Abstract
In sea urchin embryos, the apical tuft forms within the neurogenic animal plate. When FoxQ2, one of the earliest factors expressed specifically in the animal plate by early blastula stage, is knocked down, the structure of the apical tuft is altered. To determine the basis of this phenotype, we identified FoxQ2-dependent genes using microarray analysis. The most strongly down-regulated gene in FoxQ2 morphants encodes a protein with ankyrin repeats region in its N-terminal domain. We named this gene ankAT-1, Ankyrin-containing gene specific for Apical Tuft. Initially its expression in the animal pole region of very early blastula stage embryos is FoxQ2-independent but becomes FoxQ2-dependent beginning at mesenchyme blastula stage and continuing in the animal plate of 3-day larvae. Furthermore, like FoxQ2, this gene is expressed throughout the expanded apical tuft region that forms in embryos lacking nuclear β-catenin. When AnkAT-1 is knocked-down by injecting a morpholino, the cilia at the animal plate in the resulting embryos are much shorter and their motility is less than that of motile cilia in other ectoderm cells, and remains similar to that of long apical tuft cilia. We conclude that AnkAT-1 is involved in regulating the length of apical tuft cilia.
Introduction
Cilia or flagella are conserved structures observed in almost all animal cells at least once during their lives. Because of their characteristic appearances and behaviors, they have been studied extensively to examine the structural components and signaling pathways driving beating forces especially in unicellular organisms or sperm of multicellular organisms. Motile cilia assumed to produce liquid current around cells or individual organisms have elementally conserved molecular sets, such as 9 microtubules, radial spokes, and inner and outer dyneins (Inaba, 2007). The precise orientation of these fundamental sets is required for cilia to produce and control their beating behaviors. To form precisely constructed cilia, the components of axoneme are transferred towards the tip of cilia along the doublet microtubules by intraflagellar transport, IFT, dependent on kinesin-II and cytoplasmic dyneins (Kozminski et al., 1995; Pazour et al., 1998; Rosenbaum and Witman, 2002). Some of the non-motile cilia are used for sensing the external environment, for instance, at photoreceptor cells or olfactory epithelia. Chemical or mechanical receptors are transferred by the IFT system to the membrane of those non-motile cilia, and their defects cause behavioral disorders (Insinna and Besharse, 2008; Kuhara et al., 2008; Satir et al., 2010). Thus, for both motile and immotile cilia it is essential to produce the precise structure to achieve normal function.
Some invertebrate embryos living in water that use motile cilia instead of muscles to move have long immotile cilia at the anterior end of the body. These cilia, as a group, are called the apical tuft. Compared with the motile cilia normally covering the entire body of the embryo and beating synchronously to produce a water current, the apical tuft cilia are longer and almost immotile. As shown in Figure 1A, the presence of an apical tuft has been reported not only in triploblastic animals such as echinoderms (Hörstadius 1939), hemichordates (Urata and Yamaguchi, 2004), mollusca (Dictus and Damen, 1997) and annelids (Arenas-Mena et al., 2007) but also in diploblastic animals like sea anemones (Pang et al., 2004). In those embryos, the apical tuft has been thought to be part of a sensory organ because the long cilia seldom show standard beating behavior and the anterior end of the embryonic body contains many nerves (Bisgrove and Burke, 1987; Page 2002). However, very little is understood about the composition or the mechanisms of formation of these cilia.
Figure 1.
The apical tuft is present in the animal plate of the sea urchin embryos. (A) The phylogenetic position of animals that has been reported as having an apical tuft. The embryos/larvae of phyla enclosed with the square have an apical tuft at the anterior end. (B) The apical tuft observed in early gastrula. (C) Magnified image of dashed-lined rectangle in (B). (D) The apical tuft observed in prism larva. (E) The magnified image of (D) shows the apical tuft cilia increase in number and length compared to those in (C). (F) At the 72-hr pluteus stage, the embryo loses the apical tuft. (G) Magnified image of (F). (H) FoxQ2 morphant does not have a long apical tuft. Prism larva is shown here. (I) Magnified image of the rectangle region of (H). Bars in (H) and (I) = 20 µm and refer to panels (B, D, and F) and (C, E, and G), respectively.
In the sea urchin embryo, the long and almost immotile apical tuft forms during blastula stages at the animal plate, which is a neurogenic ectoderm, persists to prism stage and disappears at pluteus stage (Fig. 1B–G; supplemental movie 1). Although its existence as part of the apical organ has been known for several decades (Hörstadius 1939), its function and the molecules that confer its unusual length and lack of motility remain unknown. Dunn et al. (2007) showed that a transcription factor, NK2.1, is involved in inducing genes encoding ciliary components that are expressed around the apical tuft and NK2.1 morphants lack the long cilia and have reduced swimming activity in Strongylocentrotus purpuratus. This is the only gene known to be required for the formation of the apical tuft in sea urchin embryos so far. However, the role of NK2.1 and its target genes in the structure and function of the apical tuft is not clear. First, the NK2.1-dependent genes examined are expressed maternally and their functions related to these long cilia are unknown. Second, NK2.1 probably has many additional functions based on its expression in other parts of the embryo (Takacs et al., 2004). In this study, to understand the mechanisms controlling the formation of the long immotile cilia, we focused on identifying genes specific for the apical tuft. We used a microarray approach to identify genes whose expression depends on FoxQ2, because this factor is encoded by one of the earliest regulatory genes expressed only at the animal plate where the apical tuft forms and has been implicated in animal plate specification (Tu et al., 2006; Yaguchi et al., 2008). We show here that one of genes downstream of FoxQ2 function during mesenchyme blastula and gastrula stages is specific for apical tuft formation, and could be a key factor for further investigating the apical tuft function.
Materials and Methods
Animals and embryo culture
Adult sea urchins, Strongylocentrotus purpuratus obtained from The Cultured Abalone, Goleta, CA were maintained in seawater at 10°C and the embryos were used for microarray experiment. For all of other experiments, the embryos of the sea urchin, Hemicentrotus pulcherrimus collected around Shimoda Marine Research Center, University of Tsukuba, around Research Center for Marine Biology, Tohoku University, and around Marine and Coastal Research Center, Ochanomizu University were used. The gametes were collected by intrablastocoelar injection of 0.5 M KCl and the embryos were cultured by standard methods with filtered natural seawater (FSW) at 15°C.
Microinjection of morpholino antisense oligonucleotides (MO) and synthetic mRNAs
Dejellied eggs were arrayed in rows on a 35 mm plastic dish coated with 1% protamine sulfate (Sigma). After insemination in FSW containing 3-amino-1,2,4-triazole (Sigma), microinjection was performed with micromanipulators (Narishige) and an injector (Femtojet; Eppendorf). All control embryos shown in this study were injected with 24% glycerol. We used the following morpholinos with a concentration in final 24% glycerol in injection needles and injected them about 1% volume of egg: SpFoxQ2-MO1 (800 µM; Yaguchi et al., 2008), AnkAT-1-MO1 (1.6–2.0 mM), AnkAT-1-MO2 (0.4–3.8 mM), FoxQ2-MO (200 µM), NK2.1-MO (1.0 mM), Nodal-MO (200 µM), and Lefty-MO (400 µM). All morpholinos are designed against H. purcherrimus genes except for SpFoxQ2-MO1 against S. purpuratus FoxQ2 in microarray experiment and the morphants for which published phenotypes exist in other urchin species showed the same phenotypes (Duboc et al., 2004; Dunn et al., 2007; Duboc et al., 2008; Yaguchi et al., 2008). The morpholino sequences were the following:
AnkAT-1-MO1: 5’-AGTGACCGTCTGTATCAAATACCAT-3’,
AnkAT-1-MO2: 5’-TACGAGCTGTGTTGTCACGCAGAAC-3’,
FoxQ2-MO: 5’-TCATGATGAAATGTTGGAACGAGAG-3’,
NK2.1-MO: 5’-TGGTCTGTTTAGGGCTATATGACAT-3’,
Nodal-MO: 5’-AGATCCGATGAACGATGCATGGTTA-3’, and
Lefty-MO: 5’-AGCACCGAGTGATAATTCCATATTG-3’.
mRNAs were synthesized from linearized plasmids using mMessage mMachine kit (Life Technologies), and injected at the following concentrations in injection needles with final 24% glycerol: Δ-cadherin, 0.3–0.6 µg/µl (Logan et al., 1999), green fluorescent protein (GFP)-tagged AnkAT-1 mRNA (1.85 µg/µl), and DsRed-tagged AnkAT-1 mRNA (3.1 µg/µl).
