Abstract
Nucleolin, the most abundant nucleolar phosphoprotein of eukaryotic cells, is known primarily for its role in ribosome biogenesis and cell proliferation. It is, however, a multifunctional protein that, depending on the cellular context, can drive either cell proliferation or apoptosis. Our laboratory recently demonstrated that nucleolin can function as a repressor of c-MYC transcription by binding to and stabilizing the formation of a G-quadruplex structure in a region of the c-MYC promoter responsible for controlling 85–90% of c-MYC’s transcriptional activity. In this study, we investigate the structural elements of nucleolin that are required for c-MYC repression. The effect of nucleolin deletion mutants on the formation and stability of the c-MYC G-quadruplex, as well as c-MYC transcriptional activity, was assessed by circular dichroic spectropolarimetry (CD),1 thermal stability, and in vitro transcription. Here we report that nucleolin’s RNA binding domains (RBDs) 3 and 4, as well as the arginine-glycine-glycine (RGG) domain, are required to repress c-MYC transcription.
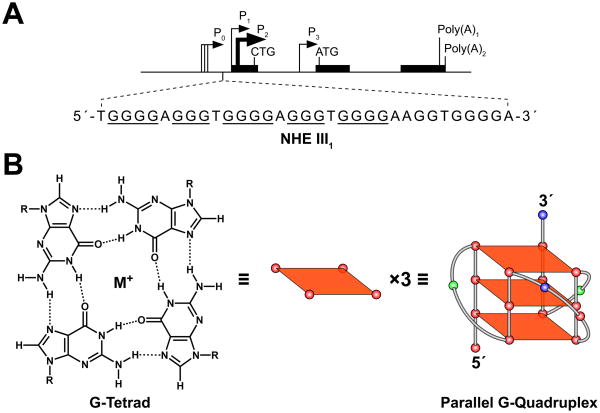
The mechanisms that regulate c-MYC transcription are complex and involve multiple promoters, start sites, and cis-elements (NHEs) (Figure 1A) (1). The NHE III1, which is located −142 to −115 base pairs upstream of the P1 promoter, has been shown to control 85–90% of c-MYC transcription. The template strand of this element consists of a G-rich sequence that can equilibrate between transcriptionally active forms (duplex and single-stranded DNA) and a silencer structure (G-quadruplex) (2).
Fig. 1.
Promoter structure of the c-MYC gene and scheme of its G-quadruplex structure. (A) Location of the NHE III1 region within the c-MYC promoter. Runs of guanines that can participate in G-quadruplex formation are underlined. (B) Scheme of a guanine tetrad and a cartoon of the c-MYC G-quadruplex structure. Left: H-bonding pattern in a G-tetrad; center: schematic diagram of a G-tetrad; right: cartoon representing a G-quadruplex structure that is found in the c-MYC promoter region.
It is well known that G-rich DNA has the ability to form G-quadruplex structures under physiological conditions (3). The fundamental structural unit of a G-quadruplex is known as a G-tetrad, which is composed of four guanines aligned in a planar ring configuration where each guanine interacts with two adjacent guanines via Hoogsteen hydrogen bonding (Figure 1B, left and center) (4–6). Two or more G-tetrads can stack to form a G-quadruplex structure (Figure 1B, right). Formation of a G-quadruplex in the c-MYC promoter has been shown to be facilitated by the negative supercoiling generated from transcription (7, 8). In addition, when the G-rich strand of the NHE III1 is assembled into a G-quadruplex, the binding sites of c-MYC transcriptional activators such as Sp1 and CNBP are masked, thus silencing c-MYC transcription (9).
Recent reports have demonstrated that putative G-quadruplex motifs are highly prevalent in human promoter regions (10–12). In addition, the presence of G-quadruplex motifs has been shown to correlate with the gene function, with oncogenes having a disproportionately high incidence of G-quadruplex motifs, whereas the promoters of tumor suppressors exhibit an extremely low potential for G-quadruplex formation (13). Importantly, G-quadruplex topological diversity arises from variations in strand directionality, loop length and sequence, and the number of G-tetrad stacks, allowing for specific structural recognition by G-quadruplex-interactive proteins (14–19).
We have previously identified nucleolin as a selective c-MYC G-quadruplex-binding protein that has the ability to induce the stable formation of the parallel c-MYC G-quadruplex from single-stranded DNA (20). In addition, we have established that nucleolin interacts with the c-MYC promoter in vivo in HeLa cells. Most importantly, our laboratory has demonstrated that nucleolin can repress c-MYC transcription in a significant and dose-dependent manner (20).
Nucleolin, one of the most abundant non-ribosomal proteins of eukaryotes, has been shown to play a role in ribosome biogenesis and cell proliferation. It is, however, a multifunctional protein whose function is dependent on cellular context. A number of stress stimuli have been shown to induce changes in the cellular localization of nucleolin (21–28), and there is strong evidence that nucleolin can function as a stress-sensitive tumor suppressor (22, 24, 29). In addition, stress- or cisplatin-activated p53 has been shown to translocate to the nucleolus where it forms a complex with nucleolin, causing nucleolin to relocate from the nucleolus to the nucleoplasm (30, 31). This binding is believed to be one of the mechanisms regulating the reactivation of the p53 protein in cisplatin-treated human cervix carcinoma cells (29, 31). Interestingly, the interaction with p53 is transient, leading to the accumulation of nucleolin in the nucleoplasm. This could allow nucleolin to subsequently interact with additional proteins or DNA targets in the nucleoplasm, which would further stimulate the execution of the apoptotic program (21–23).
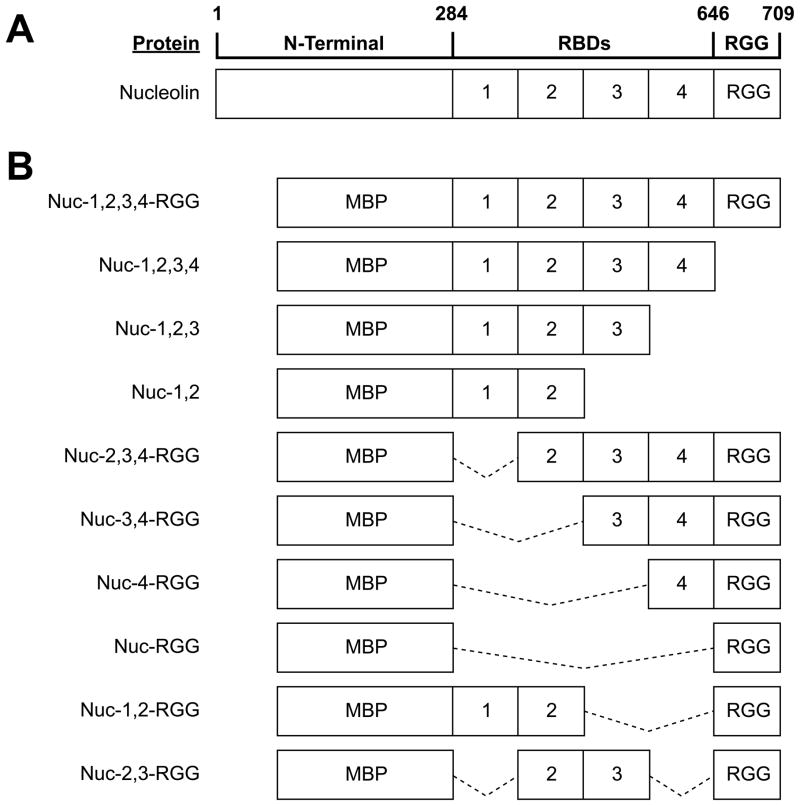
Nucleolin is a modular protein composed of an N-terminal domain rich in acidic residues, a central region containing four globular RNA binding domains (RBDs) separated by flexible linker loops, and a C-terminal domain rich in arginine and glycine residues (RGG domain) (Figure 2A) (32). This modular architecture allows for higher versatility of the protein, since by combining multiple domains, nucleolin can construct various interaction surfaces that can be assembled and disassembled as needed. Consequently, nucleolin can recognize a large number of targets. For example, by combining its first two RBDs, nucleolin interacts with the stem-loop RNA structure formed by the nucleolin recognition element (33, 34), while all four RBDs are required for binding to a single-stranded RNA motif found in pre-ribosomal RNA (35–37). In this study, we investigate the structural elements of nucleolin that are required for repression of c-MYC transcription. The effects of both N-terminal and C-terminal deletion mutants of nucleolin on c-MYC G-quadruplex formation and transcriptional repression were assessed.
