 0.142 | 0 | 0 | 0 | 0 |
| rs8187719 | 160749973 | IVS4(+30) | A>G | None | 0.01 | 0 | 0 | 0 | 0 |
| rs8187720 | 160751880 | IVS5(+12) | C>T | None | 0.1 | 0.005 | 0 | 0 | 0 |
| rs8187723 | 160777922 | IVS6(+23) | T>C | None | 0 | 0 | 0 | 0 | 0.071 |
| rs8187724 | 160777939 | IVS6(+40) | A>C | None | 0.03 | 0.01 | 0 | 0 | 0 |
| rs8187721 | 160783908 | IVS8(+17) | T>G | None | 0.005 | 0 | 0 | 0 | 0 |
ID numbers in bold indicate non-synonymous variants which were studied in this investigation. CDS (nucleotide) position is given relative to the ‘A’ in the ATG start codon for OCT3. AA, African American; AS, Asian American; EA, European American; ME, Mexican-American; PA, Pacific Islander; n = number of chromosomes examined.
Table 2.
Nonsynonymous variants of OCT3 (SLC22A3) identified in three populations in the 1000 Genomes Project
| dbSNP ID numbers |
Genomic position |
Amino Acid position |
CDS position |
Nucleotide change |
Amino Acid change |
CEU Freqency (n=57) |
CHB /JPT Freqency (n=59) |
YRI Freqency (n=56) |
|---|---|---|---|---|---|---|---|---|
| rs8187717 | 160689787 | 116 | 346 | G > T | Ala >Ser | 0 | 0 | 0.054 |
| N/A | 160748087 | 186 | 558 | G > A | Leu >Phe | 0 | 0.076 | 0.045 |
| N/A | 160778107 | 388 | 1162 | G > T | Val >Met | 0.009 | 0 | 0 |
| N/A | 160778212 | 423 | 1267 | G > T | Val >Phe | 0 | 0.068 | 0 |
ID numbers in bold indicate non-synonymous variants which were studied in this investigation. CEU, Caucasians in Utah; CHB/JPT, Chinese in Beijing and Japanese in Tokyo; YRI, Yoruba in Ibadan in Nigeria; n = number of chromosomes examined;
Functional effects of OCT3 nonsynonymous variants on model substrate and monoamines
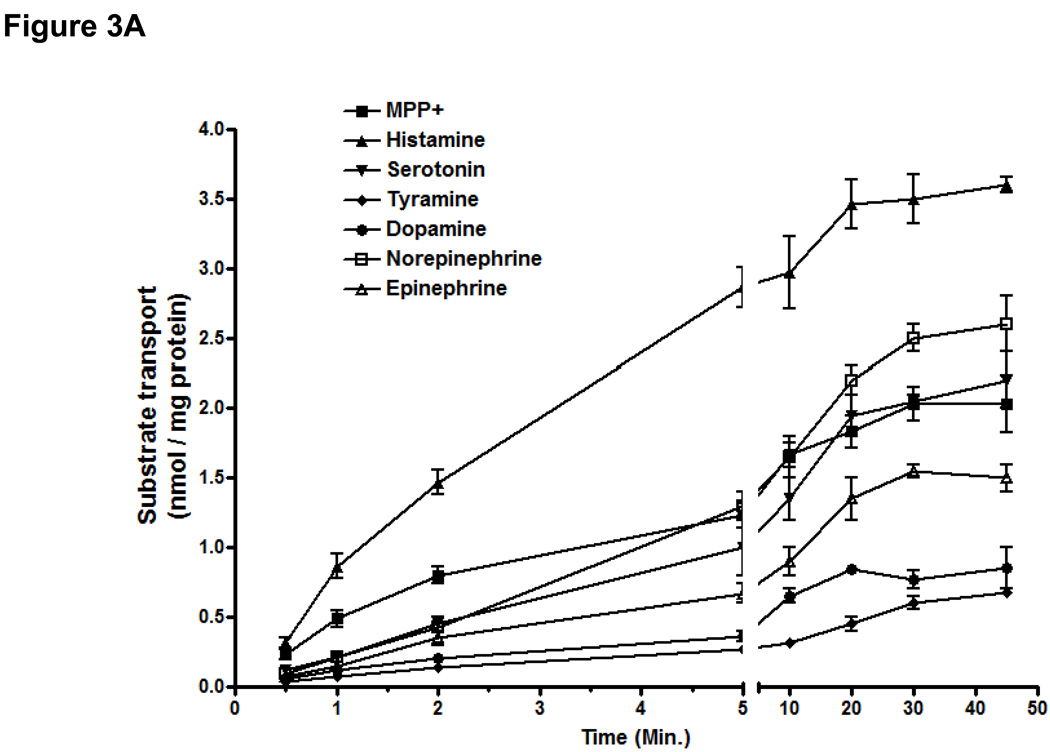
The time course of the uptakes of a diverse array of known or unknown substrates of OCT3 was examined in cells stably expressing the OCT3 reference (Fig. 3A). All the substrate uptakes were time dependent and linear at 1 minute or less, which allowed us to determine the kinetic parameters for each substrate. MPP+ and histamine had higher turnover rates than other monoamines. The uptake efficiency (the ratio of Vmax (nmol/mg protein/min) / Km (nM) were as follow: MPP+ (21.0: 1783 ± 98 / 85 ± 10)> histamine (11.0: 1498 ± 61 / 131 ± 57) > serotonin (3.8: 889 ± 61 / 236 ± 31), norepinephrine (3.4: 1051 ± 183 / 305 ± 42) > tyramine (2.3: 641 ± 61 / 281 ± 36) > dopamine (1.5: 658 ± 152 / 443 ± 56), epinephrine (1.2: 597 ± 72 / 480 ± 65) (Table 3A). OCT3 showed highest transport efficiency for histamine among the monoamines. TEA uptake at 5 minute (0.32 nmol/mg protein/min) was enhanced approximately 3-fold over empty vector and was not considered a substrate in another report (data not shown) (35).