Microarray
Sample preparation, microarray and data processing were described previously (Wei et al., 2006). The double-strand cDNAs synthesized from control and SpFoxQ2-MO1-injected embryos were labeled, hybridized and scanned by Nimblegen microarray services.
Quantitative PCR
Quantitative PCR (QPCR) was performed as described previously (Ransick, 2004) with some modifications. Total RNA from 200–300 embryos of H. pulcherrimus was isolated using TRI reagent (Sigma) and reverse transcription was performed using Superscript III (Life Technologies). iQ SYBR Green mix (Bio-Rad) was used for PCR reactions carried out with an Thermal Cycler Dice Real Time system (Takara). Primer pairs used for PCR reactions were following:
AnkAT-1-qF1; 5’-TTTGCAGAACACGACTGTCC-3’,
AnkAT-1-qR1; 5’-CTGGTCCGGTTGTAATTGCT-3’,
MT12S-rRNA-qF1; 5’-GGCCCACAACACTAAGCAAT-3’,
MT12S-rRNA-qR1; 5’-GGCACGTATTTTACCCCCTT-3’.
Relative concentrations of AnkAT-1 mRNA were normalized with mitochondrial 12S rRNA (MT12S-rRNA) Ct values.
Whole-mount in situ hybridization and immunohistochemistry
Whole-mount in situ hybridization was performed as described previously (Minokawa et al., 2004) with some modifications. Embryos were fixed with 4% paraformaldehyde-FSW overnight. To detect two different mRNAs with different fluorescent colors, we mixed the digoxygenin (Dig)-labeled and FITC-labeled probes into hybridization buffer, and detected hybrids with the tyramide signal amplification system (TSA; Perkin Elmer) as described previously (Yaguchi et al., 2008). To detect GFP and acetylated α-tubulin immunochemically, embryos were fixed with 4% paraformaldehyde-FSW for 10 min followed by 10 min treatment with 100% cold methanol. After washing with phosphate-buffered saline containing 0.5% Tween-20 (PBS-T) three times, the embryos were blocked with 5% skim milk (Difco) in PBS-T and incubated with primary antibodies at the following dilutions in the blocking solution:
mouse anti-acetylated α-tubulin (Sigma) 1:2000,
rat anti-GFP (Nacalai) 1:1000.
The primary antibodies were detected with secondary antibodies conjugated with Alexa-488 for GFP and Alexa-568 for acetylated α-tubulin (Life Technologies). The specimen were observed by confocal microscopy (Olympus, Fluoview FV10i) and the optical sections were stacked and analyzed with Volocity software (PerkinElmer) and Adobe Photoshop.
Measurement of the length and beating angle of the cilia
The ciliary movements were observed under a phase contrast microscope (Olympus, BX51) equipped with a high-speed camera (200 frames/sec; HAS-200, Ditect, Tokyo, Japan) and the length and the beating angle of cilia were analyzed with BohBoh software (Bohboh Soft, Tokyo; Shiba et al., 2002). The embryos at 39–44 hours post-fertilization were immobilized between a slide glass and a coverslip separated by a 58 µm-thickness mending tape (3M Scotch). At the apical tuft region, as shown in Figure 4D, the length and beating velocity of all cilia was determined at 40° from the center of the embryo, which was chosen based on the size of the ankAT-1 expressing region. Both parameters were also measured for the lateral cilia located orthogonally to animal plate as control. The beating angle is the degree of shift occurring within 1/100 second, as determined in two successive frames of the movie (Fig. 4E), and the degree is measured between the dashed lines drawn along the basal part (~10 µm from the base) of lateral cilia that produce power strokes. We set the lines against the base of apical tuft as well to eliminate the uncontrolled shaking of the tip of the cilia. Those angular movement data were analyzed statistically with t-test.
Figure 4.
AnkAT-1 is required for the elongation of apical tuft. (A) The graph shows the relationship between the length and beating angle of animal plate (red) and lateral (grey) cilia of AnkAT-1 morphants. (B) The graph shows the relationship between the length and beating angle of animal plate (red) and lateral (grey) cilia of control embryos. (C) Graph showing the mean ± S.D. of angular movement of cilia at animal plate and lateral regions. No difference was observed in the movements of animal plate cilia between glycerol-injected control (Gly) and AnkAT-1 morphant (Ank_MO; P>0.05). In contrast, there was a small, barely significant, difference in the movement of lateral cilia between control and AnkAT-1 morphant (*; P=0.04). (D) The image shows the positions in the embryo where the length and beating angle of cilia were measured. The size of the animal plate region (40°) was determined by the ankAT-1 in situ hybridization pattern. The lateral region is orthogonal to the animal plate. Bar = 20 µm. (E) The drawing indicates how we measured the length and beating angle of cilia. The length was from the cell surface to the tip of the cilia. The beating angle was the degree of change in cilia position during 1/100 second. The position of the cilia at each time is determined by a point at the cell surface and another within the green circle.
Results
ankAT-1 is the most strongly down-regulated gene in FoxQ2 morphants
In a previous study, we investigated the function of a transcription factor, FoxQ2, one of the earliest genes expressed exclusively at the animal plate of the sea urchin embryo (Yaguchi et al., 2008). Loss of FoxQ2 not only compromises development of serotonergic neurons but also the differentiation of long cilia, both features of animal plate development (cf. Fig. 1D, E with Fig. 1H, I). Therefore, identifying molecular pathways or components of a gene regulatory network downstream from FoxQ2 that are required for development of the long cilia will shed light on the detailed mechanisms of apical tuft formation. To isolate genes downstream of FoxQ2, we performed a microarray experiment (Wei et al., 2006) using mRNAs prepared from FoxQ2 morphants and control embryos of Strongylocentrotus purpuratus at mesenchyme blastula stage. Several genes were strongly down-regulated or up-regulated in this experiment, and among them we selected one of the most strongly affected by loss of FoxQ2, SPU_024961, whose function has not been reported in any organisms. We confirmed the microarray result, with in situ hybridization and QPCR using FoxQ2 morphants of the Japanese sea urchin, Hemicentrotus pulcherrimus, as described below in detail (Fig. 6). Based on its molecular structure, the expression pattern and the results of perturbation experiments, which will be presented below, we named the gene ankAT-1, Ankyrin-containing gene specific for Apical Tuft-1 (accession number AB569244).
Figure 6.
