Abstract
The availability of crystal structures for the ligand binding domains of ionotropic glutamate receptors, combined with their key role in synaptic function in the normal and diseased brain, offers a unique selection of targets for pharmaceutical research compared to other drug targets for which the atomic structure of the ligand binding sites is not known. Currently only a few antagonist structures have been solved, and these reveal ligand specific conformational changes that hinder rational drug design. Here we report high resolution crystal structures for three kainate receptor GluK1 antagonist complexes which reveal new and unexpected modes of binding, highlighting the continued need for experimentally determined receptor-ligand complexes.
Keywords: Glutamate Receptors, Crystal Structure, Conformational Flexibility, Halogen Bond
1. Introduction
Kainate receptors (KARs) are members of the ionotropic glutamate receptor (iGluR) family which also includes N-methyl-D-aspartate receptors (NMDARs) and (S)-2-amino-3-(3-hydroxy-5-methyl-4-isoxazolyl)propionic acid receptors (AMPARs). KARs are tetrameric assemblies of GluK1–5 subunits (Collingridge et al., 2009) and have been implicated in various functions in the central nervous system (Jane et al., 2009; Lerma, 2006; Pinheiro and Mulle, 2006). In addition, KARs and in particular those containing the GluK1 subunit have been implicated in a number of neurological conditions, such as chronic pain (Jones et al., 2006; Simmons et al., 1998), migraine (Weiss et al., 2006), epilepsy (Smolders et al., 2002) and neurodegeneration (O’Neill et al., 2000) as well as in psychiatric conditions, such as schizophrenia (Beneyto et al., 2007) and anxiety (Alt et al., 2007).
Subunit selective antagonists are required to investigate the functions of KARs in more detail and to identify new targets for drug discovery. The first KAR antagonists to be reported, particularly those based on the quinoxalinedione nucleus, also bind to AMPARs with high affinity and showed no KAR subtype selectivity (Jane et al., 2009). More recently, antagonists showing good selectivity for KARs versus AMPARs and selectivity for the GluK1 subunit within the KAR family have been reported. The most potent GluK1 selective antagonists reported to date include those based on the natural product willardiine, such as UBP310 (Dolman et al., 2007; Mayer et al., 2006) and UBP316 (Dargan et al., 2009; Dolman et al., 2007)), and a structurally distinct set of ligands such as LY466195 based on the decahydroisoquinoline nucleus (Weiss et al., 2006).
A breakthrough in our understanding of the molecular interactions between agonists and iGluRs came with the publication of high resolution crystal structures of the ligand binding domains (LBDs) of AMPAR subunits, in particular GluA2 (Armstrong and Gouaux, 2000; Hogner et al., 2002; Jin et al., 2003; Sun et al., 2002); the NMDA receptor subunits GluN1 (Furukawa and Gouaux, 2003), GluN2A (Furukawa et al., 2005), GluN3A and GluN3B (Yao et al., 2008); and the KAR GluK1 and GluK2 subunits (Frydenvang et al., 2009; Hald et al., 2007; Mayer, 2005; Nanao et al., 2005; Naur et al., 2005). By contrast, due to the difficulty in crystallizing antagonist-LBD complexes there are far fewer crystal structures available. Indeed, only two antagonist-LBD structures have been reported for the NR1 NMDAR subunit (Furukawa and Gouaux, 2003; Inanobe et al., 2005), eight for the GluA2 AMPAR subunit (Ahmed et al., 2009; Armstrong and Gouaux, 2000; Cruz et al., 2008; Hogner et al., 2003; Kasper et al., 2006; Menuz et al., 2007; Sobolevsky et al., 2009), and four for the GluK1 KAR subunit (Dargan et al., 2009; Hald et al., 2007; Mayer, 2005). However, these structures have been limited in their usefulness for drug design due to the conformational flexibility of iGluR LBDs and the dependence of the degree of domain closure on both the ligand and the iGluR subunit (Mayer, 2006). In addition, a complex network of water molecules within the LBDs of iGluRs plays a major role in receptor-ligand interactions, and this network is difficult to model without prior structural knowledge. Molecular dynamics studies on ligand-LBD complexes have made some progress in solving some of these problems (Arinaminpathy et al., 2006; Dolman et al., 2007; Lau and Roux, 2007; Postila et al.), but are not yet sufficiently accurate that they can replace data obtained for experimentally determined complexes with novel ligands.
We have previously reported that the ligand binding domains are hyper-extended in GluK1 complexes with antagonists and that consequently, the key ligand binding residue Glu723 exists in two conformations, one of which plays no direct role in antagonist binding (Mayer et al., 2006). This is unusual as this residue plays a critical role in stabilizing GluK1 and GluK2 agonist complexes (Mayer, 2005) and also GluA2 agonist and antagonist complexes (Armstrong and Gouaux, 2000; Hogner et al., 2003).
Here, we report crystal structures of the GluK1 LBD with three high affinity antagonists UBP315, UBP318 and the structurally unrelated compound LY466195. These structures reveal a much wider variation in ligand-receptor interactions and ligand binding domain closure than that found in our previous studies with the GluK1 antagonists UBP302, UBP310 and UBP316 (Dargan et al., 2009; Mayer et al., 2006). For instance, the 4-bromo group on the thiophene ring of UBP315 forms an unusual interaction with the carboxylate group of Glu426, producing a 5° greater degree of domain closure compared to the UBP302, UBP310, UBP316 and UBP318 complexes. In addition, LY466195 forms interactions with residues in the LBD of GluK1 that do not occur in the willardiine-based antagonist structures. Linked to these changes the network of ordered solvent molecules in the GluK1 binding pocket undergoes ligand specific rearrangements. These variations in the structures of antagonist-GluK1 complexes underline the necessity of obtaining X-ray crystal structures of the LBDs of iGluRs in complex with structurally unrelated antagonists to inform the drug design process.
2. Methods
The GluK1 ligand binding domain (LBD) was expressed as a soluble protein in E. coli and purified by affinity and ion exchange chromatography with no modifications from the previously reported protocol (Mayer, 2005). The construct consisted of residues N416 – K529, preceded by an 18 amino acid peptide encoding an IMAC His tag and thrombin site, and was linked via a GT dipeptide to residues P652 – E791; the affinity tag was removed by proteolysis prior to ligand binding studies and crystallization. For ligand binding assays apo protein was prepared by exhaustive dialysis of the purified GluK1 LBD with 5 buffer changes over a period of three days, with a total volume exchange of > 1010. Displacement assays were performed using 15 nM 3[H]-glutamate as reported previously (Mayer, 2005), and the data fit with a single binding site isotherm for a competitive interaction,
where Ki is the dissociation constant for the cold ligand, with the previously measured value of 57 nM used as the dissociation constant for glutamate (Kd Dlu).
Crystals were grown using the hanging drop technique at a temperature of 20°C, typically with a 1 to 1 dilution of protein with reservoir. To prepare antagonist complexes the protein was dialyzed against a 2x crystallization buffer containing 40 mM NaCl, 20 mM HEPES pH 7.0, 2 mM EDTA and 10–20 μM ligand, with up to four buffer changes for a total volume exchange of > 107; the protein was then diluted by 50% with 10 mM ligand dissolved in water adjusted to pH 7.0 with NaOH, and then concentrated to between 5–10 mg/ml. Seeding was required to obtain diffraction quality crystals. Cryopreservation was achieved by rapid serial transfers to mother liquor supplemented with increasing amounts of glycerol to a maximum concentration of 18–20 %, followed by flash cooling in liquid N2. The reservoir solution contained 100 mM Tris pH 8.5, 18–21% PEG 1K (UBP315 and UBP318), or 250 mM (NH4)2 citrate pH 5.35 and 20% PEG 3350 (LY466195).