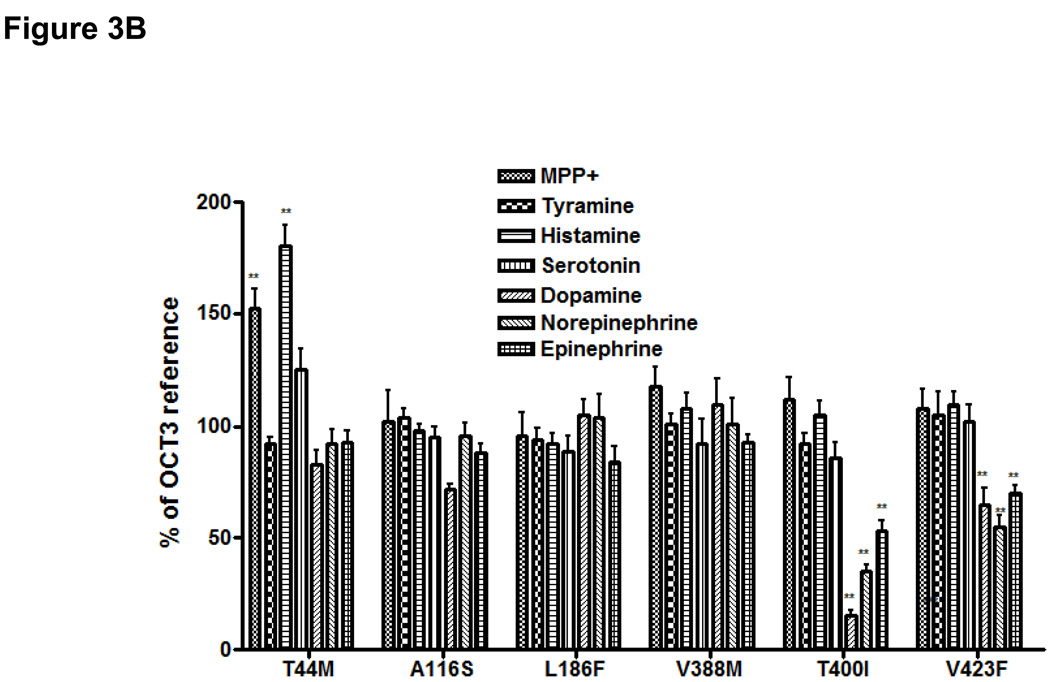
Figure 3. Uptake of various endogenous amines and MPP+ in HEK293 cells stably expressing OCT3 and 6 missense variants.
A. The time course of uptake of monoamines and MPP+. B. Substrate selectivity of genetic variants of OCT3 on model substrate and monoamines. Data are shown as mean ± SD from 3 repeated experiments and each experiment was performed in triplicate. OCT3-reference is used as control set at 100% for each substrate. **p<0.01 versus OCT3 – reference.
Table 3.
