Abstract
RSC, an essential chromatin remodeling complex in budding yeast, is involved in a variety of biological processes including transcription, recombination, repair and replication. How RSC participates in such diverse processes is not fully understood. In vitro, RSC uses ATP to carry out several seemingly distinct reactions: it repositions nucleosomes, transfers H2A/H2B dimers between nucleosomes and transfers histone octamers between pieces of DNA. This raises the intriguing mechanistic question of how this molecular machine can use a single ATPase subunit to create these varied products. Here, we use a FRET-based approach to kinetically order the products of the RSC reaction. Surprisingly, transfer of H2A/H2B dimers and histone octamers is initiated on a time scale of seconds when assayed by FRET, but formation of stable nucleosomal products occurs on a time scale of minutes when assayed by native gel. These results suggest a model in which RSC action rapidly generates an unstable encounter intermediate that contains the two exchange substrates in close proximity. This intermediate then collapses more slowly to form the stable transfer products seen on native gels. The rapid, biologically relevant time scale on which the transfer products are generated implies that such products can play key roles in vivo.
The packaging of eukaryotic DNA into chromatin introduces complex regulatory controls on DNA replication, transcription, recombination and repair. Chromatin remodeling complexes play critical roles in controlling access to the underlying DNA during these nuclear processes. While many chromatin remodeling complexes are specialized to regulate a particular type of nuclear process, the SWI/SNF family of complexes has more widespread roles. SWI/SNF complexes are involved in regulating transcription at diverse promoters, promoting cell cycle progression by regulating exit from G1, S and M phases, and assisting in DNA repair pathways (1–3). These observations raise the question of how SWI/SNF complexes can fulfill such diverse biological functions.
The known biochemical properties of SWI/SNF complexes provide some insights into its diverse functions. SWI/SNF complexes contain one core catalytic ATPase subunit and several additional non-catalytic subunits. Unlike other chromatin remodeling complexes, SWI/SNF complexes catalyze many different outputs from the same nucleosomal template. These outputs include products that result from intramolecular rearrangements, such as translationally repositioned nucleosomes and nucleosomes containing DNA loops, as well as products that result from intermolecular component exchange, such as dinucleosome-like species and nucleosomes with transferred H2A-H2B dimers and histone octamers (4–10). While these multiple outcomes might be expected from an enzyme that enables a variety of different biological processes, they also raise the fundamental question of how the single ATPase motor in SWI/SNF can generate so many different products. It has been suggested that one way in which SWI/SNF complexes generate diverse outputs is through the formation of a single unstable intermediate that can collapse into different outputs in a context dependent manner (4, 10). However, in vitro, nucleosomes with transferred histone dimers appear to form much more slowly than repositioned nucleosomes, which raises the question of whether such transfer products are off pathway outcomes (5).
Understanding how SWI/SNF generates multiple outputs would be greatly aided by the identification of reaction intermediates. However, this can be challenging when the reaction intermediates are unstable. Here we use FRET-based approaches to identify intermediates and kinetically order three different products generated by RSC, the major SWI/SNF complex in budding yeast. Surprisingly, we find that formation of dimer and octamer transfer products, as detected by FRET, is initiated at a much faster rate than inferred by gel-based assays. These results lead to a two-step model for generation of transfer products: the first step entails the rapid formation of an encounter intermediate, while the second step entails the slow collapse of this intermediate into a stable product. Our results imply that the dimer and octamer transfer products observed in vitro occur through the core mechanistic activity of RSC and are therefore feasible and biologically relevant in vivo outcomes.
Experimental Procedures
Protein Purification
RSC was purified from the strain BCY211, a kind gift from Brad Cairns, using a TAP-tag on Rsc2 as described previously (11). RSC concentration was quantified by SYPRO red staining.
Nucleosome Assembly
Recombinant Xenopus histones were purified as described previously (12). To generate fluorescently labeled octamers, a cysteine was engineered into H2A at residue 120 and H3 at residue 33. The cysteine-containing histones were labeled with Cy5 maleimide as described previous (13). The 601 positioning sequence was modified, as previously described, to contain a Pst1 site 18 bp from one end (13). DNA constructs of various lengths were generated using PCR and gel purified before use. Cy3-labeled constructs were made using an end-labeled primer. Primer sequences are available upon request. Nucleosomes were assembled using gradient salt dialysis as described previously (12). Nucleosomes were then purified away from free DNA and histones using a glycerol gradient.
