Abstract
Although it is clear that amyloid beta (Aβ) peptides play a pivotal role in the development of Alzheimer’s disease, the precise molecular model of action remains unclear. Aβ peptides form assemble both in aqueous solution and in lipid membranes. It has been proposed that deleterious effects occur when the peptides interact with membranes, possibly by forming Ca2+ permeant ion channels. In the accompanying manuscript, we propose models in which the C-terminus third of six Aβ42 peptides forms a six-stranded β-barrel in highly toxic soluble oligomers. Here we extend this hypothesis to membrane-bound assemblies. In these Aβ models, the hydrophobic β-barrel of a hexamer may either reside on the surface of the bilayer, or span the bilayer. Transmembrane pores are proposed to form between several hexamers. Once the β-barrels of six hexamers have spanned the bilayer, they may merge to form a more stable 36-stranded β-barrel. We favor models in which parallel β-barrels formed by N-terminus segments comprise the lining of the pores. These types of models explain why the channels are selective for cations and how metal ions, such as Zn2+, synthetic peptides that contain histidines, and some small organic cations may block channels or inhibit formation of channels. Our models were developed to be consistent with microscopy studies of Aβ assemblies in membranes, one of which is presented here for the first time.
Keywords: amyloid beta, Alzheimer’s disease, membrane, channels, molecular models, structure prediction, protein structure, molecular dynamics, molecular assemblies
Introduction
Aβ peptides are formed in membranes when the amyloid precursor protein (APP) is cleaved in the transmembrane region to form peptides of 39–43 residues 25. Aβ40 is the most prevalent form; however, Aβ42 is more toxic 18, 61, 73. Unfortunately, it is not known exactly which types of assemblies are responsible for the pathology of Alzheimer’s disease. Numerous studies indicate that Aβ peptides interact with membranes 9, 19, 49, that they form ion channels in some synthetic bilayers 2, 4, 5, 9, 28, 29, 31, 32, 34, 36, 43, 43, 58, that the concentration of Aβ peptides at the membrane interface is much higher than in aqueous phases, that interactions with membranes may induce fibrillogenesis 13, and that Aβ peptides cross membranes to enter the intracellular space 37, 68. It has been proposed that Aβ toxicity is due to entry of Ca2+ into cells through Aβ channels 3. This hypotheses is supported by findings that all agents shown to inhibit Aβ-induced channels in bilayers also inhibit Aβ toxicity in neurons, and some of these agents have been shown to inhibit Aβ-induced entry of Ca2+ into neurons 3, 22.
Our interest in Aβ began shortly after Arispe and Pollard 4, 5 discovered that Aβ peptides form channels in synthetic lipid bilayers. In collaboration with them, we made initial, highly tentative, models of Aβ channels 24. In these models, the N-terminus third of the peptide (called S1 here) formed a β-hairpin, the central third (called S2 here) formed an amphipathic α-helix, and the hydrophobic C-terminus third (S3) formed another α helix. Since then, several other groups have observed Aβ channels 2, 31, 32, 34, 36, 41, 65 and other effects of Aβ peptides on membranes 19, 66, and microscopy studies 42, 58 have revealed several types of membrane-bound Aβ assemblies. Solution NMR studies of Aβ peptide monomers in apolar solvents and micelles support the proposed helix-turn-helix secondary structure 15, 16, 64. However, several recent findings are inconsistent with our original helical models and with other helical models presented in the supplement that we have developed and analyzed more recently. Aβ assemblies that have been observed microscopically 42, 58 are much larger than our initial models. Here we present additional freeze fracture results that also show relatively large transmembrane (TM) assemblies. Also, the secondary structure of membrane-bound Aβ peptides have been shown to depend upon several factors. Aβ peptides appear to have a substantial amount of α-helical structure at low concentrations in membranes that contain gangliosides 9, 48, 50 or negatively charge head groups 9, 72, especially if Cu2+ or Zn2+ is present 17. However, β-secondary structure predominates at higher concentrations and/or when all of the lipids have zwitterionic head groups 14, 48, 50, 69, 72. Permeation appears to coincide with formation at higher peptide concentrations of oligomers that have predominately β secondary structure72. Channels that have relatively stable single channel conductance form much more readily when negatively charged lipids are present 2, 4, 5, 72. Solid state NMR studies of Aβ fibrils indicate that S2 and S3 form β strands, each of which binds to its counterparts in other subunits to form separate in register parallel S2 and S3 β-sheets that pack next to one another 45, 56, 59. These new data, along with recent findings that Aβ oligomers inhibit short-term memory 40 and long-term potentiation 70 and are more neurotoxic than fibrils 20, have prompted us to develop a new generation of Aβ models.