Data sets from single crystals were collected at APS beamline ID22 at 100 °K using a MAR 300 CCD detector for the UBP318 and LY466195 complexes; for the UBP315 complex data was collected using a microfocus Cu-anode sealed X-ray tube with confocal optics (Rigaku Micromax 002) and a Mar345 image plate detector in an attempt to reduce radiation damage for Br atoms. Diffraction data was indexed, scaled and merged using HKL2000 (Otwinowski and Minor, 2001). For the LY466195 complex many crystals exhibited substantial merohedral twinning, but by screening diffraction data from multiple crystals using phenix.xtriage (Adams et al., 2010) we were able to select a crystal with a twin fraction of only 0.02 % estimated by Maximum Likelihood, Britton alpha 0.015, and proceeded with standard refinement for untwinned data.
Structures for the UBP315 and UBP318 complexes were solved by Fourier difference techniques using the UBP310 complex dimer (PDB 2F34) striped of ligands, solvent, and alternative conformations as the initial model. For the LY466195 complex the structure was solved by molecular replacement with the program Phaser-1.3.1 (McCoy et al., 2007) using one monomer (2F34) as the search probe. The starting models for ligand structures were built in SYBYL 7.3 (Tripos Inc., St Louis, MO, USA) and optimized with the MMF94s force field using a dielectric constant of 78.5, and a library entry for refinement generated from these coordinates with REFMAC. Cycles of rebuilding and real space refinement in COOT (Emsley and Cowtan, 2004), alternated with cycles of restrained positional, individual B-factor, and TLS refinement using REFMAC5 (Winn et al., 2001), with TLS groups identified by TLSMD (Painter and Merritt, 2006) were performed until no interpretable features remained in Fo-Fc maps. Additional crystallographic calculations were performed using the CCP4 suite of programs (CCP4, 1994). Data collection and refinement statistics are reported in Table 1.
Table 1.
Data collection and refinement statistics
| Data Set | GluR5 UBP315 |
GluR5 UBP318 |
GluR5 LY466195 |
|---|---|---|---|
| DATA COLLECTION | |||
| Space group | C2221 | C2221 | H3 |
| Unit cell dimensions (Å) | |||
| a | 97.82 | 98.12 | 89.24 |
| b | 97.82 | 98.18 | 89.24 |
| c | 129.04 | 128.60 | 330.37 |
| α, β, γ (degrees) | 90, 90, 90 | 90, 90, 90 | 90, 90, 120 |
| Number per au a | 2 | 2 | 4 |
| Wavelength (Å) | 1.5418 | 0.91915 | 1.0000 |
| Resolution (Å) b | 40.00 – 1.80 (1.86) | 40 – 1.80 (1.86) | 40 – 1.58 (1.64) |
| Unique observations | 57347 | 58102 | 134309 |
| Mean redundancy c | 3.5 (3.4) | 7.4 (7.2) | 3.5 (3.4) |
| Completeness (%) c | 99.9 (99.8) | 93.7 (96.2) | 100 (100) |
| Rmerge (%) d | 0.040 (0.273) | 0.048 (0.338) | 0.059 (0.445) |
| I/σ(I) c | 27.9 (4.73) | 31.1 (5.22) | 18.9 (3.06) |
| Mosaicity | 0.292 | 0.179 | 0.609 |
| REFINEMENT | |||
| Resolution (Å) | 22 – 1.80 | 32 – 1.80 | 37.6 – 1.58 |
| Protein atoms (alt conf) | 4014 (154) | 4014 (171) | 8644 (623) |
| Ligand atoms | 52 | 48 | 118 |
| Chloride/ammonium ions | 2/0 | 2/0 | 0/2 |
| Water atoms | 366 | 413 | 1150 |
| PEG atoms | 32 | 32 | 0 |
| Glycerol atoms | 0 | 0 | 12 |
| Rwork/Rfree (%) e | 19.8/22.0 | 19.1/21.9 | 15.8/20.0 |
| rms deviations | |||
| Bond lengths (Å) | 0.008 | 0.008 | 0.015 |
| Bond angles (degrees) | 1.33 | 1.18 | 1.603 |
| Bonds B values MC/SC | 0.523/1.294 | 0.554/1.380 | 1.105/2.755 |
| Angles B values MC/SC | 0.867/1.900 | 0.904/1.963 | 1.635/4.164 |
| Mean B-Values (Å2) f | |||
| Protein overall | 33.93 | 28.54 | 14.76 |
| Main-chain | 33.51 | 28.05 | 11.18 |
| Side-chain | 34.34 | 29.04 | 13.89 |
| Ligand | 38.71 | 35.64 | 15.11 |
| PEG | 24.89 | 19.12 | - |
| Glycerol | - | - | 26.71 |
| Ions | 24.65 | 21.52 | 21.57 |
| Water | 40.95 | 36.21 | 31.35 |
| Ramachandran statistics g | 90.3/9.7/0/0 | 93.7/6.3/0/0 | 91.9/8.1/0/0 |
Asymmetric unit.
Values in parentheses indicate the low-resolution limit for the last shell of data.
Values in parentheses indicate statistics for the last shell of data
Rmerge = (Σ |Ii − <Ii>|)/Σi|Ii|, where <Ii> is the mean Ii over symmetry-equivalent reflections
Rwork = Σ||Fo| − |Fc||/Σ|Fo| where Fo and Fc denote observed and calculated structure factors, respectively; 5% of the reflections were excluded from refinement for calculation of the Rfree value. Values are isotropic B=values after subtraction of the TLS component in Refmac.
Percentage of residues in the most favored/allowed/generous/disallowed regions.
Subsite maps were prepared according to the nomenclature reported previously for the GluK1 glutamate and UBP310 complexes (Mayer et al., 2006) and use the same numbering scheme for conserved water molecules. The subsite maps were assembled in Photoshop based on the output from LIGPLOT (Wallace et al., 1995) and show ligands as ball and stick figures in which torsion angles were manually adjusted compared to the conformation found in the crystal structure to bring the heterocyclic rings into approximately the same plane for ease of illustration. Electron density maps and crystal structures were illustrated using scripts written for PyMOL (DeLano, 2002). Coordinates and structure factors for the UBP315, UBP318 and LY466195 complexes have been deposited in the protein data bank with codes of 2QS1, 2QS2 and 2QS4 respectively.