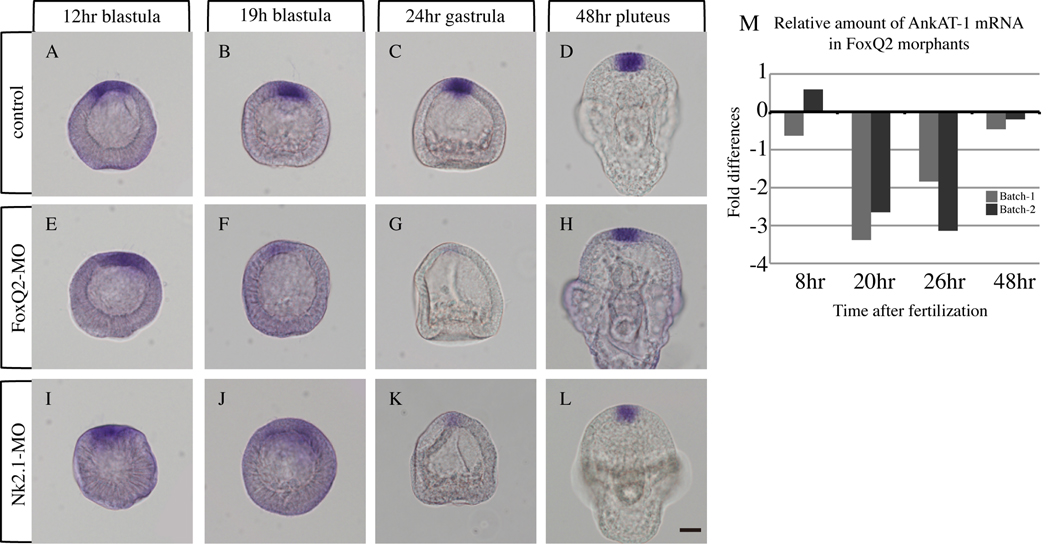
ankAT-1 expression is regulated by the FoxQ2-NK2.1 pathway temporarily during blastula and gastrula stages. (A–D) ankAT-1 is expressed at the animal plate from early blastula to early pluteus stage in normal embryos. In FoxQ2 morphants, ankAT-1 expression is down regulated during blastula (F) and gastrula (G) stages, but not at early (12-hr) blastula (E) and early (48-h) pluteus (H) stages. The temporal expression pattern of ankAT-1 in NK2.1 morphants is similar to that in FoxQ2 morphants: early blastula (I) early pluteus (L), blastula (J) gastrula (K). Comparisons of ankAT-1 mRNA levels in control and FoxQ2 morphants by QPCR supports in situ hybridization data (M). QPCR data were obtained from two independent batches. ankAT-1 mRNA levels are significantly reduced in FoxQ2 morphants only at blastula (20-hr) and gastrula (26-hr) stages. X axis = time after fertilization. Y axis = the cycle difference against control is shown as fold differences. Bar = 20 µm.
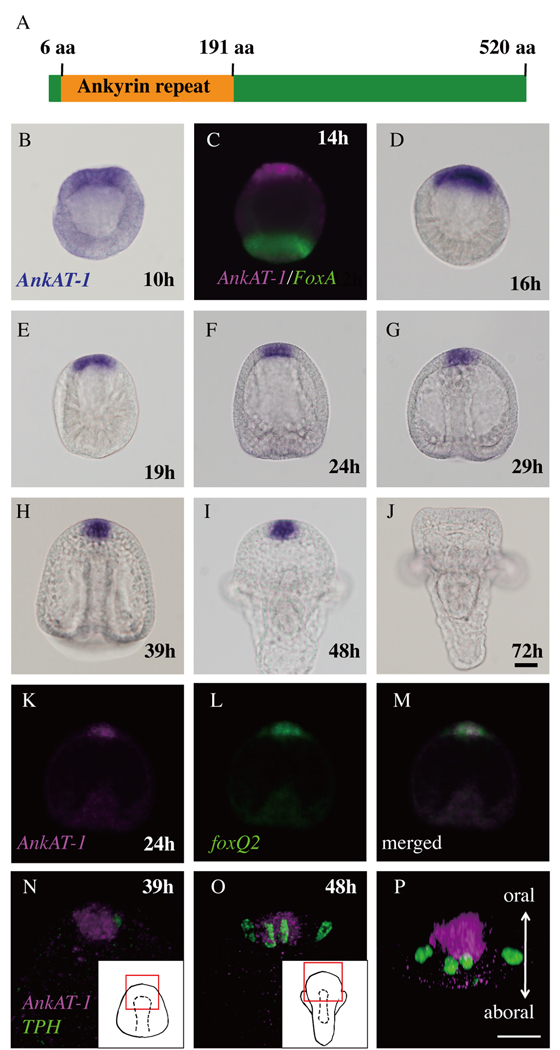
The coding region of ankAT-1 is 1,563 bps and the molecular weight of its deduced amino acid sequence is 57.7 kDa. This protein contains ankyrin repeats region at the N-terminal domain from 6aa to 191aa, revealed by domain scanning (Prosite). By Blast search, an ankyrin repeat region is found in some predicted proteins in several invertebrate genomes, although the remainder of the protein has no similarity to other proteins in GenBank (Fig. 2A). Aligned ankyrin repeat regions of these possible orthologs and the phylogenetic tree are shown in Supplemental Figure 1. The phylogenetic tree follows the reported phylogeny (Pick et al., 2010), suggesting that these proteins are AnkAT-1 orthologs.
Figure 2.
Ankyrin repeat-containing gene, ankAT-1, is expressed in the animal plate. (A) The molecular structure of AnkAT-1. Conserved ankyrin-repeat region is present at N-terminus. (B–J) in situ hybridization for ankAT-1 during development of sea urchin embryos. (B) ankAT-1 transcripts are weakly detected in the animal plate region at 10 hours (10h). (C) At the hatching blastula, ankAT-1 continues to be expressed in the animal plate, which is opposite to the region expressing foxA, a vegetal marker (Oliveri et al., 2006) (D). The animal plate expression of ankAT-1 continues until the 2-arm pluteus stage (E–I), and subsequently disappears (J). (K–M) ankAT-1 and foxQ2 double in situ hybridization. The ankAT-1 expressing region (K) corresponds to that expressing foxQ2 (L). (M) Merged image of (K) and (L). (N–P) ankAT-1 is not expressed in differentiated serotonergic neurons. (N) Double staining with ankAT-1 and tph, a marker for serotonergic neurons. Inset shows a drawing of prism stage and the red rectangle in it indicates the region shown in this panel. (O) Aboral view of pluteus stage that is drawn in inset. The red rectangle shows the region shown in this panel. (P) 90° rotation of the image in (O) to bring the oral side of the embryo up and the aboral side down. Bars in (J) and (P) = 20 µm and refer to panels (B–I and K–M) and (N and O), respectively.
ankAT-1 is expressed exclusively at the animal plate and is required for apical tuft elongation
To investigate the expression pattern of ankAT-1 during sea urchin development, we performed in situ hybridization from egg to pluteus stages. ankAT-1 is not expressed maternally as previously reported from temporal profiling by microarray (Wei et al., 2006; supplemental figure 2), and by early blastula stage (8–10 hr post-fertilization) low concentrations of AnkAT mRNA can be detected on one side of the embryo (Fig. 2B). At 14 hr, this region is opposite to the vegetal pole, as indicated by accumulation of foxA transcripts in vegetal cells (Fig. 2C; Oliveri et al., 2006). Subsequently by mesenchyme blastula stage, the entire animal plate expression becomes stronger and continues until prism stage (Fig. 2D–I), but it disappears by 2-arm pluteus stage (Fig. 2J). To examine expression within the animal plate in detail, we carried out fluorescent double in situ hybridization with ankAT-1 and either foxQ2 or tryptophan 5-hydroxylase (tph; serotonin synthase) probes, because these genes are expressed on the oral and aboral sides of the animal plate, respectively (Yaguchi and Katow, 2003; Yaguchi et al., 2008). Figure 2K–M show that ankAT-1 and foxQ2 mRNA distributions almost completely overlap at the early gastrula stage in H. pulcherrimus, near the time when the S. purpuratus FoxQ2 morphant microarray screen was done. In contrast, ankAT-1 is not expressed in tph-expressing cells, but rather in the portion of the animal plate containing cells bearing long cilia (Fig. 2N–P). tph transcripts appear at the aboral edge of the animal plate from late gastrula stage, but at that time ankAT-1 expression has disappeared from aboral side as has that of foxQ2.