Native Gel Assays
RSC remodeling reactions were performed at 30°C in reaction buffer containing 50 mM KCl, 30 mM Tris (pH 7.5), 1 mM free MgCl2, 4% glycerol and 0.02% Igepal CA-630. Saturating RSC, 107 nM, was used for all reactions. Repositioning reactions contained 7 nM nucleosome substrate. Dimer transfer reactions contained 7 nM nucleosomes with Cy5-labeled H2A and 28 nM core nucleosome. Octamer transfer reactions contained 7 nM nucleosomes with Cy5-labeled H3 and 28 nM 147 bp core 601 positioning sequence DNA. Reactions were initiated with 1 mM ATP-Mg2+ and stopped after the time indicated using excess ADP and plasmid DNA as described previously (13). Reactions were run on native 5% (v/v) polyacrylamide gels, 29 to 1 acrylamide to bis-acrylamide ratio, in 0.5 X TBE. Gels were scanned using a Typhoon Variable Mode Imager (GE Healthcare) and quantified using ImageQuant (GE Heathcare). Gels were visualized using either the fluorescent labels or SYBR gold (Invitrogen). For the octamer and dimer transfer reactions, percent transfer was calculated as the fraction of the total Cy5 intensity in the transferred band. The data were fit by a single exponential with the zero-point fixed to the transfer value at time zero using the following equation:
where p is the percent transferred, a is the percent transferred at time zero, b is the percent transferred at time infinity and k1 is the rate constant. The data were then normalized from 0 to 1 using the following equation:
where n is the normalized transfer.
For the repositioning reactions in Figure 3C, rate constants were obtained by quantifying the amount of the Cy5 signal in starting band as a percentage of the total nucleosomal Cy5 signal. The data were fit by a single exponential using the following equation:
where s is the percent of the signal in the starting band, a is the percent in the starting band at time zero, b is the percent in the starting at time infinity and k1 is the rate constant. The data were then normalized from 0 to 1 using the following equation:
where n is the normalized amount in the starting band.
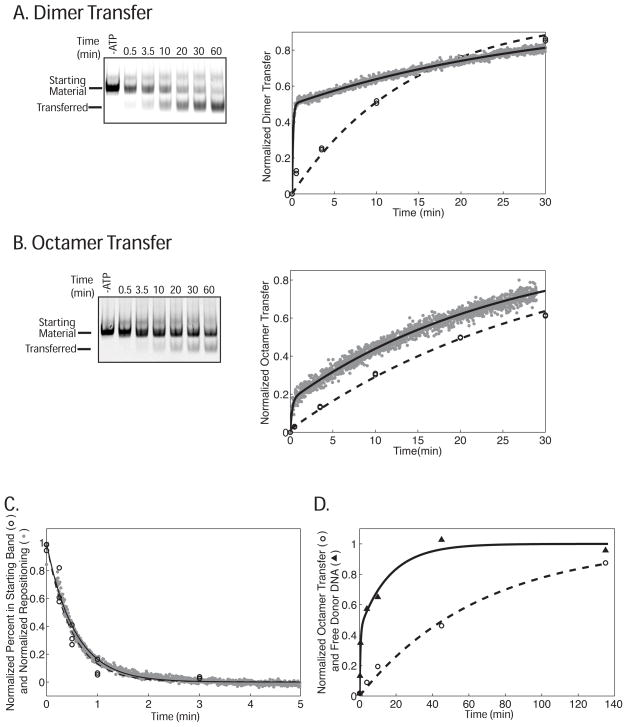
Figure 3.
Changes in FRET precede the formation of transfer products as visualized by gel for both (A) dimer transfer and (B) octamer transfer but not (C) repositioning. In (A) and (B) reactions containing the nucleosome constructs shown in Figure 2 were run on a native gel. The left panels show a representative gel scanned in the Cy5 channel. The graphs on the right show the corresponding normalized data from two separate native gel experiments (open circles with the fit shown as a dashed line) and one representative FRET-based assay (gray data points with the fit shown as a black line). The rate constant for dimer exchange and octamer transfer as measured in the gel assay were 0.072 ± 0.00012 min−1 and 0.034 ± 0.00042 min−1 respectively. These rate constants are the average and variation of two separate experiments. The rate constants for dimer and octamer transfer as measure by FRET are reported in Table 1 and are the average of three independent experiments including the ones shown in (A) and (B). Reactions (A) and (B) were preformed using 1 mM ATP. (C) The rate constant for repositioning was measured by both the FRET-based and native gel-based assays using the 0-601-55 nucleosomes used in Figure 2. To enable accurate measurement of the repositioning rate constants, the ATP concentration was lowered to 4 μM. The graph shows the corresponding normalized data from the native gel (open circles with the fit shown as a dashed line) and FRET assays (gray data points with the fit shown as a black line). The rate constants measured by FRET and native gel were 1.8 ± 0.074 min−1 and 2.1 ± 0.29 min−1 respectively. These rate constants are the average and standard error of the mean of three separate experiments. (D) Donor DNA is released more quickly than the octamer transfer product appears on the gel. Octamer transfer experiments were performed using 20-601-45 nucleosomes as an octamer donor and a 147 bp 601 containing DNA fragment as the acceptor at 1 mM ATP. The graph shows representative curves for normalized octamer transfer (open circle with a one phase fit shown as a dashed line) and normalized free donor DNA (black triangles with a two phase fit shown as a black line). The rate constant for octamer transfer was 0.037 ± 0.019 min−1. The data for acceptor DNA release fit best to two phases. However, the fast phase is too fast to accurately measure given the number of data points but we estimate a lower limit to be 3 min−1. The rate constant for the slow phase is 0.045 ± 0.011 min−1. These rate constants are the average and standard error of the mean of three separate experiments.