3. Results
3.1 High affinity binding of UBP315, UBP318 and LY466195
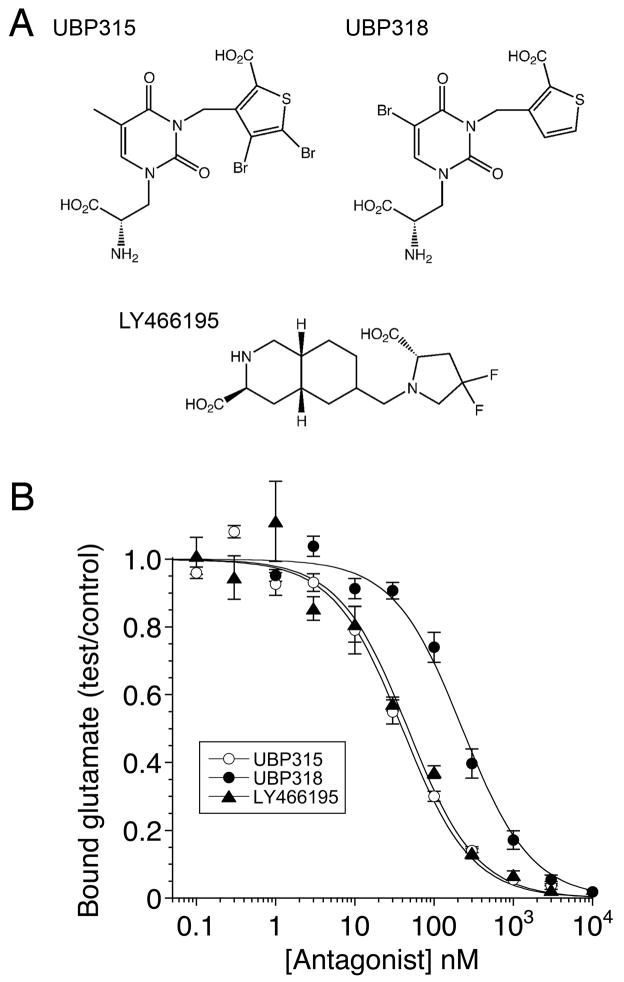
In this study we characterize the interaction of three high affinity competitive antagonists (S)-1-(2-Amino-2-carboxyethyl)-3-(2-carboxy-4,5-dibromothiophene-3-yl-methyl)-5-methylpyrimidine-2,4-dione (UBP315), (S)-1-(2-amino-2-carboxyethyl)-5-bromo-3-(2-carboxythiophene-3-yl-methyl)pyrimidine-2,4-dione (UBP318), and (3S,4aR,6S,8aR)-6-[[(2S)-2-carboxy-4,4-difluoro-1-pyrrolidinyl]methyl]decahydro-3-isoquinolinecarboxylic acid (LY466195), with the kainate receptor GluK1 ligand binding domain using X-ray crystallography and biochemical techniques. These ligands were chosen to investigate the structural effects of halogen substituents in two different positions in the willardiine series of antagonists (Dolman et al., 2007), and to explore how the binding of these ligands differs from that of a second class of competitive antagonist based on a decahydroisoquinoline backbone (Weiss et al., 2006).
We measured the affinity of UBP315, UBP318 and LY466195 for GluK1 using 3[H]-glutamate displacement assays and the genetically isolated GluK1 ligand binding domain (LBD) expressed as a soluble protein, which was purified to homogeneity and exhaustively dialyzed against a ligand free buffer as described previously (Mayer, 2005). The results reveal that UBP315 and LY466195 bind to the GluK1 LBD with similar affinities, Kd 33 ± 4 nM and 38 ± 7 nM respectively, while UBP318, Kd 186 ± 23 nM (n = 3), binds with 5-fold lower affinity (Fig. 1). For UBP315 and LY466195 these results are consistent with prior studies on full length GluK1 for which Kds of 10 ± 2 nM and 52 ± 22 nM were reported respectively, while for UBP318 we found a lower affinity than the value of 25 ± 2 nM reported in prior studies (Dolman et al., 2007; Weiss et al., 2006).
Figure 1.
Structures and binding assays for GluK1 competitive antagonists. (A) UBP315 and UBP318 are willardiine derivatives with bromine substitutions on the thiophene and uracil rings respectively; LY466195 is a decahydroisoquinoline with fluorine substitutions on the pyrrolidine ring. (B) Radioligand displacement curves for the purified GluK1 ligand binding domain showing competition between 3[H]-L-glutamate and three antagonists, fit with single binding site isotherms of 33 ± 4 nM, 186 ± 23 nM and 38 ± 7 nM for UBP315, UBP318 and LY466195, respectively; values are mean ± SEM of triplicate measurements.
3.2 Domain closure varies for GluK1 antagonists
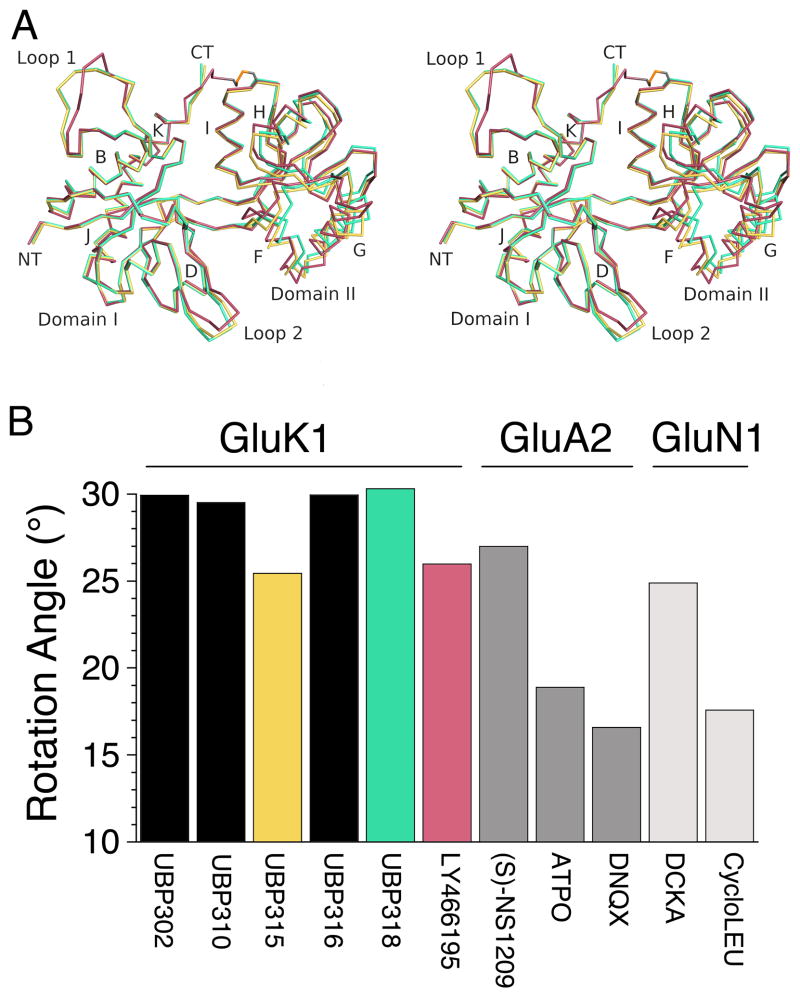
In prior work we solved crystal structures for three willardiine derivatives that act as GluK1 antagonists, UBP302, UBP310 and UBP316 (Dargan et al., 2009; Mayer et al., 2006). When compared to the closed cleft conformation for the GluK1 glutamate complex (Mayer, 2005; Naur et al., 2005) these antagonists each produced a similar, hyper extended open cleft conformation, in which, following least squares superposition using domain 1 coordinates, the rotation angle required to bring domain 2 back to the orientation found in the glutamate complex varied over a narrow range, 30.0° for UBP302, 29.5° for UBP310 and 29.8° for UBP316. In the present study, a similar extended open cleft conformation, rotation angle 30.5° was found for UBP318, while for UBP315 and LY466195, rotation angles 25.5° and 26.7° respectively, the cleft has closed by 4–5 degrees, and the rotation angle approaches the value of 26.1° found for the GluA2 complex with NS1209 (Kasper et al., 2006), and 24.9° found for the GluN1 complex with 5,7-dichlorokynurenic acid (Furukawa and Gouaux, 2003). This difference is shown in Fig. 2 and indicates that like other iGluR subtypes, the ligand binding domain of GluK1 can adopt a range of conformations when bound by competitive antagonists. However, despite the difference in domain closure the antagonist complexes have nearly identical secondary structures. Least squares superpositions, using 114 Cα atom positions in domain 1 for the UBP315 complex as a reference, excluding loops 1 and 2, the conformations of which vary due to differing intermolecular contacts in the C2221 and H3 crystal forms (Table 1), gave rmsd values of 0.10 Å for UBP310, 0.13 Å for UBP318, and 0.27 – 0.36 Å for the four subunits in the LY466195 complex. Similar calculations for 101 Cα atom positions in domain 2, excluding 6 residues at the C-terminus of helix G which is distal from the ligand binding site and involved in lattice contacts in the H3 crystal form, gave rmsds of 0.40 and 0.44 Å for UBP310 and UBP318, while for LY466195 the values were 0.32–0.37 Å for subunits A–C, and 0.62 Å for subunit D.