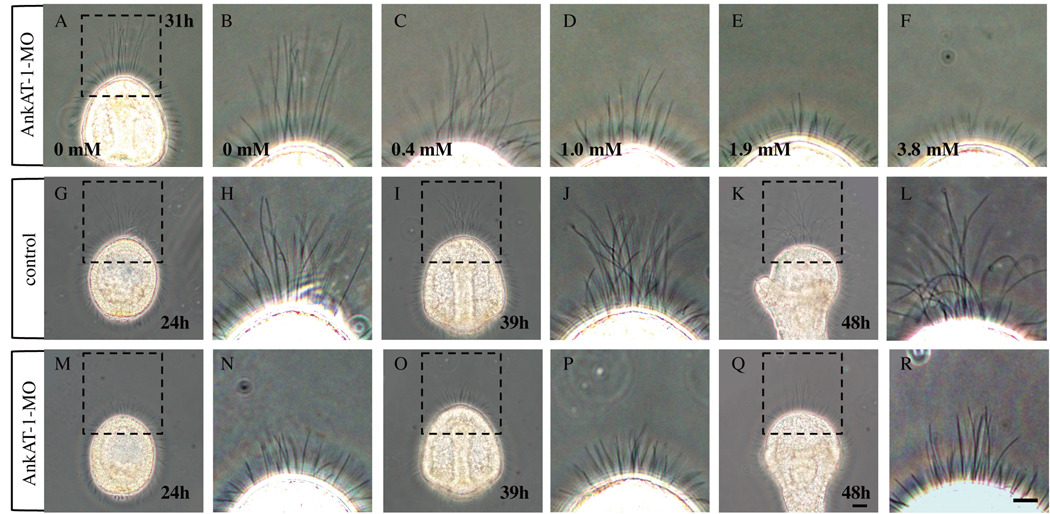
To investigate the function of AnkAT-1, we injected morpholino oligonucleotides (AnkAT-1-MO1 or AnkAT-1-MO2). Both are targeted to non-overlapping sequences in the mRNA and designed to block its translation. Both morpholinos elicit the same phenotype (supplemental figure 3), which is a dose-dependent shortening of the apical tuft-like cilia (Fig. 3A–F). The maximum effect is obtained by injecting 3.8 mM morpholino, but because some embryos injected with that concentration became unhealthy, we used 1.9 mM morpholino for the remaining experiments. To investigate in detail the effect of knockdown of AnkAT-1 during development, we compared the apical tuft of morphants with those of normal embryos at several stages. The long cilia become prominent in the animal plate a few hours after hatching in normal embryos and remain in this region until early pluteus larvae (Fig. 3G–L). In contrast, AnkAT-1-MO-injected embryos have only short cilia at the animal plate whose length is similar to that of general motile cilia (Fig. 3M–P). However, after 48 hours the long cilia at the animal plate region start to reappear in AnkAT-1 morphants (Fig. 3Q, R).
Figure 3.
AnkAT-1 morphants have short apical tuft cilia. (A–F) At 31 hours, the morphants have shorter apical tuft-like cilia at the animal pole at morpholino concentrations between 1.0 mM to 3.8 mM. As shown, there is an inverse correlation between apical tuft length and morpholino concentration indicating that the AnkAT-1-MO blocks translation of the target mRNA in a dose-dependent manner. (A) The apical tuft of control embryo at 31 hours. (B) Magnified image of dashed-lined rectangle in (A). (C–F) Apical tuft regions of 31-hr embryos injected with different concentrations of morpholino; 0.4 mM (C), 1.0 mM (D), 1.9 mM (E), and 3.8 mM (F) AnkAT-1-MO injected embryos. (G–L) The apical tuft in control embryos during development. (G, H) Early gastrula at 24 hours. (I, J) Prism larva at 39 hours. (K, L) Pluteus larva at 48 hours. (H), (J), and (L) are magnified image of rectangles of (G), (I), and (K), respectively. (M–R) The apical tuft region in AnkAT-1 morphants at the same stage as controls. Early gastrula (M, N), prism (O, P), and pluteus (Q, R) stages of morpholino- injected embryo. (N), (P), and (R) are magnified image of rectangles of (M), (O), and (Q), respectively. Bars in (Q) and (R) = 20 µm and refer to panels (A, G, I, K, M and O) and (B–F, H, J, L, N and P), respectively.
The two known properties of apical tuft cilia are that they are long and nearly immotile. AnkAT-1 is clearly involved in elongation of the cilia. To test whether it is also involved in controlling motility, we measured the length and beating angle of cilia at the animal plate region and those of motile cilia at the orthogonally lateral region as a control (Fig. 4D, E). The length of cilia at the animal plate of normal embryos is between 20 to 80 µm, and the beating angle is always below 10° within 1/100 second (Fig. 4B, red dots). In contrast, the length of lateral cilia of control embryos is about 20 µm, never longer than 26 µm, and they beat very fast (cf. supplemental movie 1) with the majority having a beating angle of more than 20° per 1/100 second (Fig. 4B, grey dots). These data indicate that the majority of cilia at the animal plate region are longer and much less motile than lateral cilia, although a few short immotile and short motile cilia are present at the same region as well. When AnkAT-1 function is knocked down, no significant difference is observed for lateral cilia, but animal plate cilia are all short, between 20 to 40 µm (Fig. 4A, red dots). These animal plate cilia in AnkAT-1 morphants show small beating angle (Fig. 4A), indicating that they are similar to the normal apical tuft in beating pattern (Fig. 4C; cf. supplemental movie 2). The angular movement of lateral cilia in AnkAT-1 morphants is only slightly significantly higher (P=0.04). These data indicate that AnkAT-1 does not confer reduced motility to apical tuft cilia, but does affect their length during normal development.