Fig. 2.
Nucleolin deletion mutants. (A) Diagram of nucleolin structure. (B) Diagram of the nucleolin deletion mutants used in this study. Solid lines indicate regions of the nucleolin peptide that have been deleted from the Nuc-1,2,3,4-RGG construct. All proteins were overexpressed in E. coli fused at the N-terminal to the maltose-binding protein (MBP).
MATERIALS AND METHODS
Plasmid Constructs
The reporter plasmid containing the NHE III1 region of the human c-MYC promoter (pDel-4) linked to the firefly luciferase gene was kindly provided by Dr. Bert Vogelstein (Johns Hopkins University) (38). Plasmids pNuc-1,2,3,4-RGG, pNuc-3,4-RGG, pNuc-1,2-RGG, and pNuc-RGG for the expression of recombinant nucleolin deletion mutants were generously provided by Dr. Leslyn A. Hanakahi (Johns Hopkins Bloomberg School of Public Health). The pNuc-1,2,3,4-RGG plasmid was used as a template for the amplification of nucleolin’s DNA coding sequence and construction of pNuc-2,3-RGG, pNuc-1,2,3, pNuc-2,3,4-RGG, and pNuc-4-RGG. Inverse PCR amplification and linearization of vector backbone were performed as previously described (39). Briefly, each PCR reaction contained 0.01 pmol template DNA (pNuc-1,2,3,4-RGG), 15 pmols of reverse primer REV: 5′-AGCCGTAGTCGGTTCTGTGCCTTCCACTTTCTGTTTCTTGGCTTCAGGAGCTGCT-3′, and forward primer FWD1: 5′-GACCACAAGCCACAAGGAAAGAAGACGAAGTTTGAATAGGAATTCCTCGACCTGC-3′ to amplify empty vector backbone for construction of pNuc-1,2,3 and pNuc-1,2,3,4; or forward primer FWD2: 5′-AAGGGTGAAGGTGGCTTCGGGGGTCGTGGTGGAGGCAGAGGCGGCTTTGGAGG AC-3′ to amplify vector backbone containing RGG domain for construction of pNuc-4-RGG, pNuc-2,3-RGG, and pNuc-2,3,4-RGG. 1× Pfu reaction buffer, 200 μM of each dNTP and 2.5 U Pfu DNA polymerase (Fermentas, Burlington, Canada) were mixed in a 50 μl final volume. The temperature cycles were as follows: 95 °C for 3 min, 18 cycles of 95 °C for 45 s, 62 °C for 1 min, and 68 °C for 2 min/kb (12 min), with a final extension at 68 °C for 2 min/kb (12 min). Linearized vectors were subsequently treated with 10 U of DpnI for 3 h at 37 °C to remove parental templates. Deletion mutants were constructed by using the In-Fusion cloning system (Clontech) according to the manufacturer’s recommendations. Briefly, each sequence of interest was amplified using Advantage HD polymerase (Clontech) for 20 cycles by PCR using the primer pairs indicated in Table 1. The amplified DNA inserts were purified with the QIAquick gel extraction kit (Qiagen) and subcloned into the linear vector using the In-Fusion cloning kit. Each cloning reaction contained 100 ng linear vector, 200 ng insert DNA, 1× In-Fusion Reaction Buffer, and 1 μL In-Fusion enzyme in a 10 μL final volume. The In-Fusion cloning reaction was incubated for 15 min at 37 °C, followed by 15 min at 50 °C and 5 min on ice. 40 μL of TE buffer (pH 8) was added to each reaction mixture. 2.5 μL of the In-Fusion mixture was then used to transform Origami B(DE3) competent cells. All clones generated by chain reaction amplification were sequenced throughout the amplified region.
Table 1.
List of primer pairs used for the construction of nucleolin deletion mutants.
| Primer Sequence |
|---|
|
Nuc-1,2,3 FWD: 5′-GAACCGACTACGGCTTTCAATCTCTTTGTTGGAAACCTAAACTTT-3′ REV: 5′-GGATGGCTGGCTTCTGGCATTAGGTGATCCTTGTGGCTTGTGGTC-3′ |
|
Nuc-1,2,3,4 FWD: 5′-GAACCGACTACGGCTTTCAATCTCTTTGTTGGAAACCTAAACTTT-3′ REV: 5′-CTTAGGTTTGGCCCAGTCCAAGGTAACTTTTTGTGGCTTGTGGTC-3′ |
|
Nuc-4-RGG FWD: 5′-GAACCGACTACGGCTAAAACTCTGTTTGTCAAAGGCCTGTCTGAG-3′ REV: 5′-CTTAGGTTTGGCCCAGTCCAAGGTAACTTTGCCACCTTCACCCTT-3′ |
|
Nuc-2,3-RGG FWD: 5′-GAACCGACTACGGCTAAAGACAGTAAGAAAGAGCGAGATGCGAGA-3′ REV: 5′-GGATGGCTGGCTTCTGGCATTAGGTGATCCGCCACCTTCACCCTT-3′ |
|
Nuc-2,3,4-RGG FWD: 5′-GAACCGACTACGGCTAAAGACAGTAAGAAAGAGCGAGATGCGAGA-3′ REV: 5′-CTTAGGTTTGGCCCAGTCCAAGGTAACTTTGCCACCTTCACCCTT-3′ |
Insert segments were generated by PCR using primers containing a fragment-specific sequence and 15-base-pair overhangs that overlap with the plasmid backbone (underlined bases) to allow for restriction-independent and ligase-free cloning.