Kinetic parameters for substrates transported by OCT3 and its nonsynonymous variants
| A | ||||
|---|---|---|---|---|
| Substrates | OCT3-Ref (Km/Vmax) | T44M (Km/Vmax) | T400I (Km/Vmax) | V423F (Km/Vmax) |
| MPP+ | 85 ± 10 /1783 ± 98 | 94 ± 16 / 2126 ± 162* | 65 ± 18 / 1865 ± 125 | 62 ± 13 / 1914 ± 125 |
| Tyramine | 281 ± 36 / 641± 61 | 242 ± 62 / 628± 85 | 303 ± 87 / 592 ± 43 | 259± 53 / 574± 82 |
| Histamine | 131 ± 57 / 1498± 104 | 164 ± 174 / 1822 ± 113* | 115± 26 / 1469 ± 164 | 129 ± 26 / 1413 ± 114 |
| Serotonin | 236 ± 31 / 889 ± 69 | 252 ± 51 / 964 ± 128 | 219 ± 21 / 997 ± 107 | 195 ± 21 / 843 ± 92 |
| Dopamine | 443± 56 / 658 ± 152 | 456 ± 140 / 507 ± 22 | 1292 ± 54* / 572 ± 44 | 1037 ± 83* / 472 ± 61 |
| Norepinephrine | 305 ± 42 / 1051 ± 183 | 445 ± 111 / 965 ± 65 | 908 ± 138* / 1080 ± 87 | 832 ± 138* / 1027 ± 103 |
| Epinephrine | 480 ± 65 / 597 ± 72 | 397 ± 85 / 507 ± 41 | 850 ± 110 */ 541 ± 56 | 712 ± 153* / 518 ± 53 |
| B | |||||||
|---|---|---|---|---|---|---|---|
| Kinetic parameter | OCT3-Ref. | T44M | A116S | L186F | V388M | T400I | V423F |
| Vmax (nmol/mg protein/min) | 4.72 ± 0.54 | 6.43 ± 0.39 * | 4.36 ± 0.37 | 4.56 ± 0.73 | 4.94 ± 0.47 | 3.36 ± 0.54 | 3.52 ± 0.53 |
| Km(mM) | 1.09 ± 0.21 | 1.15 ± 0.43 | 1.02 ± 0.23 | 1.04 ± 0.15 | 1.13 ± 0.36 | 3.81 ± 0.42* | 2.95 ± 0.21* |
For the kinetic studies, uptake rates were determined at 1 minute except metformin which was determined at 5 minutes. OCT3-Ref., OCT3-reference;
p <0.05 versus the reference. Values represent the mean ± SD calculated from 3 separate experiments and experiment was performed in triplicate. A, Kinetic parameters of MPP+ and monoamines by reference OCT3 and its nonsynonymous variants. The unit of Vmax, pmol/mg protein/min; the unit of Km, nM; B, Kinetic parameters of metformin of OCT3 and its variants.
The OCT3 genetic variants were studied for their potential differences in uptake of diverse substrates (Fig. 3B). At 1-minute, the uptake of MPP+ and histamine were enhanced more than 50% in cells expressing T44M in comparison to cells expressing the OCT3 reference (Fig. 3B). A116S, L186F and V388M exhibited a similar function to the reference for all of the substrates tested. T400I and V423F showed an interesting pattern of uptake for the various monoamines. For example, in comparison to cells expressing the reference OCT3, cells expressing T400I and V423F exhibited a greatly reduced uptake of dopamine (15% and 63% of OCT3 reference, respectively, **P<0.01), norepinephrine (41% and 50% of OCT1-reference, respectively, **P<0.01) and epinephrine (52% and 67% of OCT1-reference, respectively, **P<0.01).
Effects of OCT3 and its variants on metformin uptake and kinetics
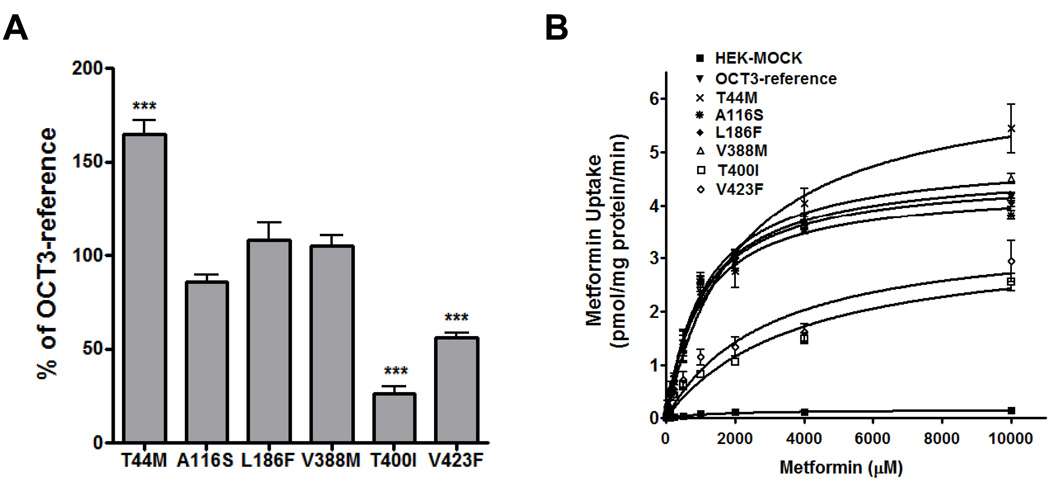
The accumulation of metformin was time dependent and substantially increased in cells stably transfected with reference OCT3 consistent with Nie et al. (18) (Fig. S2A, Supplemental Digital Content 3 http://links.lww.com/FPC/A212). OCT3 had a similar Km (1.09 ± 0.21 mM) and a slightly increased Vmax (4.72 ± 0.54 nmol/mg protein/min) to that of OCT1 (1.18 ± 0.18 mM and 3.96 ± 0.44 nmol/mg protein/min, respectively) (Table 1S, Supplemental Digital Content 4, http://links.lww.com/FPC/A213 and Fig. S2B, Supplemental Digital Content 3 http://links.lww.com/FPC/A212). As reported previously (6, 36), OCT2 had a much higher metformin uptake capacity than OCT1/3 with Vmax (10.41 ± 0.72 nmol/mg protein/min) (Fig. S2B, Supplemental Digital Content 3 http://links.lww.com/FPC/A212 and Table 1S, http://links.lww.com/FPC/A213). Metformin uptake was increased about 60% (***P<0.001) in cells expressing the T44M variant in comparison with those expressing the reference OCT3 (Fig. 4A). A116S, L186F and V388M showed a similar uptake of metformin whereas the uptake of metformin by T400I and V423F decreased about 80% and 50%, respectively (***P<0.001). Five minutes was selected for kinetic studies as the uptake was in the linear range and the assay was sufficiently sensitive to accurately detect radiolabeled metformin. As shown in Fig. 4B and Table 3B, in cells expressing T44M, metformin exhibited a similar Km (1.15 ± 0.43 mM vs 1.09 ± 0.21 mM for the OCT3-reference) and a significantly increased Vmax (6.43 ± 0.39 vs 4.72 ± 0.54 nmol/mg protein/min for the OCT3-reference, *P<0.05) in comparison to cells expressing reference OCT3. Variant A116S, L186F and V388M showed a similar Vmax and Km to the reference. The Km values of metformin were significantly increased about 2.5 to 3.5 -fold in cells expressing T400I and V423F in comparison to the reference OCT3 (3.81 ± 0.42 mM and 2.95 ± 0.21mM, respectively, *P<0.05) but the Vmax values were slightly reduced (3.36 ± 0.54 and 3.52 ± 0.53 nmol/mg protein/min, respectively) (Table 3B).