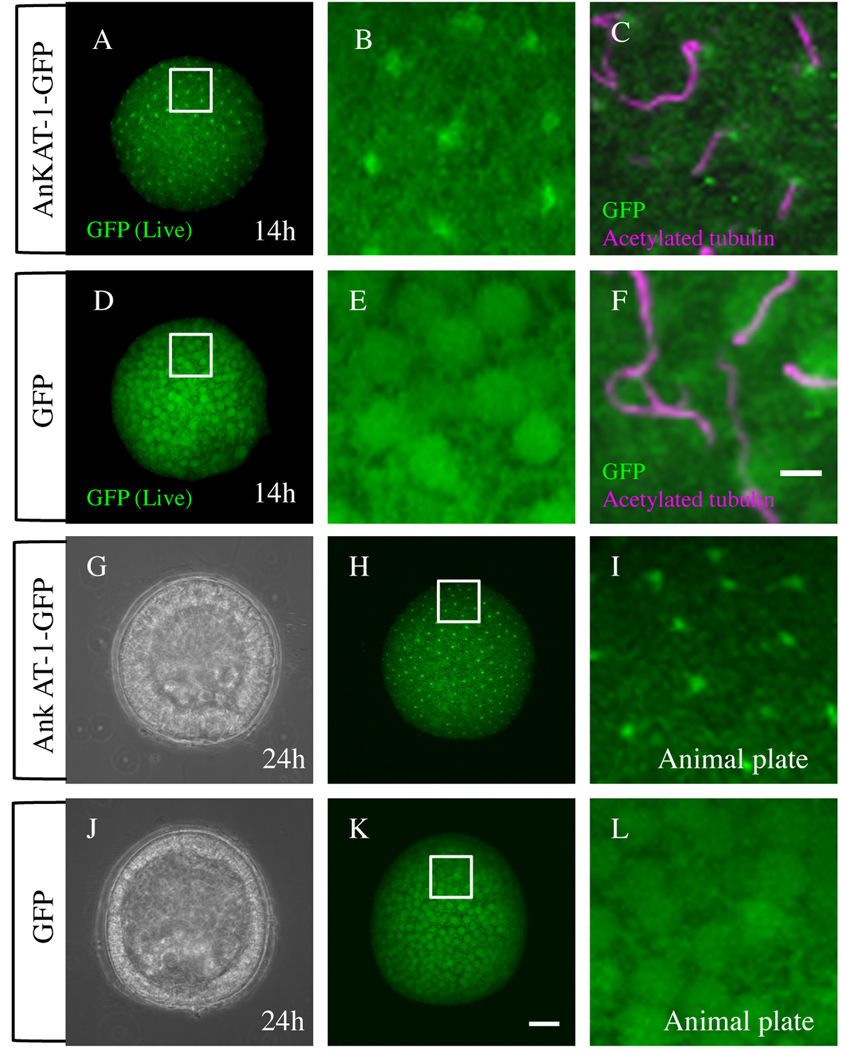
To investigate the localization of AnkAT-1 protein, we injected mRNA encoding full-length AnkAT-1 fused with green fluorescent protein (GFP). All cells of control embryos, in which only GFP mRNA was injected, are fluorescent and no specifically localized signals were observed (Fig. 5D, E). In contrast, green fluorescent puncta are present near the surface of every cell in the embryo, in which AnkAT-1-GFP mRNA was injected (Fig. 5A, B). To examine the relationship between cilia and AnkAT-1, acetylated α-tubulin and AnkAT-1/GFP distributions were compared by immunohistochemistry. At the hatching blastula stage, a strong AnkAT-1-GFP signal is observed at the base of cilia, containing acetylated tubulin, i.e. near the basal bodies (Fig. 5C), whereas GFP-alone injected embryos have no such signal (Fig. 5F). These observations were performed using hatching blastulae that have no morphological polarity. To check whether GFP-AnkAT-1 localization is observed at the apical tuft region of the embryo, we monitored the GFP signal in a 24-hr injected embryo, whose animal plate is morphologically identifiable (Fig. 5G–L). As shown in Fig 5A–C, green dots of GFP signal are observed throughout the AnkAT-1-GFP mRNA-injected embryo including the apical tuft region (Fig. 5G–I). In contrast, no spatially localized GFP signal was observed in embryos injected only with GFP mRNA (Fig. 5J–L). To eliminate the possibility that GFP fused with other proteins tends to locate at the basal body in the sea urchin embryos, we injected mRNA encoding AnkAT-1-DsRed fusion protein. The red signal in the injected embryo is also localized near the basal body as was observed in AnkAT-1-GFP mRNA injected embryos (supplemental figure 4). These results indicate that it is the AnkAT-1 part of the fusion protein that localizes it near the basal body of cilia.
Figure 5.
GFP-tagged AnkAT-1 is concentrated near the basal body. All fluorescent images shown here are captured by confocal microscopy and the optical sections were stacked. (A–C) AnkAT-1-GFP mRNA-injected embryos. (A) Live image of a 14-hr AnkAT-1-GFP mRNA-injected embryo. (B) Magnified image of rectangle region of (A). (C) Double immunohistochemistry for acetylated α-tubulin and GFP in an AnkAT-1-GFP mRNA-injected embryo. GFP was detected at the base of the cilia near the basal body. (D–F) GFP mRNA-injected embryos. (D) Live image of 14-hr GFP mRNA-injected embryo. (E) Magnified image of (D). (F) Immunohistochemistry for GFP and acetylated α-tubulin in a GFP mRNA-injected embryo. Those signals were observed at the animal plate region where the apical tuft forms (G–I). (G) DIC image of a 24-hr AnkAT-1-GFP mRNA-injected embryo viewed from animal-lateral angle. (H) Fluorescent live image of (G). Animal plate is indicated with a rectangle. (I) Magnified image of rectangle region of (H). (J–L) GFP mRNA-injected embryo. (J) DIC image of GFP mRNA-injected embryo viewed from animal-lateral angle. (K) Fluorescent live image of (J). (L) Magnified image of the animal plate (rectangle in K). Bar in (F) = 5 µm and refers to panels (B, C, E, I and L). Bar in (K) = 20 µm and refers to panels (A, D, G, H, and J).
The FoxQ2-NK2.1 pathway temporally regulates ankAT-1 expression
Expression of ankAT-1 at mesenchyme blastula stages depends strongly on FoxQ2, as shown in the microarray screen carried out on late mesenchyme blastulae. To investigate whether this relationship between FoxQ2 and ankAT-1 is constant throughout development, we examined the expression pattern of ankAT-1 in FoxQ2 morphants from blastula to pluteus stage. foxQ2 begins to be expressed in the animal half of the embryo between 16- to 32-cell stages (Tu et al., 2006; Yaguchi et al., 2008), but ankAT-1 mRNA is not detectable at these very early stages in normal development (data not shown). At hatching blastula stage, ankAT-1 is expressed in FoxQ2 morphants at the same location and at similar levels as in control embryos (Fig. 6A, E). Subsequently, ankAT-1 expression becomes much weaker at mesenchyme blastula and gastrula stages in FoxQ2 morphants than in control embryos (cf. Fig. 6B, C with F, G). Intriguingly ankAT-1 expression recovers at early pluteus stage in FoxQ2 morphants (Fig. 6D, H). QPCR data support these results. The ankAT-1 gene in FoxQ2 morphants is expressed at similar levels to those in control embryos at 8 hours (very early blastula stage) and at 48 hours (pluteus stage), but the level of gene expression in FoxQ2 morphants at both the 20-hr mesenchyme blastula and 26-hr gastrula stages is significantly decreased (Fig. 6M). Both in situ hybridization and QPCR indicate that FoxQ2 regulates ankAT-1 expression only between blastula and gastrula stages, suggesting that something else controls its expression early during cleavage and late during larval stages.
The facts that 1) FoxQ2 is upstream of AnkAT-1 (this work), 2) FoxQ2 is upstream of NK2.1 (Yaguchi et al., 2008), and 3) NK2.1 is upstream of long cilia formation (Dunn et al., 2007) strongly suggest that NK2.1 is upstream of AnkAT-1, but does not exclude the formal possibility that FoxQ2 supports apical tuft formation by two pathways, one involving AnkAT-1 and another involving NK2.1. To investigate whether NK2.1 is an intermediate regulator between FoxQ2 and AnkAT-1 at mesenchyme blastula and gastrula stages, we examined ankAT-1 expression in NK2.1 morphants. As was observed in FoxQ2 morphants, the expression levels of ankAT-1 in early (12h-blastula) and late (48h-pluteus) stages in NK2.1 morphants are same as those of control embryos (Fig. 6I, L), but much lower at 19h-mesenchyme blastula and 24h-early gastrula stages (Fig. 6J, K). Taken together, these data suggest that FoxQ2-NK2.1 pathway is required for ankAT-1 expression between mesenchyme blastula and gastrula stages.
Wnt/β-catenin and TGF-β signaling regulate ankAT-1 expression
The expression patterns of all animal plate-specific genes reported so far are not restricted to the animal pole when β-catenin nuclearization is blocked by injection of mRNA encoding the cadherin cytoplasmic domain (Δcadherin; Logan et al., 1999), in animal-half embryos (Yaguchi et al., 2006a), or in embryos animalized by Zinc treatment (Poustka et al., 2007). For example, the number of serotonergic neurons increases dramatically in Δcadherin-injected embryos radially throughout the expanded thickened ectoderm (Yaguchi et al., 2006a, 2007). ankAT-1 expression was also expanded in Δcadherin-injected embryos, covering half or more of the embryo (Fig. 7A), and the resulting embryos have long apical tuft cilia throughout the ankAT-1-positive region (Fig. 7B, C). Furthermore, this expanded apical tuft in Δcadherin-injected embryos depends on AnkAT-1 (Fig. 7D, E). ankAT-1 is expressed in almost all non-serotonergic neural cells in the thickened ectoderm of the expanded animal plate (Fig. 7F), as shown by two-color in situ hybridization with probes for ankAT-1 and tph, a marker for serotonergic neurons. Thus, without canonical Wnt signaling, ankAT-1 is expressed throughout the entire thickened animal plate region, except in the serotonergic neurons.