Purification of Recombinant Nucleolin
All recombinant nucleolin deletion mutants were fused at the N-terminal to Escherichia coli maltose-binding protein (MBP). The maltose-binding protein-fused proteins were purified on an amylose column (New England Biolabs), following the manufacturer’s protocol. The proteins were then dialyzed and concentrated in assay buffer (20 mM Tris-HCl, pH 7.4, 5 mM NaCl, 1 mM EDTA, in 50% glycerol). All purified fusion proteins migrated as single species on SDS-PAGE. Protein concentration was determined by the Bradford protein assay (Bio-Rad).
CD Spectroscopy
Oligonucleotide stocks were diluted to 5 μM in 50 mM Tris-HCl, pH 7.4. Pu47ss (single-stranded/unstructured c-MYC NHE III1 oligo) samples were incubated with either assay buffer or 5 μM of recombinant nucleolin protein at room temperature for 30 min to reach equilibrium prior to CD spectroscopy. To assemble the Pu47 oligo into the c-MYC G-quadruplex conformation, the oligo was heated at 95 °C for 10 min and left to cool gradually to room temperature. The assembled G-quadruplex was then incubated with either assay buffer or 5 μM recombinant protein at room temperature for 30 min prior to CD spectroscopy. CD spectra were recorded on a Jasco-810 spectropolarimeter (Easton, MD) at room temperature, using a quartz cell of 1-mm optical path length and an instrument scanning speed of 100 nm/min, with a response time of 1 s and over a wavelength range of 225 to 325 nm. The reported spectrum of each sample represents the average of three scans. The spectral contributions from buffer and protein were subtracted as appropriate. To determine the stability of the G-quadruplex structures induced by the different nucleolin deletion mutants, the molar ellipticity versus temperature profiles (CD melting curves) of the G-quadruplexes were measured at 262 nm for the G-quadruplex using a temperature gradient of 1 °C/min from 20 to 95 °C.
Electrophoretic Mobility Shift Assay (EMSA)
The Pu47 oligonucleotide containing the c-MYC NHE III1 region was radiolabeled by incubating the DNA oligo with [γ-32P]dATP and T4 polynucleotide kinase (Fermentas). Radiolabeled Pu47 was then pre-assembled into a G-quadruplex structure by incubating it for 10 min at 95 °C in the presence of 100 mM KCl and allowing it to gradually cool to room temperature. The radiolabeled oligo was then purified by electrophoresis on a 12% non-denaturing polyacrylamide gel. Binding of G-quadruplex DNA was carried out in 20-μL reactions containing 10 mM Tris-HCl, pH 7.4, 1 mM EDTA, 1 mM DTT, 50 ng/μL poly(dI-dC), 4 μg/mL BSA, and 25 mM KCl. Glycerol (5%) was added to each EMSA reaction immediately before loading onto a 4% non-denaturing polyacrylamide gel containing 0.5× TBE. Protein complexes were resolved by running the gel at 10 mA for 1 h at room temperature. Dried EMSA gels were exposed on a phosphor screen for 24 h; after which bound and unbound DNA was visualized and quantified using a Storm 820 phosphorimager and ImageQuant Software (Molecular Dynamics).
In Vitro Transcription
In vitro transcription assays were performed using the HeLaScribe Nuclear Extract In Vitro Transcription System (Promega) following the manufacturer’s protocol, with one exception: the DNA template used was negatively supercoiled plasmid DNA containing the c-MYC promoter region (a gift from Dr. Bert Vogelstein at Johns Hopkins University). Briefly, 25-μL reaction mixtures containing 1 mg of template DNA, 1× reaction buffer, 3 mM MgCl2, 0.4 mM rATP, 0.016 mM rUTP, 0.4 mM rCTP, 0.4 mM rGTP, and 10 μCi [α-32P]rUTP (Amersham Biosciences) were assembled on ice. Transcription was initiated by adding 8 U (50 μg) of HeLa nuclear extract and the reaction mixture was allowed to incubate for 60 min at 30 °C. The reactions were then terminated with 175 μL stop solution. After phenol/chloroform extraction, RNA transcripts were ethanol precipitated, dried, and redissolved in formamide loading buffer. RNA transcripts were resolved by denaturing gel electrophoresis (6% PAGE, 1× TBE). Radioactive transcription products were detected by autoradiography using a Storm 820 phosphorimager and ImageQuant Software (Molecular Dynamics).
RESULTS
Generation of Nucleolin Deletion Mutants
To obtain an understanding, at the molecular level, of which of the nucleolin domains contain the c-MYC G-quadruplex binding activity, and to establish the biological relevance of the various nucleolin’s domains on c-MYC repression, we designed a number of nucleolin deletion mutants (Figure 2B). Full-length nucleolin cannot be expressed in E. coli, but deletion of the N-terminal has been shown to permit adequate expression of recombinant nucleolin (40). The Nuc-1,2,3,4-RGG expression vector, which carries the coding region for RBDs 1, 2, 3, and 4, as well as the RGG, was generously provided by Dr. Hanakahi, and was used to generate the different nucleolin deletion mutants listed in Figure 2B. All recombinant proteins were purified and dialyzed in assay buffer as described above to concentrate the protein and remove excess salt.