Figure 4. Metformin uptake and kinetics in HEK293 cells expressing OCT3 and its genetic variants.
A, Metformin uptake in HEK293 cells expressing reference OCT3 and 6 missense variants. OCT3-reference is used as control for each substrate. ***p< 0.001 versus OCT3–reference. B. Metformin kinetics in HEK293 cells expressing reference OCT3 and 6 missense variants. Michaelis-Menten parameters were calculated and are shown in Table 3B.
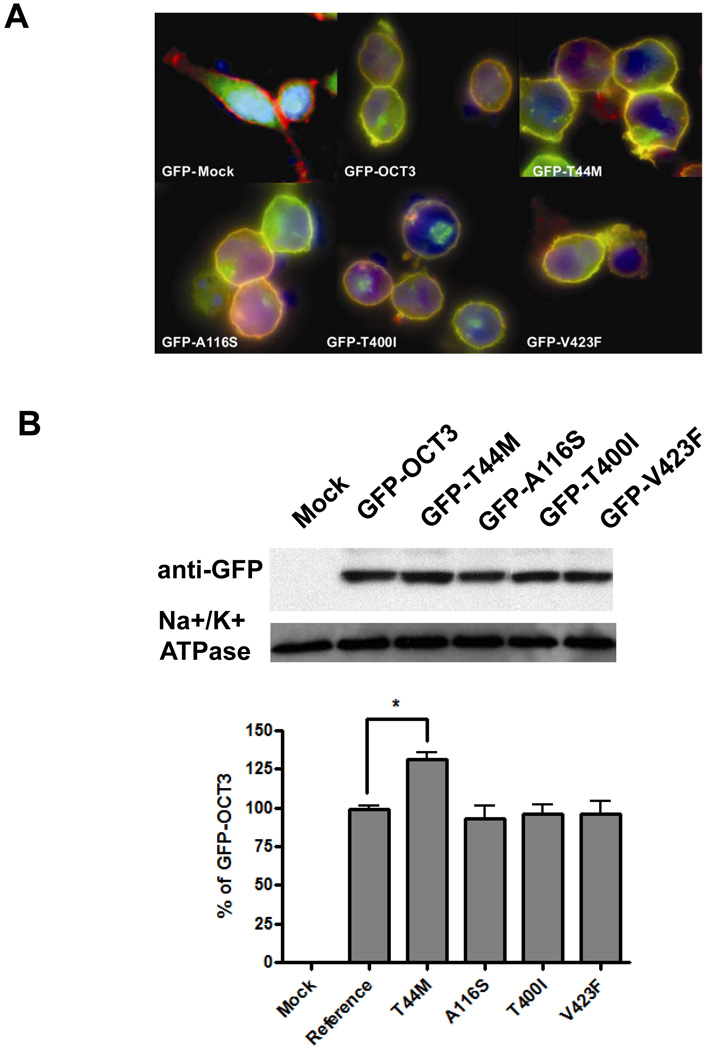
Subcellular localization and expression level of GFP-tagged fusion protein
HEK293 cells were stably transfected with GFP-tagged OCT3 reference and nonsynonymous variants (T44M, A116S, T400I and V423F). GFP-Mock (GFP vector only), was located in both the cytoplasm and nucleus, and was not co-localized with the plasma membrane (Fig.5A). Fluorescence microscopy showed that the majority of GFP-derived signal of the reference OCT3 was restricted to the plasma membrane and was colocalized with red-fluorescent labeled wheat germ agglutinin resulting in an orange color (Fig. 5A). There was also a small fraction of green fluorescence present in the cytoplasm. None of the variants appeared to affect the subcellular localization of OCT3. A slightly stronger fluorescence signal was detected in the cells expressing GFP-T44M (panel T44M) in comparison to the cells expressing GFP-OCT3 reference. The expression level of GFP-tagged OCT3 and 4 non-synonymous variants (T44M, A116S, T400I and V423F) were further assayed from biotinylated cell plasma membrane with Western blotting using GFP antibody. The expression of T44M was increased about 30% above the OCT3 reference. The rest of the variants had a similar expression level on the plasma membrane as the reference OCT3 (Fig. 5B).