Figure 7.
ankAT-1 expression is restricted by canonical Wnt signaling and can be promoted by Nodal and/or its downstream signals. (A) ankAT-1 expression is not restricted in Δcadherin-injected embryos. The apical tuft covers more than half of the embryo (B). (C) Magnified image of rectangle region in (B). (D) The embryo co-injected with AnkAT-1-MO and Δcadherin does not have long apical tuft cilia. (E) Magnified image of rectangle region in (D). (F) The distributions of mRNAs encoding ankAT-1 and tph, a serotonin synthase, are radialized in Δcadherin-injected embryos. (G, I) ankAT-1 is expressed normally at early gastrula and pluteus stages in Nodal morphants. In contrast, in Lefty morphants in which nodal signaling is enhanced, the animal plate expression is not down regulated in 72-hr embryos (J) as it is in normal embryos (Fig. 2J). (K) The long apical tuft in embryos in which Nodal signaling is enhanced persists in late 72-hr pluteus larvae. (L) Magnified image of (K). Bars in (J), (K), and (L) = 20 µm and refer to panels (A, F, and G–I), (B and D), and (C and E), respectively.
Together with foxQ2, ankAT-1 is expressed in the entire animal plate at early stages, but not on the aboral side of the animal plate after gastrulation (Fig. 2P). Therefore, both the expression of foxQ2 and ankAT-1 and the appearance of apical tuft cilia occur throughout the animal plate except where serotonergic neurons differentiate. Oral-aboral patterning of serotonergic neurogenesis in the animal plate is regulated by the Nodal pathway (Yaguchi et al., 2007), which is initiated in the oral ectoderm region (Duboc et al., 2004). Thus, to investigate whether ankAT-1 expression is regulated by the Nodal pathway as well, we injected Nodal-MO or Lefty-MO to suppress or enhance Nodal signaling, respectively, and examined ankAT-1 expression and apical tuft formation. When Nodal translation is blocked by Nodal-MO injection, the embryo is radialized with a straight archenteron and no mouth (Duboc et al., 2004), and in H. pulcherrimus the same phenotype is obtained. In this embryo, the expression pattern of ankAT-1 is similar to that of control embryos (Fig. 7G). This result indicates that Nodal and/or the consequent BMP2/4 signaling (Angerer et al., 2000; Duboc et al., 2004; Lapraz et al., 2009) are not required for ankAT-1 expression in the animal plate or for restricting its expression to this site. Lefty morphants of H. pulcherrimus, which show the same phenotype as those of P. lividus embryos (Duboc et al., 2008), have ectopic expression of Nodal that surrounds the animal plate. Nevertheless, the ankAT-1 expression pattern in the 24-hr gastrula is also similar to that of normal embryo (Fig. 7H) as was previous shown for foxQ2 (Yaguchi et al., 2008). However, in Lefty morphants, ankAT-1 expression continues after the 72-hr pluteus stage when ankAT-1 mRNA disappears in normal embryos and Nodal morphants (cf. Fig. 7J with 7I and 2J). The apical tuft is also observed at 72 hours in Lefty morphants (Fig. 7K, L). These results indicate that enhanced and/or ectopic Nodal signaling can promote ankAT-1 expression at late stages, or inhibit some unknown late suppressor of ankAT-1 expression.
Discussion
The data presented here show that ankAT-1 is a novel gene required for apical tuft elongation in the sea urchin embryo. The apical tuft is a prominent ciliary structure appearing in the animal plate region from blastula to early pluteus stages. Dunn et al. (2007) showed that NK2.1, which is expressed in the animal plate as well as in other cells in the embryo, has a role in apical tuft formation, and Poustka et al. (2007) showed the expression of genes encoding cilia components at the animal plate in Strongylocentrotus purpuratus. However, because those genes are expressed more broadly (Takacs et al., 2004; Yaguchi et al., 2006b; Poustka et al., 2007), it is difficult to identify the target genes or interacting proteins that are specifically required for apical tuft formation and function. In fact, the conserved expression pattern of nk2.1 in foregut among bilateral organisms suggests its specific role in foregut formation or function (Takacs et al., 2002). In contrast, ankAT-1 is expressed exclusively in the animal plate, and the timing of its expression is consistent with apical tuft formation. As well, AnkAT-1-knockdown results in no detectable change in morphology except for the loss of long apical tuft cilia, strongly supporting a specific function for this protein in their formation. FoxQ2 is an upstream regulator of AnkAT-1 and involved in apical tuft formation, but it functions in the differentiation of serotonergic neurons as well (Yaguchi et al., 2008). Therefore, ankAT-1 is the first identified gene specific for the apical tuft in sea urchins.
The length of short cilia observed at the animal plate region in AnkAT-1 morphants is similar to that of normal cilia present throughout the entire body that control the swimming behavior of the embryo. These shortened apical tuft cilia hardly beat, indicating that they still sustain the immobility of normal long apical tuft cilia. This suggests that AnkAT-1 functions in elongation, but does not confer the low motility property of apical tuft cilia. Several molecules required for the elongation of cilia or flagella have already been identified in many species. For instance, heterotrimeric kinesin-II is required for elongation of motile cilia in the sea urchin (Morris and Scholey, 1997), and protein kinases (Pan et al., 2004) and LF proteins (Barsel et al., 1988; Nguyen et al., 2005) are proposed to regulate flagellar length in Chlamydomonas. As well, defects in most of the IFT proteins and molecular motors cause shortening or loss of cilia (Pedersen and Rosenbaum, 2008). However, there is no similarity in amino acid sequences between these proteins and AnkAT-1. On the other hand, Ankyrin domains in many types of proteins play a role in protein-protein interactions, which is important for protein localization or transport to a specific site. For example, Ankyrin-G, ankyrin repeat containing protein, targets cyclic nucleotide-gated channels to the rod outer segment by binding the C-terminal domain of the channel β1 subunit (Kizhatil et al., 2009). The finding that AnkAT-1 appears to be located at the base of cilia raises the interesting possibility that it might help load apical tuft components onto IFT complex for transport into the cilia. For example, AnkAT-1 might function in regulating the assembly of tektin A1 that is the only axonemal structural protein known to be synthesized in quantal amounts proportional to the amount of ciliary length produced in the sea urchin embryo (Stephens, 1995). As well, it is possible that AnkAT-1 functions in specific balancing of assembly and disassembly of the cilium in the long apical tuft, as in the balance-point model described previously (Marshall et al., 2005). However, we still do not rule out the possibility that AnkAT-1 is present and functions within the apical tuft cilia because the fluorescent-protein moieties of the fusion proteins tested here could interfere with its correct trafficking, an issue that will be addressed in future studies.
To our knowledge, the mechanisms for apical tuft formation of the sea urchin have never been addressed until Dunn et al. (2007) showed NK2.1 involvement in the regulating the length of apical tuft cilia. They showed that NK2.1 positively regulates some of the cilia component genes at the animal plate region although these components are maternally distributed to the entire embryo. In addition, Yaguchi et al. (2008) showed that NK2.1 at the animal plate is positively regulated by the transcription factor, FoxQ2. In this study, we identified ankAT-1 as an apical tuft-specific gene whose expression is under control of FoxQ2 and NK2.1 functions. Therefore we conclude that there is a pathway from FoxQ2 to NK2.1 to AnkAT-1, which may be direct or indirect, between blastula and gastrula stages that is required for apical tuft elongation in the sea urchin embryo. However, it is still unclear what controls the initiation and long-term maintenance of ankAT-1 expression because it does not depend on this pathway at the very early blastula or early pluteus larva stages (Fig. 6). During both stages, ankAT-1 might be independently controlled by the same mechanism regulating foxQ2 because each of these genes is expressed at almost identical regions and when foxQ2 transcripts disappear at pluteus stage of normal embryos, ankAT-1 expression stops as well. To understand the complete set of mechanisms controlling ankAT-1 expression, further analyses focusing on the cis-regulation of this gene will be required.