C-Terminal Deletion Mutagenesis Strongly Impairs Nucleolin’s Ability to Promote G-Quadruplex Formation
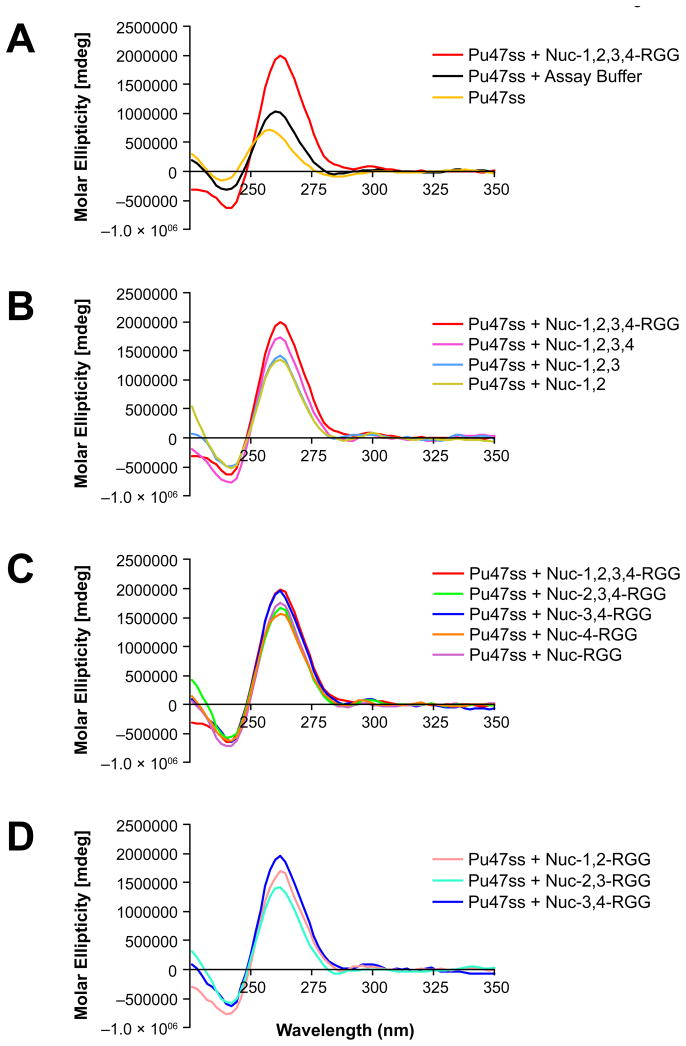
Conversions of the Pu47ss oligomer containing the c-MYC G-quadruplex-forming motif to the G-quadruplex conformation, induced by the different nucleolin deletion mutants, were monitored via CD. Each deletion mutant was incubated with Pu47ss for 1 h at room temperature before CD analysis. Consistent with our previous results (20), we found that Nuc-1,2,3,4-RGG can strongly induce the formation of a parallel c-MYC G-quadruplex structure, as observed by a shift of the maximum positive peak from 258 nm to the G-quadruplex signature peak at 262 nm (Figure 3A). Similar results were obtained from all the nucleolin deletion mutants; however, the intensity of the peaks varied among the different deletion mutants, with the C-terminal domain deletion mutants having a more dramatic decrease in their ability to induce G-quadruplex formation (Figure 3B) than the N-terminal deletion mutants (Figure 3C). Deletion of the RGG and the RBD4 domains were most detrimental, suggesting that these domains play an important role in the induction of the c-MYC G-quadruplex structure. Deletion mutant Nuc-3,4-RGG induced G-quadruplex formation to the same extent as Nuc-1,2,3,4-RGG. In addition, substitution of RBDs 3 and 4 with either RBDs 1 and 2 or RBDs 2 and 3 in Nuc-3,4-RGG did not lead to equal G-quadruplex formation, suggesting that the RBDs are not equivalent and that RBDs 3 and 4 are critical for G-quadruplex stabilization (Figure 3D). Furthermore, removal of the RGG domain from Nuc-1,2-RGG, Nuc-2,3-RGG, or Nuc-3,4-RGG completely abolished the protein’s ability to induce G-quadruplex formation (data not shown).
Fig. 3.
Effect of deletion mutagenesis on the ability of nucleolin to induce c-MYC G-quadruplex formation. (A) CD spectra of Pu47ss after incubation with Nuc-1,2,3,4-RGG, or assay buffer containing 20 mM Tris-HCl, pH 7.4, 5 mM NaCl, 1 mM EDTA, in 50% glycerol. (B) CD spectra of Pu47ss after incubation with C-terminal nucleolin deletion mutants. (C) CD spectra of Pu47ss after incubation with nucleolin’s N-terminal deletion mutants. (D) Comparison of the effect of RBD-substitution on the ability of Nuc-3,4-RGG to induce G-quadruplex formation. Formation of a parallel G-quadruplex structure is reflected by the change in wavelength from 258 nm (single-stranded DNA) to 262 nm (G-quadruplex DNA) and increased molar ellipticity at 262 nm.
The C-Terminal Region of Nucleolin Containing RBDs 3 and 4 and the RGG Domain Induces Formation of a Stable c-MYC G-Quadruplex Structure
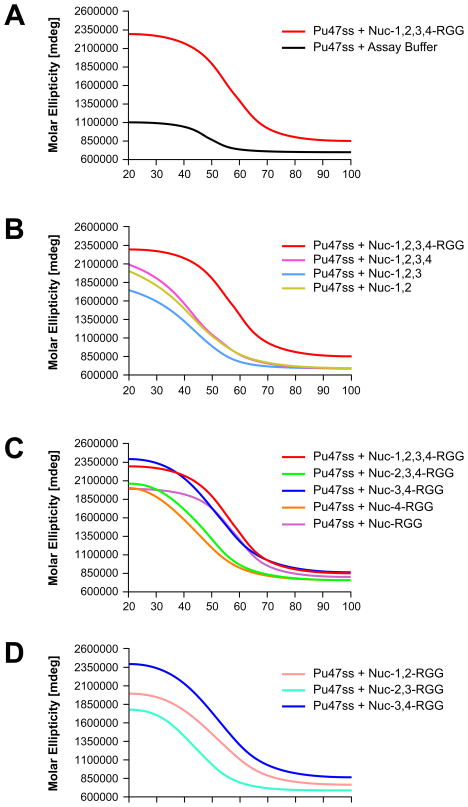
To assess the effect of each deletion mutant on the stability of the G-quadruplex structure, the thermal stability was determined by measuring the molar ellipticity of the G-quadruplex at 262 nm at increasing temperatures. Table 2 lists the effect of each deletion mutant on c-MYC G-quadruplex stability. Consistent with previous studies in our laboratory (20), we found that Nuc-1,2,3,4-RGG induces the formation of a stable c-MYC G-quadruplex structure, as observed by a shift to the right of the melting curve at 262 nm and a melting temperature of 58 °C (Figure 4A). None of the deletion mutants lacking the RGG domain were able to induce a G-quadruplex structure that was as stable as the one induced by Nuc-1,2,3,4-RGG (Figure 4B). Several of the N-terminal deletion mutants, however, were able to induce the formation of a stable G-quadruplex structure, particularly Nuc-3,4-RGG and Nuc-RGG, which induced the formation of G-quadruplexes with melting temperatures of 58 °C and 55 °C, respectively (Figure 4C). In addition, substitution of RBDs 3 and 4 with RBDs 1 and 2 or RBDs 2 and 3 in the presence of the RGG domain demonstrated that the different RBDs are not equivalent, as these proteins were not able to stabilize the c-MYC G-quadruplex to the same extent as the Nuc-3,4-RGG protein (Figure 4D). In summary, these results demonstrate that the C-terminal domain of nucleolin plays a critical role in the formation of the c-MYC G-quadruplex structure. In addition, we demonstrate that the RGG domain is essential for c-MYC G-quadruplex stabilization. Furthermore, our results confirm previous reports that show that the RGG domain plays a critical role in G-quadruplex binding (41–43).
Table 2.
Effect of nucleolin deletion mutants on c-MYC G-quadruplex thermal stability.
| Nucleolin Deletion Mutants | Tm (°C) | −ΔTm in comparison to Nuc-1,2,3,4-RGG |
|---|---|---|
| Nuc-1,2,3,4-RGG | 59 | – |
| Nuc-1,2,3,4 | 43 | 16 |
| Nuc-1,2,3 | 41 | 18 |
| Nuc-1,2 | 43 | 16 |
| Nuc-2,3,4-RGG | 51 | 8 |
| Nuc-3,4-RGG | 58 | 1 |
| Nuc-4-RGG | 43 | 16 |
| Nuc-RGG | 55 | 4 |
| Nuc-1,2-RGG | 52 | 7 |
| Nuc-2,3-RGG | 43 | 16 |
Fig. 4.