In Figure 4, the reactions were stopped by adding ADP to a final concentration of 58 mM. ADP uncontaminated by ATP was purchased from Calbiochem, cat# 117105. Reactions stopped with 0.1 units/μL apyrase, Sigma-Aldrich A6410, gave similar results as reactions stopped using ADP.
Figure 4.
Continuous RSC activity is necessary for formation of a stable transfer product. Dimer transfer reactions (A) and octamer transfer reactions (B) using the substrates from Figure 1 at 1 mM ATP were analyzed on a native gel. The left gels show the reaction with no ADP added and the right gels show reactions in which excess ADP was added after 3 min. The gels were scanned in the Cy5 channel. The graph shows the quantification of transfer for the reactions with no ADP added as black circles with a solid black line and ADP added at 3 min as open triangles with a dashed line. In (A) the values for dimer transfer are calculated from three independent experiments; error bars show the standard error of the mean. For octamer transfer one representative experiment is shown. Other experiments where ADP was added at different time points showed the same requirement for ATP (data not shown). In (B), left panel, all the time-points shown are from the same gel but lanes between the 3 min and 20 min time-points were omitted to enable a direct visual comparison with the gel in the right panel
FRET-based kinetics experiments
FRET assays were conducted using the same reaction conditions as the native gel assays. Fluorescence was measured on an ISS K2 fluorometer. Kinetics were measured as described previously (13), with samples excited at 520 nm and spectra collected at Cy3 or Cy5 peak intensity, 567 or 670 nm, respectively. All reactions contained 1 mM free MgCl2 and either 1 mM or 20 μM ATP-Mg2+ as indicated in the text. Reactions were initiated by adding ATP-Mg2+. The data were fit to either one or two exponentials using the curve fitting toolbox of MATLAB (The MathWorks). Change in FRET was followed by monitoring the change in fluorescence of either Cy5 (as in Figure 2A and B) or Cy3 (as in Figure 2C). Data were fit by two exponentials using the following equation:
where i is the fluorescence intensity, a is intensity of the starting substrate, b is the intensity of the intermediates, c is the intensity of the final product, k1 is the rate constant for the first phase and k2 is the rate constant for the second phase. The intensity at time zero, a, was fixed at a value calculated by averaging intensity for 45 s before initiating the reactions and then correcting this value for the dilution that occurred when the reaction was initiated. The data was normalized from 0 to 1 using the following equation:
where n is the normalized fluorescence intensity.
Figure 2.
FRET-based assays allow separate measurement of the rate constants for (A) dimer transfer, (B) octamer transfer and (C) repositioning. In all reactions, 0-601-55 nucleosomes were used as donor nucleosomes and either core 147 bp DNA, or core 147 bp nucleosomes were used as acceptors as indicated. Each panel shows one representative time course at 20 μM ATP. The data points are shown in gray and the fit as a black line. The data in (A), (B) and (C) right panel are best fit by two exponentials whereas the data in (C) left panel are best fit by one exponential. In the reaction schematic below, Cy3 and Cy5 labels are represented as open triangles and black circles respectively. In the schematics only the products that can be inferred by changes in FRET are shown.
In the FRET-based assay shown Figure 3C, the loss of Cy5 intensity as FRET decreased was followed and the data was fit by a single exponential using the following equation:
where i is the fluorescence intensity, a is intensity of the starting substrate, b is the intensity of the final product, and k1 is the rate constant. For this data a was not fixed. The data were then normalized from 0 to 1 using the following equation:
where n is the normalized fluorescence intensity.
Results
RSC creates three distinct products that can be visualized on native gels
It has previously been shown using native gel-based assays that RSC and other SWI/SNF complexes can reposition nucleosomes towards DNA ends, exchange H2A/H2B dimers between nucleosomes and transfer histone octamers between pieces of DNA (5–7, 14–17). We first confirmed that we could analogously visualize these three distinct products with our RSC preparation and nucleosomal constructs. To monitor changes in nucleosome position, histone H3 was labeled with Cy5 on a single cysteine residue introduced at position 33, and octamers containing this histone were assembled on a DNA fragment with 40 bp of flanking DNA on either side of a centrally placed 601 positioning sequence (40-601-40). As expected, RSC repositions these centered nucleosomes, creating a new species with a mobility consistent with end positioned nucleosomes (Figure 1A lanes 1 and 2, Supplementary Figure 1).
Figure 1.