Because ankAT-1 expression expands in embryos in which β-catenin nuclearization is blocked, Wnt/β-catenin signaling functions in restricting ankAT-1 expression (Fig. 7). Although foxQ2 expression is also restricted by a very early Wnt/β-catenin-dependent process to the animal pole region (Yaguchi et al., 2008), the restriction of ankAT-1 expression must be caused by a mechanism other than through FoxQ2 regulation because early expression of ankAT-1 is independent of FoxQ2 function. Without vegetal signaling and the consequent Nodal and BMP2/4 signals responsible for secondary axis formation (Logan et al., 1999; Duboc et al., 2004: Francois et al., 2009), ankAT-1 mRNA and the long apical tuft remain in the thickened ectoderm covering half or more of the embryo, indicating that one or more of those signals are not required for the initiation and maintenance of ankAT-1 expression. The fact that normal ankAT-1 expression occurs in Nodal morphants, which lack both Nodal and BMP2/4 signals, suggests that it is spatially regulated by a canonical Wnt-dependent process. However, correct Nodal-dependent patterning along the secondary axis is required for the late down regulation of ankAT-1 because enhanced Nodal signaling or its downstream cascade, as a result of loss of Lefty, a Nodal antagonist, leads to prolonged ankAT-1 expression and delayed apical tuft disappearance. Taken together, it is possible that Nodal and/or the downstream signaling pathways can interfere with inhibitory signals on ankAT-1 expression in normal embryos, which should be elucidated in future experiments.
Apical tuft is observed in other animal phyla, such as Mollusca, Annelid, Cnidaria, and Hemichordata (Dictus and Damen, 1997; Pang et al., 2004; Urata and Yamaguchi, 2004; Arenas-Mena et al., 2007). In all species, it is thought that the apical tuft functions as a sensory organ, but there has been no molecular characterization specifically related to its formation or function. In this study, we showed that loss of AnkAT-1 has a specific phenotype, the loss of long apical tuft cilia, and that its site of expression is exclusively in the apical tuft region, making it the first identified apical tuft-specific protein. As well, Blast searches and phylogenetic analysis suggest that organisms with apical tufts have genes similar to ankAT-1. Genes encoding ankyrin repeats similar to that of ankAT-1 were found in the genomes of hemichordatea (Saccoglossus kowalevskii; XP_002730494). Although Saccoglossus apparently lacks an apical tuft because it does not form a tornaria, Balanoglossus misakiensis and simodensis which belong to the same phylum as Saccoglossus, do have an apical tuft at the larval stage (Urata and Yamaguchi, 2004; Miyamoto et al., 2010). AnkAT-1-like ankyrin repeats were also present in placozoana (Trichoplax adhaerens; XP_002114071), cephalochordates (Branchiostoma floridae; XP_002602553), and cnidarians (Nematostella vectensis; XP_001630326), although their C-termini are completely divergent. Knockout of this gene or knockdowns of its expression will help to resolve AnkAT-1 function in these animals. It is intriguing that a gene containing AnkAT-1-like, ankyrin repeats has been identified in amphioxus and Saccoglossus in which an apical tuft has not been reported so far. Thus, it will be of interest to know how the function of this gene has changed during early deuterostome evolution. In addition, there are known differences in apical tufts among species, for instance in gastropod embryos, cells within the apical tuft are multiciliated but monociliated in the sea urchin (Page, 2002). Therefore, the complete analysis of AnkAT-1 function will reveal how the role of immotile and extraordinary long cilia has changed during evolution.
Supplementary Material
Acknowledgements
This work was supported, in part, by Special Coordination Funds for Promoting Science and Technology of the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government (MEXT), by Grant-in Aid for Young Scientists (Start-up No. 20870006 and B No. 21770227), and by Narishige Zoological Science Award to S.Y., in part by the Intramural Program of the National Institutes of Health, NIDCR to L.M.A., and in part by MEXT (No. 22370023) to K.I. We thank Robert Burke, Yoko Nakajima, and David McClay for essential reagents. We thank Masato Kiyomoto, Mamoru Yamaguchi, Hideki Katow, Masahiko Washio for providing the adult sea urchins. We thank Mrs. Y. Tsuchiya, T. Sato, H. Shinagawa, and Y. Yamada, Shimoda Marine Research Center, for collecting and keeping the adult sea urchins. We thank Dr. T. Lacalli for discussing about the amphioxus larva.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angerer LM, Oleksyn DW, Logan CY, McClay DR, Dale L, Angerer RC. A BMP pathway regulates cell fate allocation along the sea urchin animal-vegetal embryonic axis. Development. 2000;127:1105–1114. doi: 10.1242/dev.127.5.1105. [DOI] [PubMed] [Google Scholar]
- Arenas-Mena C, Wong KS, Arandi-Forosani N. Ciliary band gene expression patterns in the embryo and trochophore larva of an indirectly developing polychaete. Gene Expr Patterns. 2007;7:544–549. doi: 10.1016/j.modgep.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Barsel SE, Wexlerm DE, Lefebvre Genetic analysis of long flagellar mutants of Chlamydomonas reinhardtii. Genetics. 1988;118:637–648. doi: 10.1093/genetics/118.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove BW, Burke RD. Development of the nervous system of the pluteus larva of Strongylocentrotus drobachiensis. Cell Tissue Res. 1987;248:335–343. [Google Scholar]
- Dictus WJ, Damen P. Cell-lineage and clonal-contribution map of the trochophore larva of Patella vulgata (mollusca) Mech Dev. 1997;62:213–226. doi: 10.1016/s0925-4773(97)00666-7. [DOI] [PubMed] [Google Scholar]
- Duboc V, Lapraz F, Besnardeau L, Lepage T. Lefty acts as an essential modulator of Nodal activity during sea urchin oral-aboral axis formation. Dev Biol. 2008;320:49–59. doi: 10.1016/j.ydbio.2008.04.012. [DOI] [PubMed] [Google Scholar]
- Duboc V, Rottinger E, Besnardeau L, Lepage T. Nodal and BMP2/4 signaling organizes the oral-aboral axis of the sea urchin embryo. Dev Cell. 2004;6:397–410. doi: 10.1016/s1534-5807(04)00056-5. [DOI] [PubMed] [Google Scholar]
- Dunn EF, Moy VN, Angerer LM, Angerer RC, Morris RL, Peterson KJ. Molecular paleoecology: using gene regulatory analysis to address the origins of complex life cycles in the late Precambrian. Evol Dev. 2007;9:10–24. doi: 10.1111/j.1525-142X.2006.00134.x. [DOI] [PubMed] [Google Scholar]
- Hörstadius S. The mechanics of sea urchin development, studied by operative methods. Biol. Rev. Cambridge Phil. Soc. 1939;14:48. [Google Scholar]
- Inaba K. Molecular basis of sperm flagellar axonemes: structural and evolutionary aspects. Ann N Y Acad Sci. 2007;1101:506–526. doi: 10.1196/annals.1389.017. [DOI] [PubMed] [Google Scholar]