Effect of deletion mutagenesis on the ability of nucleolin to promote the formation of a thermally stable c-MYC G-quadruplex. (A) Melting curves obtained for Pu47 containing the c-MYC G-quadruplex motif after incubation with the various nucleolin deletion mutants, including Nuc-1,2,3,4-RGG. (B) Effect of nucleolin C-terminal deletions on c-MYC G-quadruplex stability. (C) Effect of nucleolin N-terminal deletions on c-MYC G-quadruplex stability. (D) Comparison of the effects that Nuc-1,2-RGG, Nuc-2,3-RGG, and Nuc-3,4-RGG have on c-MYC G-quadruplex stabilization.
The Minimal c-MYC G-Quadruplex Binding Domain of Nucleolin Consists of RBDs 3 and 4 and the RGG Domain
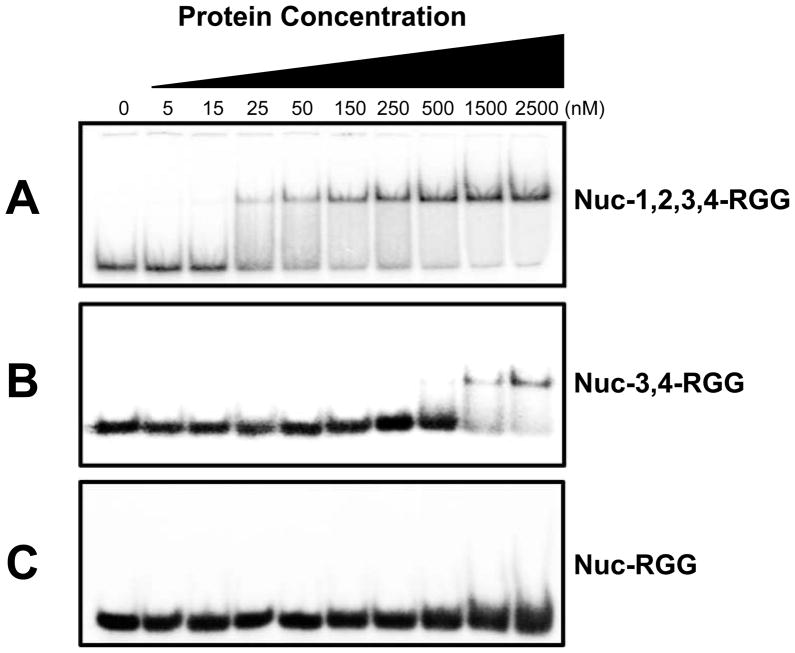
In a previous study we showed that a recombinant protein containing nucleolin’s RBDs 1, 2, 3, and 4, as well as the RGG domain, binds preferentially and with high affinity to parallel c-MYC-like G-quadruplex structures (20). In an attempt to define the smaller nucleolin sub-domains capable of interacting with the c-MYC G-quadruplex, we performed gel EMSAs on select nucleolin deletion mutants with a G-quadruplex formed from the Pu47 oligonucleotide. On the basis of our CD spectropolarimetric and thermal stability studies, we chose Nuc-1,2,3,4-RGG, Nuc-3,4-RGG, and Nuc-RGG for comparison of their c-MYC G-quadruplex binding affinity (Figure 5). The deletion proteins were expressed in E. coli as chimeric MBP-fusion proteins and purified to homogeneity. Consistent with our previous results, Nuc-1,2,3,4-RGG bound with high affinity to the c-MYC G-quadruplex structure, as determined by a strong single shift in the mobility of the radiolabeled c-MYC G-quadruplex (Figure 5A). The single mobility shift observed at both low and high protein concentrations suggests that nucleolin is binding each G-quadruplex as a monomer. In addition, we found that the minimal c-MYC G-quadruplex binding domain of nucleolin consists of RBDs 3 and 4, as well as the RGG domain (Figure 5B). Interestingly, the interaction between Nuc-3,4-RGG and the c-MYC G-quadruplex structure was significantly weaker than that of the Nuc-1,2,3,4-RGG protein with the G-quadruplex (Figure 5B). Taken together with the results from our CD and thermal stability studies, the weaker binding of Nuc-3,4-RGG indicates that while RBDs 1 and 2 are not essential for the induction of the c-MYC G-quadruplex formation, these domains are important to stabilize the interaction between nucleolin and the c-MYC G-quadruplex. It appears that the C-terminal of nucleolin is critical for the initial recognition of the c-MYC NHE III1 sequence, which in turn promotes G-quadruplex formation, while the rest of the protein may serve to further stabilize the interaction of the protein with the assembled G-quadruplex. The high affinity of nucleolin for the c-MYC G-quadruplex is most likely achieved by combining multiple weak interactions between the different protein domains with the c-MYC G-quadruplex. Accordingly, deletion of these domains would result in lower binding affinity, as shown in our EMSA studies. Furthermore, while Nuc-RGG was able to effectively induce the formation of the c-MYC G-quadruplex in solution, no binding between the Nuc-RGG deletion mutant and the c-MYC G-quadruplex was detected by this method (Figure 5C). This suggests that while the RGG domain is essential for the induction of a stable c-MYC G-quadruplex structure, it is not sufficient to achieve a stable interaction with the c-MYC G-quadruplex. It is possible that the RGG domain interacts very weakly with the c-MYC G-quadruplex structure and that these interactions are easily disrupted as a result of the stress that the sample undergoes during the electromobility separation (Figure 5C). Taken together, these results suggest that the C-terminal region of nucleolin containing RBDs 3 and 4 as well as the RGG domain is the minimal domain able to stably bind to the c-MYC G-quadruplex structure.
Fig. 5.
Comparison of the binding affinity of Nuc-1,2,3,4-RGG, Nuc-3,4-RGG, and Nuc-RGG for the c-MYC G-quadruplex. 10,000 cpm of radiolabeled oligomer was assembled into a G-quadruplex structure and incubated with Nuc-1,2,3,4-RGG (A), Nuc-3,4-RGG (B), or Nuc-1,2,3,4-RGG (C) at the indicated concentrations in a 20-μL reaction containing 10 mM Tris-HCl, pH 7.4, 1 mM EDTA, 1 mM DTT, 50 ng/μL poly(dI-dC), 4 μg/mL BSA, and 25 mM KCl for 1 h at room temperature.