Figure 5. Subcellular localization and expression quantification of GFP-tagged various OCT3.
A. Subcellular localization of GFP tagged OCT3 reference and 4 nonsynonymous variants (GFP-T44M, GFP-A116S, GFP-T400I and GFP-V423F). Various OCT3 GFP fusion constructs were stably expressed in HEK-293 cells and visualized by fluorescent microscopy. The nucleus is stained with DAPI (blue). The plasma membrane was stained using AlexaFluor 594-labeled wheat germ agglutinin (red) for co-localization staining. GFP-OCT3 protein is shown in green. B. Western blotting assay of GFP-Mock, GFP-tagged OCT3-reference and 4 variants above. Mouse monoclonal anti- Na+/K+ ATPase served as loading control. The GFP-OCT3 position is around 100 kD shifted from the 60 kD position of OCT3 itself. The bar chart represents the intensity of each band normalized to the Na+/K+ ATPase band relative to the GFP-OCT3-reference based on 3 independent assays.
Discussion
The results of this study have important implications for the role of OCT3 in response to the anti-diabetic drug, metformin. Metformin is widely used as a first-line therapy for the treatment of type 2 diabetes (33). The action of metformin appears to be related to its activation (phosphorylation) of the so-called energy sensor, AMP-activated protein kinase (AMPK), which results in suppression of glucagon-stimulated glucose production and enhancement of glucose uptake in muscle and in hepatic cells (21, 37). In hepatic cells, we previously demonstrated that the OCT3 paralog, OCT1, is a determinant of metformin activity in the liver, a major site of action of the drug (13). However, though metformin is known to have action in skeletal muscle (34), the transporter(s) responsible for metformin uptake and action in skeletal muscle is largely not known. Data in the current study suggests that OCT3, which unlike OCT1/2, has a broader expression pattern, is a one of the determinants of metformin action in skeletal muscle. First, we confirmed that metformin is a substrate of OCT3, which was also recently shown by Anne et al.(18). Further, our quantitative RT-PCR studies revealed that OCT3 is expressed at higher levels than OCT1/2 in muscle type tissues like skeletal muscle and heart. Importantly, previous studies had demonstrated that in skeletal muscle, metformin significantly increased AMPK α2 activity by increasing phosphorylation of AMPK at Thr172 (34). In this study we showed that OCT3 is an important determinant of the effects of metformin in skeletal muscle. That is, the effect of metformin on phosphorylation of AMPK in cultured skeletal muscle cells was not only greatly inhibited by cimetidine (38), but also was substantially inhibited by OCT3 shRNA suggesting that OCT3 plays a major role in the therapeutic action of metformin. Both the RT-PCR and immuostaining showed that OCT3 was expressed at high levels in cardiac myocytes. OCT3 might play an important role for the metformin uptake in heart. The MPP+ accumulation in heart of Orct3/Slc22a3-deficient mice decrease more than 70% (3) and congestive heart failure has been included as a contraindication to metformin therapy (39). Collectively, our data suggest that OCT3 is an important determinant of the peripheral effects of metformin.
Previously, we demonstrated that coding region variants of OCT1 play a role response to metformin by controlling access of the drug to AMPK in the liver (13). To determine whether genetic variants of OCT3 may also play a role in the action of metformin, we identified and functionally characterized coding region variants of OCT3. Using heterologous expression of amino acid-altering variants of OCT3, we discovered that 3 of the 6 variants had significantly altered function with respect to metformin uptake as well as other selected endogenous amines, i.e., T44M, T400I and V423F. Studies of protein expression and subcellular localization revealed that the amino-acid substitutions, T400I and V423F, did not appreciably affect expression or subcellular localization of these variants, suggesting that the impairment of transport function may result from a disruption in the structure of OCT3. T400 is highly conserved across all the species and V423 is partially conserved in mammalian OCT1 (Fig. 6A). The T400I and V423F variants showed obvious substrate selectivity with respect to monoamines. In particular, these two variants exhibited a substantial reduction in the transport of metformin and catecholamines in comparison to the other monoamines. The main structural difference among the various monoamines is the hydroxyl group in the phenyl ring, which is only present in the catecholamines (Fig. 6B). Changing from a threonine to an isoleucine by T400I may disrupt the hydrogen bonding of the catecholamines to the hydroxyl group of threonine. A larger hydrophobic replacement by phenylalanine at V423 could interfere with the hydrophobic interactions between the phenol ring of the catecholamines and V400F. We created two comparative models for OCT3 based on the crystal structures of the glycerol-3-phosphate transporter and the lactose permease (LacY) from E. coli (Fig. 3S, Supplemental Digital Content 1, http://links.lww.com/FPC/A210) (29, 30). The templates have similar fold and belong to the Major Facilitator Superfamily. Both models place T400 in the 8th transmembrane helix while V423 is plaed in the 9th (LacY, 2CFQ) and 10th (GlpT, 1PW4) transmembrane helix, respectively, in close proximity to the extracellular loop between the 9th and the 10th transmembrane helices. This is in agreement with a previous study describing a comparative model of rOCT1 based on a structure of the lactose permease LacY from E. coli (30). Helix 8th is predicted to be one of the helixes (H2, H5, H8 and H11) lining the substrate-transporting pore (26), suggesting that it may interact with substrates of OCT3. However, the possibility that the amino acid residue substitutions may alter the structure of OCT3 and indirectly affect the interaction of substrates with the transporter cannot be excluded. The effect of V423F on the substrate selectivity, which was not among the helixes lining the pore, might be indirect and remain elusive (Fig. 3S, Supplemental Digital Content 1, http://links.lww.com/FPC/A210). Further information including a high resolution crystal structure of a mammalian organic cation transporter is clearly needed to identify residues involved in the interaction of various substrates with OCTs.