Figure 2.
GluK1 antagonists produce different extents of domain closure. (A) Cα traces for crystal structures of GluK1 complexes with UBP315 (yellow), UBP318 (green) and LY466195 (red), superimposed using domain 1 Cα coordinates; with the exception of loop 1 and loop 2 the structures are essentially identical, but show different extents of domain closure, illustrated by movements of helices F, G and H in domain 2. (B) Extent of domain opening relative to glutamate (GluK1 and GluA2) and glycine (GluN1) bound crystal structures for a series of competitive antagonists illustrates that the ligand binding domain adopts a range of conformations for all three iGluR families.
3.3 Crystal structure of the UBP315 complex
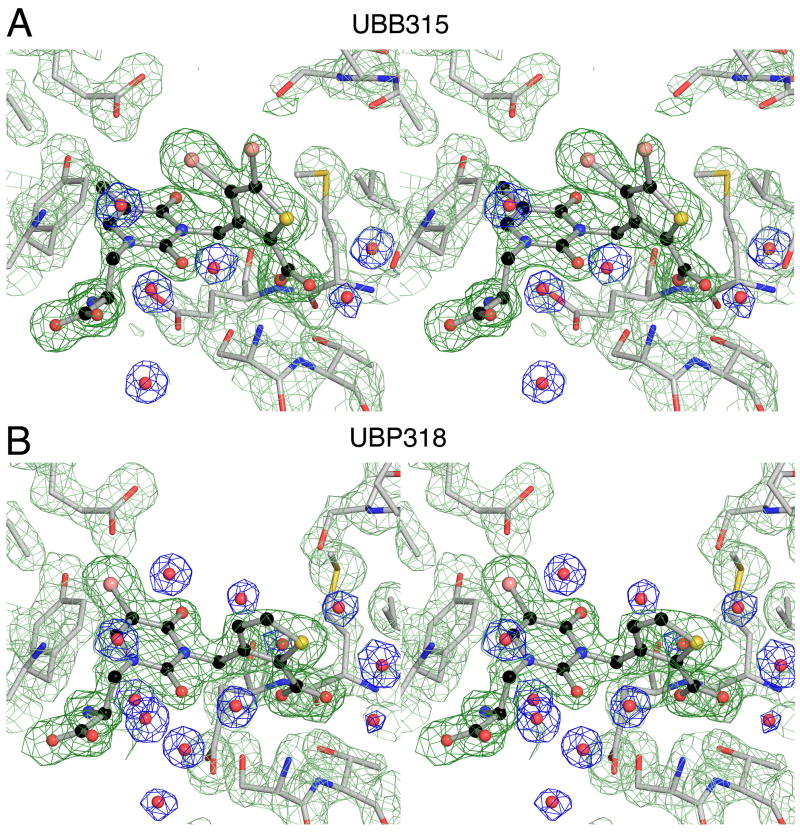
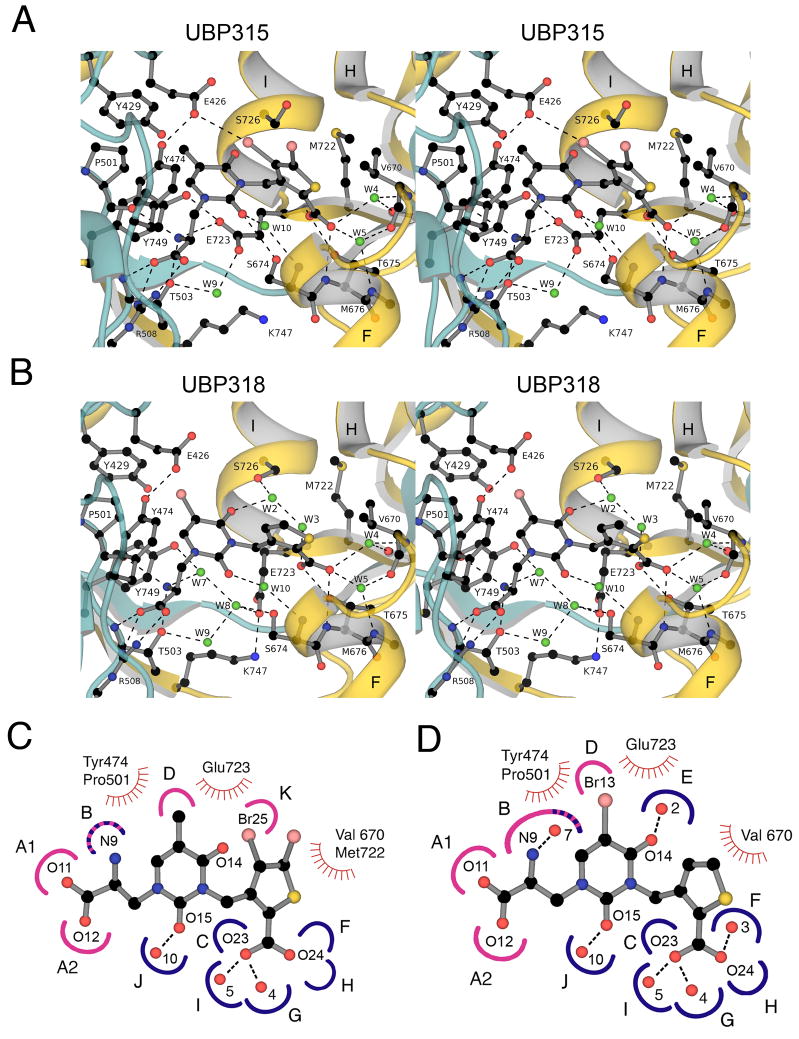
The GluK1 UBP315 complex was solved with data to Bragg spacings of 1.8 Å and refined to an Rfree value of 22% (Table 1). The resulting electron density maps were of sufficient quality to reveal side ligand and chain conformations, and solvent content in the ligand binding site with little ambiguity (Fig. 3A), allowing a detailed analysis of UBP315 interactions with the GluK1 ligand binding site (Fig. 4). The α-carboxylate and α-amino groups of UBP315 interact with highly conserved residues in domain 1, and the mechanism of binding is nearly identical to that reported previously for a large range of KAR agonists and antagonists, including the parent thiophene willardiine derivative UBP310 (Mayer, 2005), and will not be discussed in detail. However, different from the UBP310 complex, there are subtle changes in the orientation of the thiophene group and its interaction with domain 2 in the UBP310, UBP315, UBP316 and UBP318 complexes. Most striking is a ligand specific change in rotamer for Glu723, which has been proposed to play a role in the activation of iGluRs by agonists (Armstrong and Gouaux, 2000). In the UBP315 complex the side chain of Glu723 adopts a single extended conformation which is clearly resolved in electron density maps (Fig. 3A), resulting in displacement of two water molecules from the binding site, and formation of an ion pair contact of the side chain carboxylate group with the ligand α-NH2 group (Fig. 4A). This extended conformation of Glu723 is also found in GluK1 agonist complexes (Mayer, 2005), and differs from the conformation seen in the UBP310, UBP316 and UBP318 complexes (Mayer et al., 2006).
Figure 3.