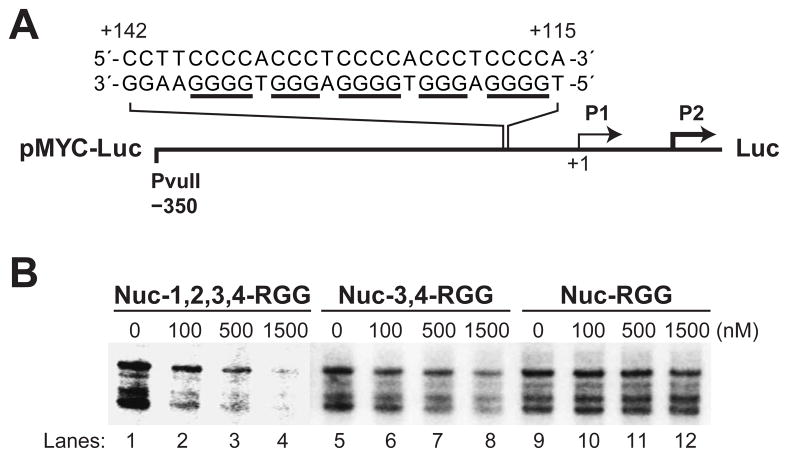
Nuc-3,4-RGG Inhibits c-MYC Promoter Activity In Vitro
Our laboratory has previously demonstrated by luciferase assay that transient expression of full-length nucleolin in mammalian cells can exert a strong and dose-dependent inhibitory effect on the luciferase activity of a c-MYC promoter-driven construct (20). In this study we used the same c-MYC reporter plasmid (Figure 6A) to investigate by in vitro transcription whether the Nuc-1,2,3,4-RGG recombinant protein retains the dose-dependent inhibitory activity on c-MYC promoter activity. The multiple bands observed in the gel correspond to the transcripts from the several transcription initiation sites found in the luciferase coding region (44). Here we demonstrate that Nuc-1,2,3,4-RGG strongly inhibits c-MYC promoter activity in a dose-dependent manner, as shown by the substantial decrease in the amount of radiolabeled luciferase transcript synthesized after incubation of the reporter plasmid with increasing concentrations of Nuc-1,2,3,4-RGG (Figure 6B, lanes 1–4). Similarly, we show that Nuc-3,4-RGG is able to inhibit c-MYC promoter activity in a dose-dependent manner, although this repression is significantly weaker than that exerted by Nuc-1,2,3,4-RGG (Figure 6B, lanes 5–8). Furthermore, we report that while the RGG domain is essential to induce a stable c-MYC G-quadruplex formation, this domain is not sufficient to affect c-MYC promoter activity (Figure 6B, lanes 9–12). Consequently, it appears that the minimal c-MYC G-quadruplex binding region retaining the transcription repression activity of nucleolin consists of RBDs 3 and 4 and the RGG domain.
Fig. 6.
Effect of Nuc-1,2,3,4-RGG, Nuc-3,4-RGG, and Nuc-RGG on c-MYC promoter activity as determined by in vitro transcription. (A) Diagram of the pMYC-Luc reporter construct containing the c-MYC NHE III1 region that can assemble into a G-quadruplex structure. (B) Effect of Nuc-1,2,3,4-RGG (lanes 1–4), Nuc-3,4-RGG (lanes 5–8), and Nuc-RGG (lanes 9–12) on c-MYC promoter activity as determined by in vitro transcription assay. Reactions contained 1 μg of pMYC-WT (Del-4) plasmid and the indicated concentrations of nucleolin per reaction. Plasmid and the recombinant protein were incubated for 1 h at 4 °C prior to the addition of HeLa nuclear extract. In vitro transcription reactions were carried out at 42 °C for exactly 60 min, after which all reactions were stopped by the addition of stop buffer, as described in Materials and Methods. The multiple bands correspond to the transcripts from the several transcription initiation sites found in the luciferase coding region (44).
DISCUSSION
A growing number of G-quadruplex-interactive proteins are being identified in diverse organisms (45). The existence of proteins that interact with high affinity and selectivity to G-quadruplex structures to modulate or stabilize them provides strong arguments for the biological relevance of G-quadruplex structures in vivo. Our laboratory has previously identified nucleolin as a c-MYC G-quadruplex-binding protein that represses c-MYC promoter activity by inducing the formation of the c-MYC G-quadruplex structure (20). Here we report that the C-terminal region of nucleolin containing RBDs 3 and 4 and the RGG domain is essential for c-MYC G-quadruplex binding and stabilization. Furthermore, we show that this segment of the protein is the minimal region able to repress c-MYC transcription.
Nucleolin is a modular protein that is found in organisms ranging from yeast to mammals (32). The N-terminal domain of nucleolin comprises long acidic stretches interspersed with basic repeats, and its length is quite variable among the different species (32). This domain is highly phosphorylated, and it is believed to assume a non-globular extended structure (46). The N-terminal domain of nucleolin has been shown to play a role in chromatin condensation, protein–protein interactions, and nucleolin functional regulation (32, 47). On the other hand, the four RBDs found in the central region of nucleolin are highly conserved in human, rat, mouse, hamster, chicken, and Xenopus laevis (32). Interestingly, the RBDs found in these species are less conserved within the same protein than between RBDs from the different species (32). The RGG domain of nucleolin is defined by spaced Arg-Gly-Gly repeats interspersed by aromatic amino acids. CD and homology studies of this domain suggest that this region can adopt a highly flexible helical conformation made up of repeated β-turns with arginine and phenylalanine side chains projecting outside the spiral structure (48). It was originally thought that this domain would create electrostatic and hydrophobic ridges prone to interact nonspecifically with RNA and DNA (48); however, a number of recent studies have shown that this structure is able to confer substrate specificity (42, 49, 50). These findings are further supported by our previous EMSA and filter binding studies where we show that recombinant nucleolin containing the RGG domain, as well as all four RBDs, can discriminate between different G-quadruplex structures and bind preferentially to parallel c-MYC-like G-quadruplex structures (20).
While the results of the EMSA and effect on transcription of the 3,4-RGG and 1,2,3,4-RGG are in reasonable accord, i.e., the 3,4-RGG is much less effective as a transcriptional inhibitor than the 1,2,3,4-RGG and EMSA shows a similar loss of binding of 3,4-RGG to the MYC G-quadruplex relative to the 1,2,3,4-RGG, there is little difference between the Tms of the MYC G-quadruplex in the presence of the two proteins. There are a number of possible reasons for this discrepancy. While in the CD experiment the concentrations of the DNA and protein are equivalent, in the EMSA and transcription assays they are very different, with the protein greatly in excess by several orders of magnitude. However, there are other possibilities for this discrepancy, including limitation in the window of change that can occur in the ΔTm after the G-quadruplex is formed. If the 3,4-RGG and 1,2,3,4-RGG are both effective in facilitating the formation of the G-quadruplex, then the Tms are likely to be about the same. However, if additional interactions due to the RBDs 1 and 2 are required to form a stable complex that will survive EMSA and competition with other proteins in cells, then the apparent discrepancy between the results can be explained. Another problem in correlating the cell-free data with the cellular data is that in cells the G-quadruplex is formed under negative superhelicity (7), whereas the cell-free system uses a single-stranded template. Thus, the dynamics are quite different in addition to competition in cells with other proteins, such as NM23-H2, Sp1, and CNBP, which also bind to this element. Thus, the more dynamic state of the EMSA may be a better measure of the effect on transcription than a Tm that measures just the ability of the protein to capture the folded species. The first identified G-quadruplex inducer protein is the beta subunit of the Oxytricha telomere end-binding protein β (51, 52). This protein has been reported to promote the formation of a G-quadruplex structure in telomeric DNA in a cell cycle–dependent manner where the telomere end-binding protein β phosphorylation state is directly linked with G-quadruplex formation (53). Since nucleolin function is also regulated by phosphorylation, it would be interesting to investigate whether changes in nucleolin phosphorylation in these parameters would affect nucleolin–c-MYC promoter interaction as well as c-MYC G-quadruplex formation.