The endogenous role of OCT3 may be versatile due to its role in the uptake of multiple monoamines (8, 40–42). The uptake data showed OCT3 especially favors histamine over other monoamines. The discovery of genetic variants with functional changes may have some implications for the regulation of tissue levels of endogenous substrates such as histamine as well as to the pharmacologic action of the important anti-diabetic drug, metformin. The study suggests that in addition to OCT1, OCT2 and MATE1 (6, 14–17, 36, 43, 44), OCT3 should be considered as an important candidate gene for the uptake of metformin in muscle type cells and its variation may modulate the response to metformin.
Supplementary Material
Acknowledgements
This study was supported by the National Institutes of Health [GM61390, GM74929 and GM36780]. Ligong Chen is partially supported by [GM74929]. Avner Schlessinger is supported by NIH F32 GM088991-01A1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declared no conflict of interest.
References
- 1.Koepsell H, Lips K, Volk C. Polyspecific organic cation transporters: Structure, function, physiological roles, and biopharmaceutical implications. Pharmcol Res. 2007;24:1227–1251. doi: 10.1007/s11095-007-9254-z. [DOI] [PubMed] [Google Scholar]
- 2.Jonker JW, Schinkel AH. Pharmacological and physiological functions of the polyspecific organic cation transporters: OCT1, 2, and 3 (SLC22A1-3) J Pharmcol Exp Ther. 2004;308:2–9. doi: 10.1124/jpet.103.053298. [DOI] [PubMed] [Google Scholar]
- 3.Zwart R, Verhaagh S, Buitelaar M, Popp-Snijders C, Barlow DP. Impaired activity of the extraneuronal monoamine transporter system known as uptake-2 in Orct3/Slc22a3-deficient mice. Mol Cell Biol. 2001;21:4188–4196. doi: 10.1128/MCB.21.13.4188-4196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grundemann D, Schechinger B, Rappold GA, Schomig E. Molecular identification of the corticosterone-sensitive extraneuronal catecholamine transporter. Nat Neurosci. 1998;1:349–351. doi: 10.1038/1557. [DOI] [PubMed] [Google Scholar]
- 5.Ogasawara M, Yamauchi K, Satoh Y, Yarnaji R, Inui K, Jonker JW, Schinkel AH, Maeyama K. Recent advances in molecular pharmacology of the histamine systems: Organic cation transporters as a histamine transporter and histamine metabolism. J Pharmacol Sci. 2006;101:24–30. doi: 10.1254/jphs.fmj06001x6. [DOI] [PubMed] [Google Scholar]
- 6.Kimura N, Masuda S, Katsura T, Inui K. Transport of guanidine compounds by human organic cation transporters, hOCT1 and hOCT2. Biochem Pharmcol 2009. 2009;77:1429–1436. doi: 10.1016/j.bcp.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 7.Vialou V, Amphoux A, Zwart R, Giros B, Gautron S. Organic cation transporter 3 (Slc22a3) is implicated in salt-intake regulation. J Neurosci. 2004;24:2846–2851. doi: 10.1523/JNEUROSCI.5147-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baganz NL, Horton RE, Calderon AS, Owens WA, Munn JL, Watts LT, Koldzic-Zivanovic N, Jeske NA, Koek W, Toney GM, Daws LC. Organic cation transporter 3: Keeping the brake on extracellular serotonin in serotonin-transporter-deficient mice. Proc Natl Acad Sci USA. 2008;105:18976–18981. doi: 10.1073/pnas.0800466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ, Shah RB, Chinnaiyan AM. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 10.Trégouët DA, König IR, Erdmann J, Munteanu A, Braund PS, Hall AS, Grosshennig A, Linsel-Nitschke P, Perret C, DeSuremain M, Meitinger T, Wright BJ, Preuss M, Balmforth AJ, Ball SG, Meisinger C, Germain C, Evans A, Arveiler D, Luc G, Ruidavets JB, Morrison C, van der Harst P, Schreiber S, Neureuther K, Schäfer A, Bugert P, El Mokhtari NE, Schrezenmeir J, Stark K, Rubin D, Wichmann HE, Hengstenberg C, Ouwehand W, Ziegler A, Tiret L, Thompson JR, Cambien F, Schunkert H, Samani NJ Wellcome Trust Case Control Consortium; Cardiogenics Consortium. Genome-wide haplotype association study identifies the SLC22A3-LPAL2-LPA gene cluster as a risk locus for coronary artery disease. Nat Genet. 2009;41:283–285. doi: 10.1038/ng.314. [DOI] [PubMed] [Google Scholar]