Electron density maps for crystal structures of the GluK1 UBP315 and UBP318 complexes. (A) Stereoview at 1.8 Å resolution shows an Fo-Fc omit map contoured at 3.5 sigma (dark green) for which UBP315 atoms were omitted from the Fc calculation, and 2mFo-DFc maps for side chains (pale green) and water molecules (blue). (B) Stereoview of electron density maps at 1.8 Å resolution for the UBP318 complex prepared as described above for UBP315,
Figure 4.
Crystal structures and subsite maps for the GluK1 UBP 315 and UBP318 complexes. (A) Stereoview of the GluK1 UBP315 complex; the ribbon diagram showing secondary structure elements for domains 1 and 2 is colored cyan and gold, respectively; waters are shown as green spheres; hydrogen-bonds and ion pair interactions are represented as dashed lines. The side chain of Glu426 forms a halogen bond with Br25 of the ligand. (B) Stereoview of the GluK1 UBP318 complex colored as above; note the different conformation of the Glu723 and S726 side chains, rearrangement of solvent networks, and the change in orientation of the thiophene ring compared to the UBP315 complex. (C) Subsite map showing schematic representations of UBP315 with the GluK1 binding pocket; hydrogen bond and ion pair sites generated by domains 1 and 2 are colored pink and blue respectively; stripes indicate sites generated by both domains; sites of van der Waals contacts are indicated by hatched curved red lines. To make this figure, torsion angles in the UBP315 ligand were adjusted compared to the conformation found in the crystal structure to bring the heterocyclic rings into approximately the same plane for ease of illustration. (D) Subsite map showing schematic representations of UBP318 with the GluK1 binding pocket.
This switch in conformation of Glu723 appears to result from the 4° increase in domain closure found for the UBP315 complex compared to the more open conformation found for UBP310, UBP316 and UBP318 (Fig. 2). This closure likely results from an unusual halogen bond interaction of the UBP315 thiophene ring 4 position Br substituent with the carboxylate group of the Glu426 side chain located at the entrance to the ligand binding site in domain 1. The 2 position carboxylate of the thiophene ring is by contrast embedded in domain 2, as a result of interactions with sites C, F, G, H and I, located at the ends of helices F and I (Fig. 4C). The additional interaction of the Br atom with Glu723 in domain 1, due to the halogen bond, pulls the ligand binding cleft closed by a rigid body motion of domain 2. As a result the top of helix H moves 2.2 Å towards domain 1, measured from movement of the Cα atom of Ser706, and accompanying this are additional changes in solvent content (W2 and W3) and ligand orientation in the UBP315 and UBP318 complexes (Fig. 4).
3.4 Crystal structure of the UBP318 complex
The GluK1 UBP318 complex was also solved with data to Bragg spacings of 1.8 Å and refined to an Rfree value of 22% (Table 1) with good quality electron density maps (Fig. 3B). The mode of binding of UBP318 is different in several aspects from that for UBP315. Notably the side chain of Glu723 adopts a single well-ordered downward conformation (Fig. 3B) to form an ion pair contact with the side chain of K747 at the base of the ligand binding domain (Fig. 4B). Linked to the downward movement of Glu723, two additional water molecules, W7 and W8, enter the ligand binding site to form a network linking the side chain of Glu723 in domain 2 with the ligand α-amino group and the hydroxyl group of Tyr749 in domain 1. In the UBP310 and UBP316 complexes there is disorder in the region, with alternate conformations for the Glu723 side chain, and partial occupancy for W7 and W8. The single conformation observed for Glu723 and full occupancy for W7 and W8 found in the UBP318 complex probably occurs because the ligand binding domain opens by an additional 1° and 0.8° compared to the UBP310 and UBP316 complexes.
The uracil ring 5-position Br atom in UBP 318 projects into a hydrophobic pocket formed by the methylene groups of Tyr429, Pro501 and Glu426, which is flanked by the hydroxyl group of Tyr 474, all of which makes van der Waals contacts with the halogen atom. In UBP310 the methyl group which replaces the Br atom in UBP318 makes similar contacts, but due to the larger size of the Br atom there is a 0.5 Å displacement of the Tyr474 hydroxyl group away from the ligand in the UBP318 versus UBP310 complex. The mode of binding of the thiophene 2-carboxylate group of UBP318 with residues in domain 2 matches that found for UBP310, and includes contacts made by four conserved water molecules, W2, W3, W4 and W5 (Fig. 4B) two of which are absent in the UBP315 complex (Fig. 4A).
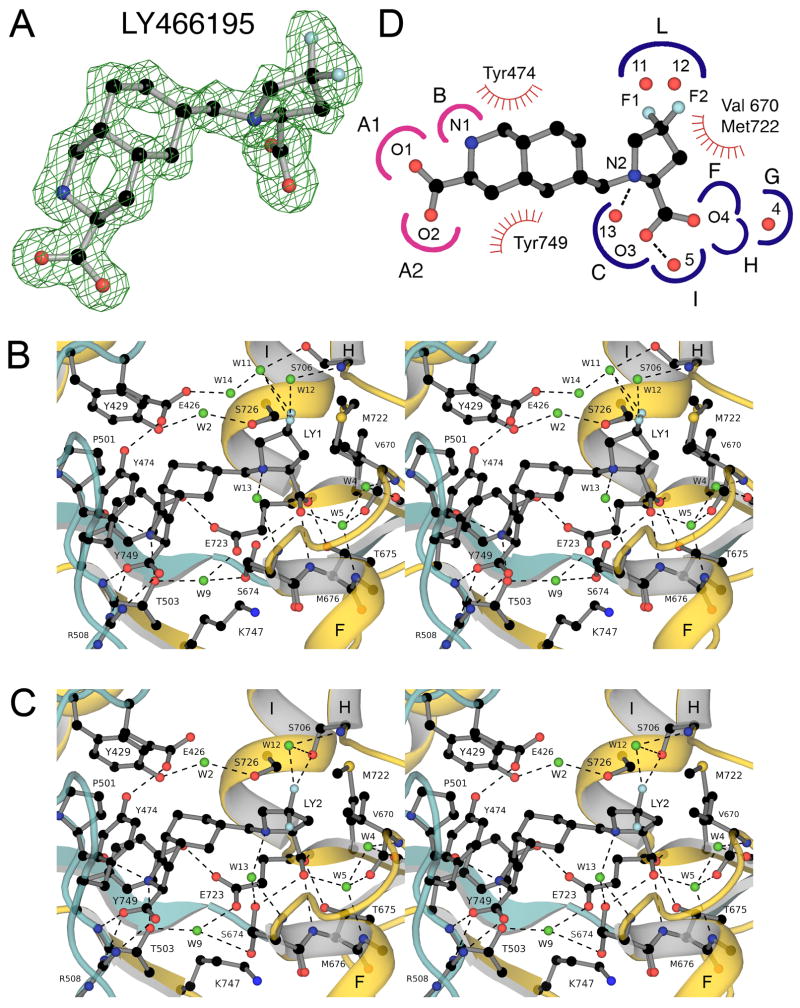
3.5 Crystal structure of the LY466195 complex
Crystal structures have not yet been reported for ligands from the extensive series of decahydroisoquinolines which have been developed as glutamate receptor antagonists (Clarke et al., 1997; Ornstein et al., 1996; Ornstein et al., 1992; Weiss et al., 2006). Of this series we chose to study LY466195, one of the most potent GluK1 antagonists reported to date. The crystal form for the GluK1 LY466195 complex, space group H3 was solved with data to Bragg spacings of 1.64 Å, refined to an Rfree value of 20% (Table 1), and contained 4 molecules in the asymmetric unit arranged as two GluK1 dimers formed by chains A/C and B/D respectively. Each of the four antagonist molecules showed strong electron density, which revealed puckering of the 4,4-difluoro-pyrrolidine group towards either helix H, which we named conformation LY1 (Fig. 5B), or helix F, named conformation LY2 (Fig 5C). This conformational freedom leads to a movement of the fluorine atoms by up to 3.5 Å, with subsequently different interactions with residues and solvent molecules in domain 2 (Fig. 5). In chain A both ligand conformations were present and were refined with equal occupancy; in chain B two up conformations were present, but with slightly different extents of puckering; chain C had only the up conformation; chain D only the down conformation.