Furthermore, a number of stress stimuli have been shown to modulate nucleolin localization and function (28). For example, nucleoplasmic localization of nucleolin has been associated with the pro-apoptotic effects of a number of anticancer drugs (30, 54, 55). Specifically, the mechanisms of action of cisplatin and Quarfloxin are linked with the redistribution of nucleolin from the nucleoli to the nucleoplasm (30, 54). Consequently, it has been suggested that nucleolin can function as a stress-sensitive tumor suppressor (28). If this is the case, it is possible that stress-induced nuclear localization would allow nucleolin to interact with the c-MYC promoter to induce G-quadruplex formation. Since nucleolin is a bona fide target of the c-MYC proto-oncogene (56), it is not unreasonable to suggest that nucleolin may form part of a negative feedback mechanism to prevent aberrant c-MYC expression.
In summary, there is a large body of evidence supporting the function of nucleolin as a tumor suppressor. Our findings that nucleolin represses c-MYC expression by inducing the formation of a G-quadruplex structure provide an explanation for the importance of nucleolin translocation from the nucleolus to the nucleoplasm after cellular stress or drug treatment.
Acknowledgments
We are grateful to Dr. Leslyn A. Hanakahi for generously providing plasmids pNuc-1,2,3,4-RGG, pNuc-3,4-RGG, pNuc-1,2-RGG, and pNuc-RGG and to Dr. Bert Vogelstein for kindly providing pDel-4 reporter plasmid. We thank Dr. David Bishop for preparing, proofreading, and editing the final version of the manuscript and figures.
Footnotes
This work was supported, in whole or in part, by the National Institutes of Health (GM085585 and Under-Represented Minority Supplements to CA94166 and CA95060)
Abbreviations: RBD, RNA binding domain; RBD1, RNA binding domain 1; RBD2, RNA binding domain 2; RBD3, RNA binding domain 3; RBD4, RNA binding domain 4; NHE, nuclease hypersensitive element; CD, circular dichroism; RGG, arginine-glycine-glycine; MBP, maltose-binding protein; Pu47ss, purine-rich 47-mer containing the c-MYC NHE III1; pDel-4, reporter plasmid containing the NHE III1 region of the c-MYC promoter; EMSA, electromobility shift assay.
References
- 1.Wierstra I, Alves J. The c-myc promoter: still MysterY and Challenge. Adv Cancer Res. 2008;99:113–333. doi: 10.1016/S0065-230X(07)99004-1. [DOI] [PubMed] [Google Scholar]
- 2.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis JT. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew Chem Int Ed Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 4.Arnott S, Chandrasekaran R, Marttila CM. Structures for polyinosinic acid and polyguanylic acid. Biochem J. 1974;141:537–543. doi: 10.1042/bj1410537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghosal G, Muniyappa K. Hoogsteen base-pairing revisited: resolving a role in normal biological processes and human diseases. Biochem Biophys Res Commun. 2006;343:1–7. doi: 10.1016/j.bbrc.2006.02.148. [DOI] [PubMed] [Google Scholar]
- 6.Zimmerman SB, Cohen GH, Davies DR. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J Mol Biol. 1975;92:181–192. doi: 10.1016/0022-2836(75)90222-3. [DOI] [PubMed] [Google Scholar]
- 7.Brooks TA, Hurley LH. The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nat Rev Cancer. 2009;9:849–861. doi: 10.1038/nrc2733. [DOI] [PubMed] [Google Scholar]
- 8.Sun D, Hurley LH. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: implications for drug targeting and control of gene expression. J Med Chem. 2009;52:2863–2874. doi: 10.1021/jm900055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.González V, Hurley LH. The c-MYC NHE III1: function and regulation. Annu Rev Pharmacol Toxicol. 2010;50:111–129. doi: 10.1146/annurev.pharmtox.48.113006.094649. [DOI] [PubMed] [Google Scholar]
- 10.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verma A, Halder K, Halder R, Yadav VK, Rawal P, Thakur RK, Mohd F, Sharma A, Chowdhury S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J Med Chem. 2008;51:5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- 13.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai J, Chen D, Jones RA, Hurley LH, Yang D. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006;34:5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsugami A, Okuizumi T, Uesugi S, Katahira M. Intramolecular higher order packing of parallel quadruplexes comprising a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad of GGA triplet repeat DNA. J Biol Chem. 2003;278:28147–28153. doi: 10.1074/jbc.M303694200. [DOI] [PubMed] [Google Scholar]
- 17.Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J Am Chem Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simonsson T. G-quadruplex DNA structures—variations on a theme. Biol Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 19.Yang D, Hurley LH. Structure of the biologically relevant G-quadruplex in the c-MYC promoter. Nucleosides Nucleotides Nucleic Acids. 2006;25:951–968. doi: 10.1080/15257770600809913. [DOI] [PubMed] [Google Scholar]
- 20.González V, Guo K, Hurley L, Sun D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J Biol Chem. 2009;284:23622–23635. doi: 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daniely Y, Borowiec JA. Formation of a complex between nucleolin and replication protein A after cell stress prevents initiation of DNA replication. J Cell Biol. 2000;149:799–810. doi: 10.1083/jcb.149.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daniely Y, Dimitrova DD, Borowiec JA. Stress-dependent nucleolin mobilization mediated by p53-nucleolin complex formation. Mol Cell Biol. 2002;22:6014–6022. doi: 10.1128/MCB.22.16.6014-6022.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim K, Dimitrova DD, Carta KM, Saxena A, Daras M, Borowiec JA. Novel checkpoint response to genotoxic stress mediated by nucleolin-replication protein A complex formation. Mol Cell Biol. 2005;25:2463–2474. doi: 10.1128/MCB.25.6.2463-2474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klibanov SA, O’Hagan HM, Ljungman M. Accumulation of soluble and nucleolar-associated p53 proteins following cellular stress. J Cell Sci. 2001;114:1867–1873. doi: 10.1242/jcs.114.10.1867. [DOI] [PubMed] [Google Scholar]
- 25.Martelli AM, Robuffo I, Bortul R, Ochs RL, Luchetti F, Cocco L, Zweyer M, Bareggi R, Falcieri E. Behavior of nucleolar proteins during the course of apoptosis in camptothecin-treated HL60 cells. J Cell Biochem. 2000;78:264–277. [PubMed] [Google Scholar]