- Insinna C, Besharse JC. Intraflagellar transport and the sensory outer segment of vertebrate photoreceptors. Dev Dyn. 2008;237:1982–1992. doi: 10.1002/dvdy.21554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizhatil K, Baker SA, Arshavsky VY, Bennett V. Ankyrin-G promotes cyclic nucleotide-gated channel transport to rod photoreceptor sensory cilia. Science. 2009;323:1614–1617. doi: 10.1126/science.1169789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Beech PL, Rosenbaum JL. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J Cell Biol. 1995;131:1517–1527. doi: 10.1083/jcb.131.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhara A, Okumura M, Kimata T, Tanizawa Y, Takano R, Kimura KD, Inada H, Matsumoto K, Mori I. Temperature sensing by an olfactory neuron in a circuit controlling behavior of C. elegans. Science. 2008;320:803–807. doi: 10.1126/science.1148922. [DOI] [PubMed] [Google Scholar]
- Lapraz F, Besnardeau L, Lepage T. Patterning of the dorsal-ventral axis in echinoderms: insights into the evolution of the BMP-chordin signaling network. PLoS Biol. 2009;7:e1000248. doi: 10.1371/journal.pbio.1000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CY, Miller JR, Ferkowicz MJ, McClay DR. Nuclear beta-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development. 1999;126:345–357. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- Marshall WF, Rosenbaum JL. Intraflagellar transport balances continuous turnover of outer doublet microtubules; implications for flagellar length control. J Cell Biol. 2001;155:405–414. doi: 10.1083/jcb.200106141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Qin H, Rodrigo Brenni M, Rosenbaum JL. Flagellar length control system: testing a simple model based on intraflagellar transport and turnover. Mol Biol Cell. 2005;16:270–278. doi: 10.1091/mbc.E04-07-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minokawa T, Rast JP, Arenas-Mena C, Franco CB, Davidson EH. Expression patterns of four different regulatory genes that function during sea urchin development. Gene Expr Patterns. 2004;4:449–456. doi: 10.1016/j.modgep.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Miyamoto N, Nakajima Y, Wada H, Saito Y. Development of the nervous system in the acorn worm Balanoglossus simodensis: insights into nervous system evolution. Evol Dev. 2010;12:416–424. doi: 10.1111/j.1525-142X.2010.00428.x. [DOI] [PubMed] [Google Scholar]
- Morris RL, Scholey JM. Heterotrimeric kinesin-II is required for the assembly of motile 9 ciliary axonemes on sea urchin embryos. J Cell Biol. 1997;138:1009–1022. doi: 10.1083/jcb.138.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen RL, Tam L, Lefebvre PA. The LF1 gene of Chlamydomonas reinhardtii encodes a novel protein required for flagellar length control. Genetics. 2005;169:1415–1425. doi: 10.1534/genetics.104.027615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveri P, Walton KD, Davidson EH, McClay DR. Repression of mesodermal fate by foxa, a key endoderm regulator of the sea urchin embryo. Development. 2006;133:4173–4181. doi: 10.1242/dev.02577. [DOI] [PubMed] [Google Scholar]
- Page LR. Larval and metamorphic development of the foregut and proboscis in the caenogastropod Marsenina (Lamellaria) stearnsii. J Morphol. 2002;252:202–217. doi: 10.1002/jmor.1099. [DOI] [PubMed] [Google Scholar]
- Pan J, Wang Q, Snell WJ. An aurora kinase is essential for flagellar disassembly in Chlamydomonas. Dev Cell. 2004;6:445–451. doi: 10.1016/s1534-5807(04)00064-4. [DOI] [PubMed] [Google Scholar]
- Pang K, Matus DQ, Martindale MQ. The ancestral role of COE genes may have been in chemoreception: evidence from the development of the sea anemone, Nematostella vectensis (Phylum Cnidaria; Class Anthozoa) Dev Genes Evol. 2004;214:134–138. doi: 10.1007/s00427-004-0383-7. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Wilkerson CG, Witman GB. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT) J Cell Biol. 1998;141:979–992. doi: 10.1083/jcb.141.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick KS, Philippe H, Schreiber F, Erpenbeck D, Jackson DJ, Wrede P, Wiens M, Alié A, Morgenstern B, Manuel M, Wörheide G. Improved phylogenomic taxon sampling noticeably affects nonbilaterian relationships. Mol Biol Evol. 2010;27:1983–1987. doi: 10.1093/molbev/msq089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poustka AJ, Kuhn A, Groth D, Weise V, Yaguchi S, Burke RD, Herwig R, Lehrach H, Panopoulou G. A global view of gene expression in lithium and zinc treated sea urchin embryos: new components of gene regulatory networks. Genome Biol. 2007;8:R85. doi: 10.1186/gb-2007-8-5-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransick A. Detection of mRNA by in situ hybridization and RT-PCR. Methods Cell Biol. 2004;74:601–620. doi: 10.1016/s0091-679x(04)74024-8. [DOI] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Satir P, Pedersen LB, Christensen ST. The primary cilium at a glance. J Cell Sci. 2010;123:499–503. doi: 10.1242/jcs.050377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba K, Mogami Y, Baba SA. Ciliary movement of sea-urchin embryos. (Ochanomizu Univ, Tokyo) Nat Sci Rep. 2002;53:6. [Google Scholar]
- Sea Urchin Genome Sequencing Consortium. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens RE. Ciliogenesis in sea urchin embryos—a subroutine in the program of development. BioEssay. 1995;17:331–340. doi: 10.1002/bies.950170409. [DOI] [PubMed] [Google Scholar]
- Takacs CM, Amore G, Oliveri P, Poustka AJ, Wang D, Burke RD, Peterson KJ. Expression of an NK2 homeodomain gene in the apical ectoderm defines a new territory in the early sea urchin embryo. Dev Biol. 2004;269:152–164. doi: 10.1016/j.ydbio.2004.01.023. [DOI] [PubMed] [Google Scholar]
- Takacs CM, Moy VN, Peterson KJ. Testing putative hemichordate homologues of the chordate dorsal nervous system and endostyle: expression of NK2.1 (TTF-1) in the acorn worm Ptychodera flava (Hemichordata, Ptychoderidae) Evol Dev. 2002;4:405–417. doi: 10.1046/j.1525-142x.2002.02029.x. [DOI] [PubMed] [Google Scholar]
- Tu Q, Brown CT, Davidson EH, Oliveri P. Sea urchin forkhead gene family: phylogeny and embryonic expression. Dev Biol. 2006;300:49–62. doi: 10.1016/j.ydbio.2006.09.031. [DOI] [PubMed] [Google Scholar]
- Urata M, Yamaguchi M. The development of the enteropneust hemichordate Balanoglossus misakiensis KUWANO. Zoolog Sci. 2004;21:533–540. doi: 10.2108/zsj.21.533. [DOI] [PubMed] [Google Scholar]
- Wei Z, Angerer RC, Angerer LM. A database of mRNA expression patterns for the sea urchin embryo. Dev Biol. 2006;300:476–484. doi: 10.1016/j.ydbio.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi S, Katow H. Expression of tryptophan 5-hydroxylase gene during sea urchin neurogenesis and role of serotonergic nervous system in larval behavior. J Comp Neurol. 2003;466:219–229. doi: 10.1002/cne.10865. [DOI] [PubMed] [Google Scholar]
- Yaguchi S, Yaguchi J, Burke RD. Specification of ectoderm restricts the size of the animal plate and patterns neurogenesis in sea urchin embryos. Development. 2006a;133:2337–2346. doi: 10.1242/dev.02396. [DOI] [PubMed] [Google Scholar]
- Yaguchi S, Nakajima Y, Wang D, Burke RD. Embryonic expression of engrailed in sea urchins. Gene Expr Patterns. 2006b;6:566–571. doi: 10.1016/j.modgep.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Yaguchi S, Yaguchi J, Burke RD. Sp-Smad2/3 mediates patterning of neurogenic ectoderm by nodal in the sea urchin embryo. Dev Biol. 2007;302:494–503. doi: 10.1016/j.ydbio.2006.10.010. [DOI] [PubMed] [Google Scholar]
- Yaguchi S, Yaguchi J, Angerer RC, Angerer LM. A Wnt-FoxQ2-nodal pathway links primary and secondary axis specification in sea urchin embryos. Dev Cell. 2008;14:97–107. doi: 10.1016/j.devcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.