Figure 5.
Crystal structure of the GluK1 LY466195 complex. (A) Fo-Fc map at 1.58 Å resolution contoured at 3.5 sigma; atoms for LY466195 from subunit D were omitted from the Fc calculation. (B) Stereoview of the GluK1 LY466195 complex for subunit D colored as for Fig. 3; the 4,4-difluoro-pyrrolidine adopts a down conformation, and both halogen atoms project towards helix H in the back of ligand binding pocket. (C) Stereoview of the GluK1 LY466195 complex for subunit A in which the ligand 4,4-difluoro-pyrrolidine group adopts an up conformation and projects out of the ligand binding pocket. (D) Subsite map showing schematic representations of LY466195 with the GluK1 binding pocket of subunit D colored and oriented to match the representation shown in Figs. 3 and 4.
Linked to these different conformations there were small differences in the extent of domain closure, such that the base of helix F, defined by the CA position of Lys 681 moved towards domain 1 by as much as 3 Å in the down versus up ligand conformations. Overall however, the extent of domain closure for the LY466195 complexes, range 25.5° to 26.7° with down the most closed, was close to that found for the two subunits in the UBP315 complex, range 25.5° to 26.0°, and several degrees more closed than for the UBP310, UBP316 and UBP318 complexes (Fig. 2). In all four subunits E723 adopts an extended conformation (Fig. 5B, C), and makes a direct hydrogen bond contact with the hydroxyl group of Y749, as found in the UBP315 complex.
The similar extent of domain closure for LY466195 and UBP315 occurs because the decahydroisoquinoline ring acts as a rigid scaffold that positions the pyrrolidine ring 2-position carboxylate substituent adjacent to sites F, G H and I in the ligand binding site pocket (Fig. 5D). These sites contain two conserved water molecules, W4 and W5, which are present in all antagonist complexes. Additional water molecules play roles in the binding of LY466195 due to its unique chemistry compared to the UBP complexes. A water molecule (W13) links the pyrrolidine ring with the backbone of Ser674 at the terminus of helix F, while two additional molecules, W11 and W12, link the pyrrolidine group fluorine atoms with the side chain hydroxyl group and main chain amide of Ser706 in helix H. LY466195 is the only antagonist studied in this series which interacts with residues in helix G.
4. Discussion
In this study we describe three new crystal structures for GluK1 competitive antagonist complexes. The results reveal conformational flexibility for the ligand binding domain and, surprisingly the ligands, and give insight into the binding mechanisms and pharmacology of GluK1-specific ligands. Together with data from prior crystallographic studies on GluK1 agonist, partial agonist and antagonist complexes, our results reveal that the ligand-binding domain can adopt a wide range of twist angles or domain closure. There is not just one agonist or partial agonist bound conformation, nor a single antagonist bound conformation. Extensive MD simulations on the structurally related GluA2 ligand binding domain, using umbrella sampling, provide some insight into the underlying cause, and reveal that there are multiple open cleft conformations connected by a relatively flat energy landscape (Lau and Roux, 2007). It is not yet clear whether the lowest energy apo state of the GluK1 LBD corresponds to the most open conformation observed for the UBP310, UBP316 and UBP318 complexes, or to the more closed conformations seen for UBP315 and LY466195, or for the conformation found for ATPO which is intermediate between these extremes (Hald et al., 2007). Also unknown is the difference in free energy of these conformations in GluK1. Conformational flexibility was also observed for the pyrrolidine group in LY466195, which flexes to adopt a range of different puckers, and for the thiophene ring in the UBP310, UBP315, UBP316 and UBP318 complexes, where the values for the N3 (uracil), C (attached to N3), C3 (thiophene), C4 (thiophene) torsion are −20.33°, −41.90°, −26.98°, −17.03° respectively. This rotation results in a movement of the 2-position carboxyl group oxygen atoms by up to 2 Å (Supplementary Fig. 1A), and changes the strength of hydrogen bonds made with the pocket formed by the N-terminus of helix F.
The high resolution of the structures reported here reveals numerous subtle differences in the binding mechanism for the three ligands which are analyzed using the subsite maps shown in Figs 4 and 5. In the discussion below we focus only on a subset of sites which differ in the 3 antagonist complexes when compared to the closed cleft glutamate complex reported previously (Mayer et al., 2006). Site B formed by the main-chain carbonyl oxygen of Pro501 and the hydroxyl group of Thr503 in domain 1 binds the ligand α-amino group or its equivalent in LY466195. Due to differences in domain closure site B is modified in the glutamate and UBP315 complexes and includes in addition a carboxylate group oxygen atom from Glu723 in domain 2. In the UBP310, UBP316 and UBP318 complexes a solvent network formed by W7 and W8 links the ligand α-amino group with the side chain of Glu723, while in the LY466195 complex W7 and W8 are displaced by the bulky decahydroisoquinoline group.
Site D is formed by side chains of Glu426, Tyr429, Pro501 and Tyr474 and in the glutamate complex binds W1. In all 4 UBP ligand complexes W1 is displaced by the 5-position substituent which makes a van der Waals contact with site D. In the LY466195 complex W1 is displaced by the decahydroisoquinoline ring. Site E is formed by the side chain of Ser726 and binds W2, which makes a hydrogen bond with the 4-position carbonyl group on the uracil ring of UBP310, UBP316 and UBP318 complexes. In the UBP315 complex W2 is displaced, and the side chain of Ser726 adopts a different rotamer. In the LY466195 complex W2 is present, but does not interact with the ligand, and instead links the hydroxyl groups of Tyr429 in domain 1 and Ser726 in domain 2. Site F is formed by the main chain amide of Glu723, which binds W3. In the glutamate and UBP310, UBP316 and UBP318 complexes W3 makes a hydrogen bond with the gamma-carboxylate or thiophene 2-position carboxylate. In the UBP315 and LY466195 complexes W3 is displaced and there is a direct hydrogen bond connecting the main chain amide of Glu723 to the ligand gamma carboxyl group. Site I is formed by the main chain carbonyl oxygen of Val670 and the main chain amide of Met676 and binds water W5 which is present in the glutamate complex and all antagonist complexes.
Site K is utilized only in the UBP315 complex and is formed by one of the carboxylate group oxygen atoms of Glu426 which makes a halogen bond to the 5-position Br substituent. This is a previously unexploited region of the GluK1 binding pocket, and is the only domain 1 contact made with the thiophene ring. The formation of halogen bonds in biological molecules is an under appreciated aspect of ligand binding best documented for iodinated thyroid hormones (Auffinger et al., 2004; Howard et al., 2004; Voth et al., 2007).
Site M is formed by the hydroxyl group and the main chain amide of S706. In the glutamate complex the serine hydroxyl group binds W7, which makes a hydrogen bond to W4. In the LY466195 complex this site interacts both directly and indirectly via two water molecules with the 4,4-difluoro-pyrrolidine group. The side chain of S706 adopts at least 3 different conformations, suggesting that site M is not a particularly strong contact. This is also supported by the observed multiple conformations of the ring pucker of the pyrrolidine group.