RSC catalyzes (A) movement of the DNA relative to the octamer, (B) transfer of H2A/H2B dimers between nucleosomes and (C) transfer of histone octamers between two pieces of DNA. RSC remodeling reactions were run on native gels and scanned in the Cy5 channel. In the reaction schematic shown below each gel, the black circle indicates the position of the Cy5 label and the length of the DNA template is indicated below each substrate. All reactions contained 107 nM RSC, 1 mM ATP and 7 nM Cy5 labeled nucleosome. The reactions shown in B and C contain 28 nM of unlabeled core nucleosomes or free DNA, respectively.
To detect dimer transfer, we took advantage of the fact that nucleosomes formed on DNA of different lengths can be resolved on a native gel. To track the histone dimer, H2A was labeled with Cy5 on a single cysteine residue introduced at position 120. Histone octamers containing this labeled H2A were assembled on the 39-601-39 DNA fragment. RSC acts on these nucleosomes in the presence of unlabeled core nucleosomes to generate two Cy5 labeled products (Figure 1B lanes 3 and 4, Supplementary Figure 1): a repositioned nucleosome and a core nucleosome. These data indicate that the H2A/H2B dimer is transferred to the unlabeled core nucleosome as observed previously (5).
To track octamer transfer, nucleosomes with Cy5-labeled H3 were assembled on the 40-601-40 DNA fragment. These nucleosomes were mixed with a 147 bp fragment of DNA containing the 601 positioning sequence. We find that in the presence of RSC and ATP, a new product appears on the gel that migrates with the same mobility as a core nucleosome (Figure 1C lanes 5 and 6 and Supplementary Figure 1). Additionally as the octamer is transferred to the 147 bp DNA fragment, free 227 bp DNA fragment is created (Supplementary Figure 1B lane 2). These data indicate that RSC transfers the Cy5 labeled octamer from the 227 bp DNA to the 147 bp DNA fragment, consistent with previous observations (6,7).
A FRET-based approach reveals a rapid initial phase in the transfer of dimers and octamers
While gel-based assays are very powerful in separating out different remodeled products, these assays cannot always identify transient nucleosomal intermediates. To better understand how RSC generates transfer products we searched for reaction intermediates using a FRET-based assay that allows us to track each process individually. All the experiments in this section were done at 20 μM ATP, a subsaturating concentration that made it possible to capture the fast phases of the reactions. Dimer transfer was tracked using two nucleosomes containing differentially labeled histones. Nucleosomes containing Cy5-labeled H2A were mixed with nucleosomes containing Cy3-labeled H3. In this assay, the Cy5-labeled nucleosomes act as a donor of labeled H2A/H2B dimers while the Cy3-labeled nucleosomes acts as a dimer acceptor. When the Cy5-labeled H2A/H2B dimer from the donor nucleosome is transferred to the acceptor nucleosome containing Cy3-labeled H3, the two fluorophores are brought together, causing an increase in FRET (Figure 2A). To measure FRET efficiency we excited the nucleosomes at the Cy3 absorption maximum and measured Cy5 emission over time. Interestingly, we found that FRET increases in two phases: a rapid phase with a rate constant of 1.9 ± 0.098 min−1, followed by a slower phase with a rate constant of 0.41 ± 0.028 min−1 (Table 1). The two phase nature of the reaction is even more clear at 1 mM ATP (Figure 3A) where the difference between the rate constants is larger.
Table 1.
RSC remodeling rate constants
| Rate Constants (min−1) | 1 mM ATP | 20 μM ATP | ||
|---|---|---|---|---|
| k1 | k2 | k1 | k2 | |
| Repositioning Alone | ≥ 10 | ≥ 10 | ||
| Dimer Transfer | 8.2 ± 0.14 | 0.027 ± 0.00057 | 1.9 ± 0.098 | 0.41 ± 0.028 |
| Repositioning Under Dimer Transfer Reaction Conditions | ≥ 10 | ≥ 9 | ||
| Octamer Transfer | 3.7 ± 1.4 | 0.033 ± 0.0084 | 0.66 ± 0.062 | 0.082 ± 0.015 |
| Repositioning Under Octamer Transfer Reaction Conditions | ≥ 10 | 0.80 ± 0.017 | ≥ 9 | 0.83 ± 0.043 |
Each value is the average of three independent experiments ± standard error of the mean. k1 and k2 are the rate constants of the first and second phase, respectively.
To monitor the transfer of octamers from one piece of DNA to another, nucleosomes containing Cy5-labeled H3 were mixed with Cy3-labeled DNA fragments (Figure 2B). In this assay, the Cy5-labeled nucleosome acts as a donor of labeled octamer while the Cy3-labeled DNA acts as an octamer acceptor. Transfer of the labeled octamer from the donor nucleosome to the acceptor DNA brings the two fluorophores into close proximity causing an increase in FRET. Analogous to dimer transfer, we observed that FRET increased in two phases: a rapid phase with a rate constant of 0.66 ± 0.062 min−1, followed by a slower phase with a rate constant of 0.082 ± 0.015 min−1 (Table 1).