- 26.Mi Y, Thomas SD, Xu X, Casson LK, Miller DM, Bates PJ. Apoptosis in leukemia cells is accompanied by alterations in the levels and localization of nucleolin. J Biol Chem. 2003;278:8572–8579. doi: 10.1074/jbc.M207637200. [DOI] [PubMed] [Google Scholar]
- 27.Mongelard F, Bouvet P. Nucleolin: a multiFACeTed protein. Trends Cell Biol. 2007;17:80–86. doi: 10.1016/j.tcb.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 28.Storck S, Shukla M, Dimitrov S, Bouvet P. Functions of the histone chaperone nucleolin in diseases. Subcell Biochem. 2007;41:125–144. doi: 10.1007/1-4020-5466-1_7. [DOI] [PubMed] [Google Scholar]
- 29.Saxena A, Rorie CJ, Dimitrova D, Daniely Y, Borowiec JA. Nucleolin inhibits Hdm2 by multiple pathways leading to p53 stabilization. Oncogene. 2006;25:7274–7288. doi: 10.1038/sj.onc.1209714. [DOI] [PubMed] [Google Scholar]
- 30.Kito S, Morimoto Y, Tanaka T, Haneji T, Ohba T. Cleavage of nucleolin and AgNOR proteins during apoptosis induced by anticancer drugs in human salivary gland cells. J Oral Pathol Med. 2005;34:478–485. doi: 10.1111/j.1600-0714.2005.00346.x. [DOI] [PubMed] [Google Scholar]
- 31.Wesierska-Gadek J, Schloffer D, Kotala V, Horky M. Escape of p53 protein from E6-mediated degradation in HeLa cells after cisplatin therapy. Int J Cancer. 2002;101:128–136. doi: 10.1002/ijc.10580. [DOI] [PubMed] [Google Scholar]
- 32.Ginisty H, Sicard H, Roger B, Bouvet P. Structure and functions of nucleolin. J Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- 33.Serin G, Joseph G, Faucher C, Ghisolfi L, Bouche G, Amalric F, Bouvet P. Localization of nucleolin binding sites on human and mouse pre-ribosomal RNA. Biochimie. 1996;78:530–538. doi: 10.1016/0300-9084(96)84759-6. [DOI] [PubMed] [Google Scholar]
- 34.Serin G, Joseph G, Ghisolfi L, Bauzan M, Erard M, Amalric F, Bouvet P. Two RNA-binding domains determine the RNA-binding specificity of nucleolin. J Biol Chem. 1997;272:13109–13116. doi: 10.1074/jbc.272.20.13109. [DOI] [PubMed] [Google Scholar]
- 35.Ginisty H, Amalric F, Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 1998;17:1476–1486. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ginisty H, Amalric F, Bouvet P. Two different combinations of RNA-binding domains determine the RNA binding specificity of nucleolin. J Biol Chem. 2001;276:14338–14343. doi: 10.1074/jbc.M011120200. [DOI] [PubMed] [Google Scholar]
- 37.Ginisty H, Serin G, Ghisolfi-Nieto L, Roger B, Libante V, Amalric F, Bouvet P. Interaction of nucleolin with an evolutionarily conserved pre-ribosomal RNA sequence is required for the assembly of the primary processing complex. J Biol Chem. 2000;275:18845–18850. doi: 10.1074/jbc.M002350200. [DOI] [PubMed] [Google Scholar]
- 38.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 39.Williams M, Louw AI, Birkholtz LM. Deletion mutagenesis of large areas in Plasmodium falciparum genes: a comparative study. Malar J. 2007;6:64. doi: 10.1186/1475-2875-6-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanakahi LA, Dempsey LA, Li MJ, Maizels N. Nucleolin is one component of the B cell-specific transcription factor and switch region binding protein, LR1. Proc Natl Acad Sci USA. 1997;94:3605–3610. doi: 10.1073/pnas.94.8.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanakahi LA, Sun H, Maizels N. High affinity interactions of nucleolin with G-G-paired rDNA. J Biol Chem. 1999;274:15908–15912. doi: 10.1074/jbc.274.22.15908. [DOI] [PubMed] [Google Scholar]
- 42.Ramos A, Hollingworth D, Pastore A. G-quartet-dependent recognition between the FMRP RGG box and RNA. RNA. 2003;9:1198–1207. doi: 10.1261/rna.5960503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaeffer C, Bardoni B, Mandel JL, Ehresmann B, Ehresmann C, Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vopálenský V, Mašek T, Horváth O, Vicenová B, Mokrejš M, Pospíšek M. Firefly luciferase gene contains a cryptic promoter. RNA. 2008;14:1720–1729. doi: 10.1261/rna.831808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fry M. Tetraplex DNA and its interacting proteins. Front Biosci. 2007;12:4336–4351. doi: 10.2741/2391. [DOI] [PubMed] [Google Scholar]
- 46.Sapp M, Richter A, Weisshart K, Caizergues-Ferrer M, Amalric F, Wallace MO, Kirstein MN, Olson MO. Characterization of a 48-kDa nucleic-acid-binding fragment of nucleolin. Eur J Biochem. 1989;179:541–548. doi: 10.1111/j.1432-1033.1989.tb14581.x. [DOI] [PubMed] [Google Scholar]
- 47.Erard M, Lakhdar-Ghazal F, Amalric F. Repeat peptide motifs which contain β-turns and modulate DNA condensation in chromatin. Eur J Biochem. 1990;191:19–26. doi: 10.1111/j.1432-1033.1990.tb19088.x. [DOI] [PubMed] [Google Scholar]
- 48.Ghisolfi L, Joseph G, Amalric F, Erard M. The glycine-rich domain of nucleolin has an unusual supersecondary structure responsible for its RNA-helix-destabilizing properties. J Biol Chem. 1992;267:2955–2959. [PubMed] [Google Scholar]
- 49.Corbin-Lickfett KA, Chen IH, Cocco MJ, Sandri-Goldin RM. The HSV-1 ICP27 RGG box specifically binds flexible, GC-rich sequences but not G-quartet structures. Nucleic Acids Res. 2009;37:7290–7301. doi: 10.1093/nar/gkp793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 51.Fang G, Cech TR. The β subunit of Oxytricha telomere-binding protein promotes G-quartet formation by telomeric DNA. Cell. 1993;74:875–885. doi: 10.1016/0092-8674(93)90467-5. [DOI] [PubMed] [Google Scholar]
- 52.Fang G, Cech TR. Characterization of a G-quartet formation reaction promoted by the β-subunit of the Oxytricha telomere-binding protein. Biochemistry. 1993;32:11646–11657. doi: 10.1021/bi00094a022. [DOI] [PubMed] [Google Scholar]
- 53.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 54.Drygin D, Siddiqui-Jain A, O’Brien S, Schwaebe M, Lin A, Bliesath J, Ho CB, Proffitt C, Trent K, Whitten JP, Lim JK, Von Hoff D, Anderes K, Rice WG. Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Cancer Res. 2009;69:7653–7661. doi: 10.1158/0008-5472.CAN-09-1304. [DOI] [PubMed] [Google Scholar]
- 55.Teng Y, Girvan AC, Casson LK, Pierce WM, Jr, Qian M, Thomas SD, Bates PJ. AS1411 alters the localization of a complex containing protein arginine methyltransferase 5 and nucleolin. Cancer Res. 2007;67:10491–10500. doi: 10.1158/0008-5472.CAN-06-4206. [DOI] [PubMed] [Google Scholar]
- 56.Greasley PJ, Bonnard C, Amati B. Myc induces the nucleolin and BN51 genes: possible implications in ribosome biogenesis. Nucleic Acids Res. 2000;28:446–453. doi: 10.1093/nar/28.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]