We also noted additional changes in solvent networks not described above. In the GluK1 glutamate complex there is an interdomain hydrogen bond between the hydroxyl groups of Tyr429 and Thr725. This contact is broken in the UBP310, UBP316 and UBP318 complexes, and the space filled by a water molecule. However, in UBP315 and LY466195 complexes, the two antagonists which produce increased domain closure, there is a new interdomain hydrogen bond formed between the hydroxyl groups of Tyr749 and Thr725. This is the only new interdomain contact that results from the increased domain closure and it is also present in the ATPO complex.
In conclusion, the available antagonist GluK1 LBD complexes and those reported here reveal a wide variation in ligand-receptor interactions and ligand binding domain closure. Given the variation in ligand binding domain closure these complexes will be useful for ligand docking and in silico screening of compound libraries. It is interesting that the degree of domain closure is very similar for UBP315 and LY466195, two structurally unrelated ligands that bind with distinct mechanisms and with high affinity. This suggests that energetic minima do exist in the conformational landscape of large-scale domain rearrangements, yet diverse ligand-binding configurations are compatible with the same domain arrangement. Just how diverse are the ligand binding conformations is shown by a superposition of the UBP315 and LY466195 ligands in Supplemental Fig. 1B. Our understanding of the rules underlying both protein conformational bias and the surprising chemical plasticity of the ligand-binding pocket of iGluRs will benefit from continued crystallographic studies of subtype-selective ligands as they become available.
Supplementary Material
Acknowledgments
GMA was a postbac IRTA fellow of the NIH. Synchrotron diffraction data was collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser-cat.org/members.html. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. This work was supported by the intramural research program of NICHD, NIH, DHHS. Work leading to the synthesis of UBP315 and UBP318 was supported by the BBSRC. We thank Dr. David Bleakman for the gift of LY466195 and Dr. Jinjin Zhang for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed AH, Thompson MD, Fenwick MK, Romero B, Loh AP, Jane DE, Sondermann H, Oswald RE. Mechanisms of antagonism of the GluR2 AMPA receptor: structure and dynamics of the complex of two willardiine antagonists with the glutamate binding domain. Biochemistry. 2009;48:3894–3903. doi: 10.1021/bi900107m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt A, Weiss B, Ornstein PL, Gleason SD, Bleakman D, Stratford RE, Jr, Witkin JM. Anxiolytic-like effects through a GLUK5 kainate receptor mechanism. Neuropharmacology. 2007;52:1482–1487. doi: 10.1016/j.neuropharm.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Arinaminpathy Y, Sansom MS, Biggin PC. Binding site flexibility: Molecular simulation of partial and full agonists with a glutamate receptor. Mol Pharmacol. 2006;69:5–12. doi: 10.1124/mol.105.016691. [DOI] [PubMed] [Google Scholar]
- Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: Crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- Auffinger P, Hays FA, Westhof E, Ho PS. Halogen bonds in biological molecules. Proc Natl Acad Sci U S A. 2004;101:16789–16794. doi: 10.1073/pnas.0407607101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beneyto M, Kristiansen LV, Oni-Orisan A, McCullumsmith RE, Meador-Woodruff JH. Abnormal glutamate receptor expression in the medial temporal lobe in schizophrenia and mood disorders. Neuropsychopharmacology. 2007;32:1888–1902. doi: 10.1038/sj.npp.1301312. [DOI] [PubMed] [Google Scholar]
- CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallog sect D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Clarke VR, Ballyk BA, Hoo KH, Mandelzys A, Pellizzari A, Bath CP, Thomas J, Sharpe EF, Davies CH, Ornstein PL, Schoepp DD, Kamboj RK, Collingridge GL, Lodge D, Bleakman D. A hippocampal GluR5 kainate receptor regulating inhibitory synaptic transmission. Nature. 1997;389:599–603. doi: 10.1038/39315. [DOI] [PubMed] [Google Scholar]
- Collingridge GL, Olsen RW, Peters J, Spedding M. A nomenclature for ligand-gated ion channels. Neuropharmacology. 2009;56:2–5. doi: 10.1016/j.neuropharm.2008.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz LA, Estebanez-Perpina E, Pfaff S, Borngraeber S, Bao N, Blethrow J, Fletterick RJ, England PM. 6-Azido-7-nitro-1,4-dihydroquinoxaline-2,3-dione (ANQX) forms an irreversible bond to the active site of the GluR2 AMPA receptor. J Med Chem. 2008;51:5856–5860. doi: 10.1021/jm701517b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dargan SL, Clarke VR, Alushin GM, Sherwood JL, Nistico R, Bortolotto ZA, Ogden AM, Bleakman D, Doherty AJ, Lodge D, Mayer ML, Fitzjohn SM, Jane DE, Collingridge GL. ACET is a highly potent and specific kainate receptor antagonist: characterisation and effects on hippocampal mossy fibre function. Neuropharmacology. 2009;56:121–130. doi: 10.1016/j.neuropharm.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]
- Dolman NP, More JC, Alt A, Knauss JL, Pentikainen OT, Glasser CR, Bleakman D, Mayer ML, Collingridge GL, Jane DE. Synthesis and pharmacological characterization of N3-substituted willardiine derivatives: role of the substituent at the 5-position of the uracil ring in the development of highly potent and selective GLUK5 kainate receptor antagonists. J Med Chem. 2007;50:1558–1570. doi: 10.1021/jm061041u. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Frydenvang K, Lash LL, Naur P, Postila PA, Pickering DS, Smith CM, Gajhede M, Sasaki M, Sakai R, Pentikainen OT, Swanson GT, Kastrup JS. Full domain closure of the ligand-binding core of the ionotropic glutamate receptor iGluR5 induced by the high affinity agonist dysiherbaine and the functional antagonist 8,9-dideoxyneodysiherbaine. J Biol Chem. 2009;284:14219–14229. doi: 10.1074/jbc.M808547200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H, Gouaux E. Mechanisms of activation, Inhibition and Specificity: Crystal Structures of NR1 Ligand-Binding Core. Embo J. 2003;22:1–13. doi: 10.1093/emboj/cdg303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- Hald H, Naur P, Pickering DS, Sprogoe D, Madsen U, Timmermann DB, Ahring PK, Liljefors T, Schousboe A, Egebjerg J, Gajhede M, Kastrup JS. Partial agonism and antagonism of the ionotropic glutamate receptor iGLuR5: structures of the ligand-binding core in complex with domoic acid and 2-amino-3-[5-tert-butyl-3-(phosphonomethoxy)-4-isoxazolyl]propionic acid. J Biol Chem. 2007;282:25726–25736. doi: 10.1074/jbc.M700137200. [DOI] [PubMed] [Google Scholar]
- Hogner A, Greenwood JR, Liljefors T, Lunn ML, Egebjerg J, Larsen IK, Gouaux E, Kastrup JS. Competitive antagonism of AMPA receptors by ligands of different classes: crystal structure of ATPO bound to the GluR2 ligand-binding core, in comparison with DNQX. J Med Chem. 2003;46:214–221. doi: 10.1021/jm020989v. [DOI] [PubMed] [Google Scholar]