- 11.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, Wang Z, Welch R, Staats BJ, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Gelmann EP, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover R, Hunter DJ, Chanock SJ, Thomas G. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 12.Ashiya M, Smith RE. Non-insulin therapies for type 2 diabetes. Nat Rev Drug Discov. 2007;6:777–778. doi: 10.1038/nrd2354. [DOI] [PubMed] [Google Scholar]
- 13.Shu Y, Sheardown SA, Brown C, Owen RP, Zhang S, Castro RA, Ianculescu AG, Yue L, Lo JC, Burchard EG, Brett CM, Giacomini KM. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J Clin Invest. 2007;117:1422–1431. doi: 10.1172/JCI30558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becker ML, Visser LE, van Schaik RH, Hofman A, Uitterlinden AG, Stricker BH. Interaction between polymorphisms in the OCT1 and MATE1 transporter and metformin response. Pharmacogenet Genomics. 2010;20:38–44. doi: 10.1097/FPC.0b013e328333bb11. [DOI] [PubMed] [Google Scholar]
- 15.Becker ML, Visser LE, van Schaik RH, Hofman A, Uitterlinden AG, Stricker BH. Genetic variation in the organic cation transporter 1 is associated with metformin response in patients with diabetes mellitus. Pharmacogenomics J. 2009;9:242–247. doi: 10.1038/tpj.2009.15. [DOI] [PubMed] [Google Scholar]
- 16.Tsuda M, Terada T, Ueba M, Sato T, Masuda S, Katsura T, Inui K. Involvement of Human Multidrug and Toxin Extrusion 1 in the Drug Interaction between Cimetidine and Metformin in Renal Epithelial Cells. J Pharmacol Exp Ther. 2009;329:185–191. doi: 10.1124/jpet.108.147918. [DOI] [PubMed] [Google Scholar]
- 17.Tsuda M, Terada T, Mizuno T, Katsura T, Shimakura J, Inui K. Targeted Disruption of the Multidrug and Toxin Extrusion 1 (Mate1) Gene in Mice Reduces Renal Secretion of Metformin. Mol Pharmacol. 2009;75:1280–1286. doi: 10.1124/mol.109.056242. [DOI] [PubMed] [Google Scholar]
- 18.Nies AT, Koepsell H, Winter S, Burk O, Klein K, Kerb R, Zanger UM, Keppler D, Schwab M, Schaeffeler E. Expression of Organic Cation Transporters OCT1 (SLC22A1) and OCT3 (SLC22A3) Is Affected by Genetic Factors and Cholestasis in Human Liver. Hepatology. 2009;50:1227–1240. doi: 10.1002/hep.23103. [DOI] [PubMed] [Google Scholar]
- 19.Lazar A, Walitza S, Jetter A, Gerlach M, Warnke A, Herpertz-Dahlmann B, Gründemann D, Grimberg G, Schulz E, Remschmidt H, Wewetzer C, Schömig E. Novel mutations of the extraneuronal monoamine transporter gene in children and adolescents with obsessive-compulsive disorder. Int J Neuropsychopharmacol. 2008;11:35–48. doi: 10.1017/S1461145707007742. [DOI] [PubMed] [Google Scholar]
- 20.Lazar A, Grundemann D, Berkels R, Taubert D, Zimmermann T, Schomig E. Genetic variability of the extraneuronal monoamine transporter EMT (SLC22A3) J Hum Genet. 2003;48:226–230. doi: 10.1007/s10038-003-0015-5. [DOI] [PubMed] [Google Scholar]
- 21.Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, Wu M, Ventre J, Doebber T, Fujii N, Musi N, Hirshman MF, Goodyear LJ, Moller DE. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108:1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zang MW, Zuccollo A, Hou XY, Nagata D, Walsh K, Herscovitz H, Brecher P, Ruderman NB, Cohen RA. AMP-activated protein kinase is required for the lipid-lowering effect of metformin in insulin-resistant human HepG2 cells. J Biol Chem. 2004;279:47898–47905. doi: 10.1074/jbc.M408149200. [DOI] [PubMed] [Google Scholar]
- 23.Urban TJ, Yang C, Lagpacan LL, Brown C, Castro RA, Taylor TR, Huang CC, Stryke D, Johns SJ, Kawamoto M, Carlson EJ, Ferrin TE, Burchard EG, Giacomini KM. Functional effects of protein sequence polymorphisms in the organic cation/ergothioneine transporter OCTN1 (SLC22A4) Pharmacogenet Genomics. 2007;17:773–782. doi: 10.1097/FPC.0b013e3281c6d08e.. [DOI] [PubMed] [Google Scholar]
- 24.Pieper U, Eswar N, Braberg H, Madhusudhan MS, Davis FP, Stuart AC, Mirkovic N, Rossi A, Marti-Renom MA, Fiser A, Webb B, Greenblatt D, Huang CC, Ferrin TE, Sali A. MODBASE, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2009;37:347–354. doi: 10.1093/nar/gkh095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 26.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 27.Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- 28.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 30.Popp C, Gorboulev V, Muller TD, Gorbunov D, Shatskaya N, Koepsell H. Amino acids critical for substrate affinity of rat organic cation transporter 1 line the substrate binding region in a model derived from the tertiary structure of lactose permease. Mol Pharmacol. 2005;67:1600–1611. doi: 10.1124/mol.104.008839. [DOI] [PubMed] [Google Scholar]