Repositioning occurs more quickly than either dimer or octamer transfer
Previous work using native gel-based assays has suggested that nucleosomes with transferred dimers are formed more slowly than repositioned nucleosomes (5). To test whether we observe the same phenomenon using our FRET-based assay, we compared the rates of repositioning to that of dimer and octamer transfer (Figure 2C). We examined the kinetics of nucleosome repositioning using histone octamers containing Cy5-labeled H3 assembled on a Cy3–labeled DNA fragment containing the 601 positioning sequence flanked by 55 bp of linker DNA on one end. The resulting end-positioned nucleosomes start out with high FRET efficiency that decreases as the nucleosome is repositioned. The decrease in FRET was measured by following the unquenching of Cy3 fluorescence over time.
Comparing the rate of repositioning with that of dimer and octamer transfer is complicated by the fact that transfer reactions are intermolecular reactions between two substrates while repositioning is an intramolecular reaction with a single substrate. Therefore to allow more meaningful comparison between the two types of reactions, we used RSC concentrations that were saturating and in excess of both substrates. These conditions help ensure that the second substrate in the transfer reactions does not inhibit the remodeling reaction. To further control for the effects of the second substrate the rate of dimer transfer was compared to the rate of repositioning in the presence of core nucleosomes. We found that, at 20 μM ATP, the rate of repositioning is too fast to measure using manual mixing. However, under these conditions we are able to estimate a lower limit for the repositioning rate constant of 9 min−1 (Table 1). Based on this limit the fast phase of dimer transfer is at least 5-fold slower than the fast phase of repositioning. Thus although a fast phase is present in the dimer transfer reaction, the repositioning reaction still occurs at a faster rate.
Similarly, the rate of repositioning in the presence of core DNA was too fast to accurately measure and a lower limit on the rate constant of 9 min−1 was also estimated (Table 1). When the rate of octamer transfer is compared to the rate of repositioning in the presence of core DNA, the fast phase of octamer transfer is at least ten-fold slower than the fast phase of repositioning (Table 1). We also noticed that when DNA was added to the repositioning reaction, there was an additional slower phase in the repositioning kinetics. This slower phase could reflect an additional decrease in FRET caused by the transfer of the Cy5-labeled octamer to the unlabeled core DNA.
The formation of the dimer and octamer transfer intermediates observed by FRET precedes the formation of the stable transferred products observed by native gel
The two phases seen in the dimer and octamer transfer FRET kinetics experiments could either represent (i) two populations of nucleosomes that are converted at different rates or (ii) the rapid formation of an intermediate that is converted more slowly to the final product. To further explore these two models, we measured the kinetics of the two transfer reactions using a native gel assay under saturating ATP, 1mM. Under these reaction conditions, for dimer exchange, we find that the product seen on the native gel is formed at least 100-fold more slowly than the fast phase seen in the FRET reactions (Figure 3A, Table 1). Also, by FRET, 52% of the change occurs in the fast phase, whereas by gel if a fast phase is present no more than 9% of the change occurs in that phase. When octamer transfer is examined by native gel we do not find any evidence for a fast phase, whereas by FRET 16% of the change occurs in the fast phase. Further octamer transfer products are formed at least 100-fold more slowly in the gel assay than the fast phase observed by FRET (Figure 3B, Table 1). In contrast, the rate constants for repositioning as measured by the native gel and FRET-based assays are the same within error (Figure 3C, Supplementary Figure 2A).
The absence of a large fast phase in the gel-based assays for dimer and octamer transfer rules out the first model. Together with the fast phase observed by FRET, these data then strongly suggest the presence of a transient intermediate that is formed very rapidly but cannot survive on a gel. The rate constants for dimer and octamer transfer as measured by the native gel assay are similar to the respective rate constants of the second phase observed by FRET (Figure 3, Table 1). This is consistent with a model in which the slower phase represents the transition from a transient intermediate to a stable product that is visible on the native gel.
In the context of octamer transfer we were also able to follow the appearance of free DNA released from the donor nucleosome. A large fraction (>45%) of free donor DNA appears at least 50-fold faster than the stable transferred product on a gel (Figure 3D, Supplementary Figure 2B and C). The free DNA is also observed to appear with similar rates in the absence of acceptor DNA (data not shown). The implications of this result are discussed in the later sections.