- Hogner A, Kastrup J, Jin R, Liljefors T, Mayer M, Egebjerg J, Larsen I, Gouaux E. Structural Basis for AMPA Receptor Activation and Ligand Selectivity: Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding Core. J Mol Biol. 2002;322:93. doi: 10.1016/s0022-2836(02)00650-2. [DOI] [PubMed] [Google Scholar]
- Howard EI, Sanishvili R, Cachau RE, Mitschler A, Chevrier B, Barth P, Lamour V, Van Zandt M, Sibley E, Bon C, Moras D, Schneider TR, Joachimiak A, Podjarny A. Ultrahigh resolution drug design I: details of interactions in human aldose reductase-inhibitor complex at 0.66 A. Proteins. 2004;55:792–804. doi: 10.1002/prot.20015. [DOI] [PubMed] [Google Scholar]
- Inanobe A, Furukawa H, Gouaux E. Mechanism of partial agonist action at the NR1 subunit of NMDA receptors. Neuron. 2005;47:71–84. doi: 10.1016/j.neuron.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Jane DE, Lodge D, Collingridge GL. Kainate receptors: pharmacology, function and therapeutic potential. Neuropharmacology. 2009;56:90–113. doi: 10.1016/j.neuropharm.2008.08.023. [DOI] [PubMed] [Google Scholar]
- Jin R, Banke TG, Mayer ML, Traynelis SF, Gouaux E. Structural basis for partial agonist action at ionotropic glutamate receptors. Nature Neurosci. 2003;6:803–810. doi: 10.1038/nn1091. [DOI] [PubMed] [Google Scholar]
- Jones CK, Alt A, Ogden AM, Bleakman D, Simmons RM, Iyengar S, Dominguez E, Ornstein P, Shannon HE. Anti-allodynic and anti-hyperalgesic effects of selective competitive GLUK5 (GluR5) ionotropic glutamate receptor antagonists in the capsaicin and carrageenan models in rats. J Pharmacol Exp Ther. 2006 doi: 10.1124/jpet.106.105601. [DOI] [PubMed] [Google Scholar]
- Kasper C, Pickering DS, Mirza O, Olsen L, Kristensen AS, Greenwood JR, Liljefors T, Schousboe A, Watjen F, Gajhede M, Sigurskjold BW, Kastrup JS. The structure of a mixed GluR2 ligand-binding core dimer in complex with (S)-glutamate and the antagonist (S)-NS1209. J Mol Biol. 2006;357:1184–1201. doi: 10.1016/j.jmb.2006.01.024. [DOI] [PubMed] [Google Scholar]
- Lau AY, Roux B. The free energy landscapes governing conformational changes in a glutamate receptor ligand-binding domain. Structure. 2007;15:1203–1214. doi: 10.1016/j.str.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerma J. Kainate receptor physiology. Curr Opin Pharmacol. 2006;6:89–97. doi: 10.1016/j.coph.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Mayer ML. Crystal Structures of the GluR5 and GluR6 Ligand Binding Cores: Molecular Mechanisms Underlying Kainate Receptor Selectivity. Neuron. 2005;45:539–552. doi: 10.1016/j.neuron.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Mayer ML. Glutamate receptors at atomic resolution. Nature. 2006;440:456–462. doi: 10.1038/nature04709. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Ghosal A, Dolman NP, Jane DE. Crystal structures of the kainate receptor GluR5 ligand binding core dimer with novel GluR5-selective antagonists. J Neurosci. 2006;26:2852–2861. doi: 10.1523/JNEUROSCI.0123-06.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menuz K, Stroud RM, Nicoll RA, Hays FA. TARP auxiliary subunits switch AMPA receptor antagonists into partial agonists. Science. 2007;318:815–817. doi: 10.1126/science.1146317. [DOI] [PubMed] [Google Scholar]
- Nanao MH, Green T, Stern-Bach Y, Heinemann SF, Choe S. Structure of the kainate receptor subunit GluR6 agonist-binding domain complexed with domoic acid. Proc Natl Acad Sci U S A. 2005:1708–1713. doi: 10.1073/pnas.0409573102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naur P, Vestergaard B, Skov LK, Egebjerg J, Gajhede M, Kastrup JS. Crystal structure of the kainate receptor GluR5 ligand-binding core in complex with (S)-glutamate. FEBS Lett. 2005;579:1154–1160. doi: 10.1016/j.febslet.2005.01.012. [DOI] [PubMed] [Google Scholar]
- O’Neill MJ, Bogaert L, Hicks CA, Bond A, Ward MA, Ebinger G, Ornstein PL, Michotte Y, Lodge D. LY377770, a novel iGlu5 kainate receptor antagonist with neuroprotective effects in global and focal cerebral ischaemia. Neuropharmacology. 2000;39:1575–1588. doi: 10.1016/s0028-3908(99)00250-6. [DOI] [PubMed] [Google Scholar]
- Ornstein PL, Arnold MB, Allen NK, Bleisch T, Borromeo PS, Lugar CW, Leander JD, Lodge D, Schoepp DD. Structure-activity studies of 6-substituted decahydroisoquinoline-3-carboxylic acid AMPA receptor antagonists. 2. Effects of distal acid bioisosteric substitution, absolute stereochemical preferences, and in vivo activity. J Med Chem. 1996;39:2232–2244. doi: 10.1021/jm950913h. [DOI] [PubMed] [Google Scholar]
- Ornstein PL, Schoepp DD, Arnold MB, Augenstein NK, Lodge D, Millar JD, Chambers J, Campbell J, Paschal JW, Zimmerman DM, et al. 6-substituted decahydroisoquinoline-3-carboxylic acids as potent and selective conformationally constrained NMDA receptor antagonists. J Med Chem. 1992;35:3547–3560. doi: 10.1021/jm00097a012. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Denzo and Scalepack. International Tables for Crystallography F. 2001:226–235. [Google Scholar]
- Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- Pinheiro P, Mulle C. Kainate receptors. Cell Tissue Res. 2006;326:457–482. doi: 10.1007/s00441-006-0265-6. [DOI] [PubMed] [Google Scholar]
- Postila PA, Swanson GT, Pentikainen OT. Exploring kainate receptor pharmacology using molecular dynamics simulations. Neuropharmacology. 58:515–527. doi: 10.1016/j.neuropharm.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons RM, Li DL, Hoo KH, Deverill M, Ornstein PL, Iyengar S. Kainate GluR5 receptor subtype mediates the nociceptive response to formalin in the rat. Neuropharmacology. 1998;37:25–36. doi: 10.1016/s0028-3908(97)00188-3. [DOI] [PubMed] [Google Scholar]
- Smolders I, Bortolotto ZA, Clarke VR, Warre R, Khan GM, O’Neill MJ, Ornstein PL, Bleakman D, Ogden A, Weiss B, Stables JP, Ho KH, Ebinger G, Collingridge GL, Lodge D, Michotte Y. Antagonists of GLU(K5)-containing kainate receptors prevent pilocarpine-induced limbic seizures. Nat Neurosci. 2002;5:796–804. doi: 10.1038/nn880. [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechamism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- Voth AR, Hays FA, Ho PS. Directing macromolecular conformation through halogen bonds. Proc Natl Acad Sci U S A. 2007;104:6188–6193. doi: 10.1073/pnas.0610531104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Weiss B, Alt A, Ogden AM, Gates M, Dieckman DK, Clemens-Smith A, Ho KH, Jarvie K, Rizkalla G, Wright RA, Calligaro DO, Schoepp D, Mattiuz EL, Stratford RE, Johnson B, Salhoff C, Katofiasc M, Phebus LA, Schenck K, Cohen M, Filla SA, Ornstein PL, Johnson KW, Bleakman D. Pharmacological characterization of the competitive GLUK5 receptor antagonist decahydroisoquinoline LY466195 in vitro and in vivo. J Pharmacol Exp Ther. 2006;318:772–781. doi: 10.1124/jpet.106.101428. [DOI] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- Yao Y, Harrison CB, Freddolino PL, Schulten K, Mayer ML. Molecular mechanism of ligand recognition by NR3 subtype glutamate receptors. Embo J. 2008;27:2158–2170. doi: 10.1038/emboj.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.