- 31.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bleasby K, Castle JC, Roberts CJ, Cheng C, Bailey WJ, Sina JF, Kulkarni AV, Hafey MJ, Evers R, Johnson JM, Ulrich RG, Slatter JG. Expression profiles of 50 xenobiotic transporter genes in humans and pre-clinical species: A resource for investigations into drug disposition. Xenobiotica. 2006;36:963–988. doi: 10.1080/00498250600861751. [DOI] [PubMed] [Google Scholar]
- 33.Kirpichnikov D, McFarlane SI, Sowers JR. Metformin: An update. Ann Intern Med. 2002;137:25–33. doi: 10.7326/0003-4819-137-1-200207020-00009. [DOI] [PubMed] [Google Scholar]
- 34.Musi N, Hirshman MF, Nygren J, Svanfeldt M, Bavenholm P, Rooyackers O, Zhou G, Williamson JM, Ljunqvist O, Efendic S, Moller DE, Thorell A, Goodyear LJ. Metformin increases AMP-activated protein kinase activity in skeletal muscle of subjects with type 2 diabetes. Diabetes. 2002;51:2074–2081. doi: 10.2337/diabetes.51.7.2074. [DOI] [PubMed] [Google Scholar]
- 35.Grundemann D, Liebich G, Kiefer N, Koster S, Schomig E. Selective substrates for non-neuronal monoamine transporters. Mol Pharmacol 1999. 1999;56:1–10. doi: 10.1124/mol.56.1.1. [DOI] [PubMed] [Google Scholar]
- 36.Chen Y, Li S, Brown C, Cheatham S, Castro RA, Leabman MK, Urban TJ, Chen L, Yee SW, Choi JH, Huang Y, Brett CM, Burchard EG, Giacomini KM. Effect of genetic variation in the organic cation transporter 2 on the renal elimination of metformin. Pharmacogenet Genomics. 2009;19:497–504. doi: 10.1097/FPC.0b013e32832cc7e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abbud W, Habinowski S, Zhang JZ, Kendrew J, Elkairi FS, Kemp BE, Witters LA, Ismail-Beigi F. Stimulation of AMP-activated protein kinase (AMPK) is associated with enhancement of Glut1-mediated glucose transport. Arch Biochem Biophys. 2000;380:347–352. doi: 10.1006/abbi.2000.1935. [DOI] [PubMed] [Google Scholar]
- 38.Somogyi A, Stockley C, Keal J, Rolan P, Bochner F. Reduction of Metformin Renal Tubular Secretion by Cimetidine in Man. Br J Clin Pharmacol. 1987;23:545–551. doi: 10.1111/j.1365-2125.1987.tb03090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Misbin RI, Green L, Stadel BV, Gueriguian JL, Gubbi A, Fleming GA. Lactic acidosis in patients with diabetes treated with metformin. New Eng J Med. 1998;338:265–266. doi: 10.1056/NEJM199801223380415. [DOI] [PubMed] [Google Scholar]
- 40.Vialou V, Balasse L, Callebert J, Launay JM, Giros B, Gautron S. Altered aminergic neurotransmission in the brain of organic cation transporter 3-deficient mice. J Neurochem. 2008;106:1471–1482. doi: 10.1111/j.1471-4159.2008.05506.x. [DOI] [PubMed] [Google Scholar]
- 41.Mooney JJ, Samson JA, Hennen J, Pappalardo K, McHale N, Alpert J, Koutsos M, Schildkraut JJ. Enhanced norepinephrine output during long-term desipramine treatment: A possible role for the extraneuronal monoamine transporter (SLC22A3) J Psychiatr Res. 2008;42:605–611. doi: 10.1016/j.jpsychires.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider E, Machavoine F, Pleau JM, Bertron AF, Thurmond RL, Ohtsu H, Watanabe T, Schinkel AH, Dy M. Organic cation transporter 3 modulates murine basophil functions by controlling intracellular histamine levels. J Exp Med. 2005;202:387–393. doi: 10.1084/jem.20050195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Becker MS, Visser LE, van Schaik RH, Hofman A, Uitterlinden AG, Stricker BH. Genetic Variation in the Multidrug and Toxin Extrusion 1 Transporter Protein Influences the Glucose-Lowering Effect of Metformin in Patients With Diabetes: A Preliminary Study. Diabetes. 2009;58:745–749. doi: 10.2337/db08-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang ZJ, Yin OQ, Tomlinson B, Chow MS. OCT2 polymorphisms and in-vivo renal functional consequence: studies with metformin and cimetidine. Pharmacogenet Genomics. 2008;18:637–645. doi: 10.1097/FPC.0b013e328302cd41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.