Formation of stable transfer products requires continuous ATP-dependent activity by RSC
Previous work examining octamer transfer has shown that ATP hydrolysis is necessary for the formation of the octamer transfer product (6, 7). Additionally it has been shown that the acceptor DNA must be present during ATP hydrolysis (6, 7). In these experiments, ATP-dependent nucleosome remodeling was allowed to occur in the absence of acceptor DNA. If acceptor DNA was then added after removal of ATP, no octamer transfer was observed (7). These previous results are consistent with the requirement of ATP for the formation of the unstable intermediate formed by the donor nucleosome and acceptor DNA. To further investigate if ATP is also required for the collapse of the intermediate, we took advantage of our ability to temporally separate the two steps of the transfer reaction. We stopped RSC reactions after most of the fast phase was completed but before the products of dimer or octamer transfer had accumulated. We then used a gel-based assay to determine if stable transferred products could be detected. To stop the reactions, we used excess ADP, which, when added before RSC, is able to completely block repositioning (data not shown). Addition of ADP after 3 minutes prevented the formation of stable transfer products as did the addition of apyrase, which hydrolyzes ATP (Supplemental Figure 3). These results indicate that formation of both the transient intermediate and the stable products require the ATP dependent activity of RSC.
Discussion
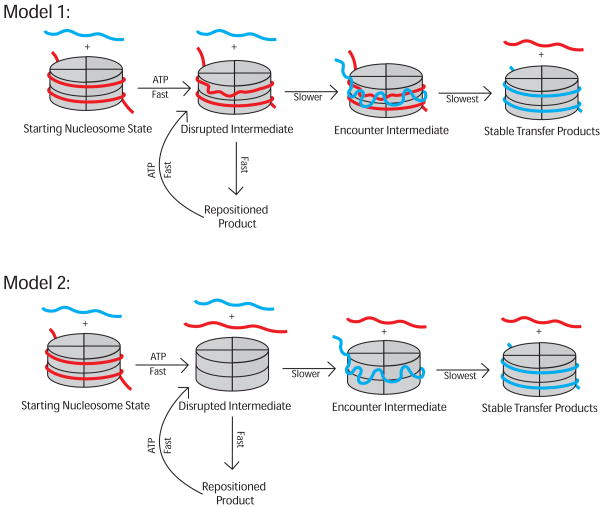
Previous work has suggested that SWI/SNF complexes generate an unstable reaction intermediate that contains disrupted histone-DNA contacts (4, 6, 8, 18–20). It has also been proposed that SWI/SNF complexes generate and inter-convert products with different regions of DNA exposed via the formation of such an intermediate (4–7, 10). Our results add to this reaction framework by capturing, for the first time, a transient reaction intermediate generated during the octamer and dimer transfer reactions catalyzed by the major SWI/SNF complex in budding yeast, RSC. As discussed below, we hypothesize that this intermediate entails a close encounter between the two exchange substrates and is formed after the disruption of histone-DNA contacts (Figure 5).
Figure 5.
Models for RSC catalyzed octamer transfer. In these schematic the acceptor DNA is shown in blue. A disrupted intermediate is inferred based on previous work (4, 6, 18, 19) whereas the encounter intermediate is inferred based on this work. It is assumed that repositioning is limited by the formation of the disrupted intermediate, the fast phase of octamer transfer is limited by the formation of the encounter intermediate, and formation of the stable transfer product is limited by the conversion of the encounter intermediate to the stable transfer product. ATP is shown for ATP-dependent steps.
Comparison of the FRET-based data and gel-based data suggests that dimer and octamer transfer occur via at least two steps: (i) a fast step in which the histones from the donor nucleosome are brought in close vicinity of the acceptor nucleosome or DNA to generate an unstable intermediate species, and (ii) a slow step in which this unstable species is converted to a stable nucleosome containing histone components deriving from the donor nucleosome. There are two extreme models to explain these observations. Below we discuss these models in the context of octamer transfer, but a similar analysis would apply to the dimer exchange reaction.
In the first model, complete dissociation of the donor DNA from the histone octamer and association of acceptor DNA with the histone octamer are coupled. The unstable intermediate contains, in close proximity, a donor nucleosome with disrupted histone-DNA contacts and an acceptor DNA (Figure 5A). In such an encounter intermediate, the acceptor DNA could be partially associated with the exposed regions of the histone octamer in the donor nucleosome. The slow step would then involve complete transfer of the octamer from the donor DNA to the acceptor DNA. The disrupted intermediate could arise due to globally disrupted histone-DNA contacts as recently proposed (19). Alternatively the disrupted intermediate could be a consequence of the histone octamer translocating beyond the DNA ends and thereby generating exposed octamer surfaces that can interact with another piece of DNA. In the context of dimer exchange, the encounter intermediate would resemble a dinucleosome. Indeed previous work has shown that SWI/SNF family complexes can generate dinucleosome-like species from mononucleosomes (7, 8, 10, 21–25). If the encounter intermediate involves movement of the donor octamer beyond a DNA end, then it would be expected to form less readily in the context of nucleosomal arrays. However, previous work has shown that RSC can catalyze exchange of dimers between nucleosomes within arrays, suggesting that the dimer exchange reaction does not rely solely on the histone octamer moving off the DNA end (5).
In the second model, dissociation of the donor DNA from the histone octamer and association of acceptor DNA with the histone octamer occur in two separate steps (Figure 5B). The unstable encounter intermediate contains only the histone octamer from the donor nucleosome in close proximity to the acceptor DNA, while the donor DNA is completely removed in a prior step. The slow step would then involve association of the donor DNA with the histone octamer to form a stable nucleosomal complex. In the first model, a slightly positively charged RSC active site could facilitate global histone-DNA disruption as well as accommodate the acceptor DNA. In the second model, the RSC active site has to be compatible with binding a nucleosome as well as the separated histone octamer and DNA, and each imposes very different electrostatic requirements on the RSC active site. In such a case, it is possible, as discussed below, that other RSC subunits participate in binding the different intermediate histone and DNA states.
Interestingly, we find that free DNA from the donor nucleosome appears faster than the octamer transfer product when visualized by native gel. Further the donor free DNA appears with similar kinetics in the absence of the acceptor DNA. These observations are initially more consistent with the second model, in which the disrupted intermediate entails complete dissociation of the nucleosomal DNA. However, it is also possible that the less stably bound DNA in the disrupted intermediate of Model 1 dissociates readily on a native gel.
In both the above models, collapse of the encounter intermediate into a stable nucleosome is not ATP-dependent. The ATP requirement that we observe in Figure 4 arises because, only a small fraction of the disrupted intermediate gets converted to the encounter intermediate in each remodeling cycle. As a result, ATP-dependent RSC activity is necessary to constantly replenish the pool of disrupted intermediate (Figure 5). By itself, the much slower formation of the stable transfer products, compared to repositioned products, could be interpreted to mean that the transfer products are off-pathway products. However, the observation that the substrates of the transfer reactions come together much faster suggests that the exchange products can be on-pathway RSC products. Further, the time-scale on which the encounter intermediate is formed is comparable to the in vivo time-scales associated with RNA and DNA polymerase translocation on 150 bp of DNA (26, 27), raising the possibility that these exchange products can have biologically relevant roles.
Both models (Figure 5) raise the possibility that RSC has binding sites for more than one nucleosomal or DNA substrate. These additional binding sites may be provided by non-catalytic RSC subunits. Precedence for such a mechanism comes from previous work on the SWR and SWI/SNF complexes that identified specific subunits required for dimer transfer (28, 29). These subunits are believed to facilitate efficient dimer transfer by directly binding to H2A/H2B dimers. Although a homologous subunit in RSC is not readily apparent, it is possible that one of RSC’s many non-catalytic subunits may be involved in binding additional substrates to facilitate transfer. RSC function may also be additionally regulated by other chromatin factors that bias RSC towards forming predominantly dimer exchange or octamer transfer products.
Supplementary Material
Acknowledgments
We thank B. Cairns for the BCY211 yeast strain. We would also like to thank members of the Narlikar laboratory for technical advice and helpful comments on the manuscript; Coral Zhou for preparation of the nucleosomes used in Figure 3C and D; the anonymous reviewers of this work for experimental suggestions that helped strengthen the manuscript.
This work was supported by grants from the NIH and the Sandler Family Foundation. C.E.R was supported by a NSF Graduate Research Fellowship.
Abbreviations
- FRET
fluorescence resonance energy transfer
- TAP-tag
Tandem Affinity Purification
Footnotes
Supporting Information Available
The full gels used to create Figure 1 including nucleosome positioning standards are shown in Supplementary Figure 1. Supplementary Figure 2 shows representative gels from Figure 3C and D. Supplementary Figure 3 shows the effect of apyrase on dimer and octamer transfer. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Cao Y, Cairns BR, Kornberg RD, Laurent BC. Sfh1p, a component of a novel chromatin-remodeling complex, is required for cell cycle progression. Mol Cell Biol. 1997;17:3323–3334. doi: 10.1128/mcb.17.6.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fyodorov DV, Kadonaga JT. The many faces of chromatin remodeling: SWItching beyond transcription. Cell. 2001;106:523–525. doi: 10.1016/s0092-8674(01)00478-0. [DOI] [PubMed] [Google Scholar]
- 3.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 4.Narlikar GJ, Phelan ML, Kingston RE. Generation and interconversion of multiple distinct nucleosomal states as a mechanism for catalyzing chromatin fluidity. Mol Cell. 2001;8:1219–1230. doi: 10.1016/s1097-2765(01)00412-9. [DOI] [PubMed] [Google Scholar]
- 5.Bruno M, Flaus A, Stockdale C, Rencurel C, Ferreira H, Owen-Hughes T. Histone H2A/H2B dimer exchange by ATP-dependent chromatin remodeling activities. Mol Cell. 2003;12:1599–1606. doi: 10.1016/s1097-2765(03)00499-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lorch Y, Zhang M, Kornberg RD. Histone octamer transfer by a chromatin-remodeling complex. Cell. 1999;96:389–392. doi: 10.1016/s0092-8674(00)80551-6. [DOI] [PubMed] [Google Scholar]
- 7.Phelan ML, Schnitzler GR, Kingston RE. Octamer transfer and creation of stably remodeled nucleosomes by human SWI-SNF and its isolated ATPases. Mol Cell Biol. 2000;20:6380–6389. doi: 10.1128/mcb.20.17.6380-6389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorch Y, Cairns BR, Zhang M, Kornberg RD. Activated RSC-nucleosome complex and persistently altered form of the nucleosome. Cell. 1998;94:29–34. doi: 10.1016/s0092-8674(00)81218-0. [DOI] [PubMed] [Google Scholar]
- 9.Saha A, Wittmeyer J, Cairns BR. Mechanisms for nucleosome movement by ATP-dependent chromatin remodeling complexes. Results Probl Cell Differ. 2006;41:127–148. doi: 10.1007/400_005. [DOI] [PubMed] [Google Scholar]
- 10.Schnitzler G, Sif S, Kingston RE. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- 11.Wittmeyer J, Saha A, Cairns B. DNA translocation and nucleosome remodeling assays by the RSC chromatin remodeling complex. Methods Enzymol. 2004;377:322–343. doi: 10.1016/S0076-6879(03)77020-7. [DOI] [PubMed] [Google Scholar]
- 12.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 13.Yang JG, Madrid TS, Sevastopoulos E, Narlikar GJ. The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat Struct Mol Biol. 2006;13:1078–1083. doi: 10.1038/nsmb1170. [DOI] [PubMed] [Google Scholar]
- 14.Flaus A, Owen-Hughes T. Dynamic properties of nucleosomes during thermal and ATP-driven mobilization. Mol Cell Biol. 2003;23:7767–7779. doi: 10.1128/MCB.23.21.7767-7779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 16.Bouazoune K, Miranda TB, Jones PA, Kingston RE. Analysis of individual remodeled nucleosomes reveals decreased histone-DNA contacts created by hSWI/SNF. Nucleic Acids Res. 2009;37:5279–5294. doi: 10.1093/nar/gkp524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol. 2005;12:747–755. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- 18.Lorch Y, Maier-Davis B, Kornberg RD. Chromatin remodeling by nucleosome disassembly in vitro. Proc Natl Acad Sci U S A. 2006;103:3090–3093. doi: 10.1073/pnas.0511050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorch Y, Maier-Davis B, Kornberg RD. Mechanism of chromatin remodeling. Proc Natl Acad Sci U S A. 2010;107:3458–3462. doi: 10.1073/pnas.1000398107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dechassa ML, Sabri A, Pondugula S, Kassabov SR, Chatterjee N, Kladde MP, Bartholomew B. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol Cell. 2010;38:590–602. doi: 10.1016/j.molcel.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schnitzler GR, Cheung CL, Hafner JH, Saurin AJ, Kingston RE, Lieber CM. Direct imaging of human SWI/SNF-remodeled mono- and polynucleosomes by atomic force microscopy employing carbon nanotube tips. Mol Cell Biol. 2001;21:8504–8511. doi: 10.1128/MCB.21.24.8504-8511.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulyanova NP, Schnitzler GR. Inverted factor access and slow reversion characterize SWI/SNF-altered nucleosome dimers. J Biol Chem. 2007;282:1018–1028. doi: 10.1074/jbc.M609473200. [DOI] [PubMed] [Google Scholar]
- 23.Ulyanova NP, Schnitzler GR. Human SWI/SNF generates abundant, structurally altered dinucleosomes on polynucleosomal templates. Mol Cell Biol. 2005;25:11156–11170. doi: 10.1128/MCB.25.24.11156-11170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engeholm M, de Jager M, Flaus A, Brenk R, van Noort J, Owen-Hughes T. Nucleosomes can invade DNA territories occupied by their neighbors. Nat Struct Mol Biol. 2009;16:151–158. doi: 10.1038/nsmb.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorch Y, Zhang M, Kornberg RD. RSC unravels the nucleosome. Mol Cell. 2001;7:89–95. doi: 10.1016/s1097-2765(01)00157-5. [DOI] [PubMed] [Google Scholar]
- 26.O’Farrell PH. Developmental biology. Big genes and little genes and deadlines for transcription. Nature. 1992;359:366–367. doi: 10.1038/359366a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol. 2009;16:1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu WH, Wu CH, Ladurner A, Mizuguchi G, Wei D, Xiao H, Luk E, Ranjan A, Wu C. N terminus of Swr1 binds to histone H2AZ and provides a platform for subunit assembly in the chromatin remodeling complex. J Biol Chem. 2009;284:6200–6207. doi: 10.1074/jbc.M808830200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X, Zaurin R, Beato M, Peterson CL. Swi3p controls SWI/SNF assembly and ATP-dependent H2A-H2B displacement. Nat Struct Mol Biol. 2007;14:540–547. doi: 10.1038/nsmb1238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.