A major difficulty in structural studies of Aβ assemblies is polymorphism; e.g., in membranes Aβ secondary structures, dimensions of Aβ assemblies, and effects on membrane permeabilities of Aβ peptides all vary and can be influenced by factors such as peptide concentration, nature of the soluble assembly to which the membrane is exposed, and lipid composition of the membrane. Thus, it is unreasonable to propose a single, unique model for membrane-bound Aβ assemblies. Instead, we have focused on identifying structural motifs that are stable in a membrane environment, that can form a variety of assemblies, and that can account for much of the experimental data. We have focused particularly on processes in which peptides interact with membranes to form small assemblies, which in turn interact to form a variety of channels. Our efforts have involved construction and evaluation of scores of models of both soluble and membrane-bound Aβ assemblies. Here we present only those models that best satisfy our modeling criteria and are most consistent with experimental findings. We have concentrated on Aβ42 because it is more toxic than Aβ40 18, 61, 73 and has a greater tendency to form hexamers in solution 8.
Material and Methods
An ideal computational approach to analyzing the formation of Aβ assemblies in membranes would be to begin the simulations with many Aβ of randomized structures on or in a large patch of lipid membrane and then simulate the entire folding and aggregation over time until an equilibrium has been obtained, analyze and compare the free energies of the assemblies that are formed, and determine which, if any, of the assemblies are consistent with experimental results. Unfortunately, this approach is far beyond the present capacity for all atom MD simulations because the time scale of the actual processes (minutes to days) is many orders of magnitude longer than possible for simulating such large systems (typically less than a microsecond). Conformational searches can be accelerated by procedures such as replica exchange or other tempering methods, coarse-graining of the lipid/peptide system, implicit solvent models of membranes and water, and discrete MD. However, to our knowledge such methods have never been demonstrated to correctly fold and approximate experimentally determined structures of known membrane channel proteins, and we have little confidence that they would succeed in this case. Additionally, free energies cannot be calculated accurately for membrane protein systems. Instead, we have adopted a long-term, iterative approach to modeling membrane channel proteins that has been successful for us. Initial models, such as our first models of voltage-gated channels27 or Aβ channels 24, are often highly speculative and with little experimental support. As these initial models are tested and new data are obtained and as computational methods improve, we typically generate a new generation of models, such as those presented here. We hope that in continuing this iterative process, accurate models supported by a wealth of experimental results will develop. Historically, this approach is best exemplified by our studies of voltage-gated channels (see Shafrir et al 62, 63 for latest versions of these models and discussion of how well the modeling effort predicted subsequently determined crystal structures). Early stage models, such as those presented here, have substantial uncertainties and should be treated as hypotheses to be tested experimentally. In this project we developed a large variety of alternative models that satisfy a series of initial modeling criteria, perform simulations on these models, and then determine which models are more stable, form the highest number of non-aqueous H-bonds per monomer, and are most consistent with available experimental results. Here we have presented only those models that best meet these conditions. A few of the more plausible alternative models, and why we chose not to include them in the main text, are described in the supplement.
Energetics and Symmetry
The concept that chemical moieties have energetically favorable interactions is fundamental to our approach. We develop models in which hydrophobic groups tend to be in hydrophobic environments either inside the protein or exposed to lipid alkyl chains, hydrophilic moieties tend to be exposed to water and/or interact with other polar moieties, and charged moieties tend to interact with oppositely charged moieties. In developing a strategy to model membrane-bound Aβ oligomers, it was important to understand that Aβ peptides have an amphiphilic sequence. Our nomenclature divides the Aβ42 peptide into three segments of equal length, S1, S2, and S3. The N-terminus S1 segment (1DAEFRHDSGYEVHH14) is very hydrophilic, the middle S2 segment (15QKLVFFAEDVGSNK28) has about the same number of hydrophobic and hydrophilic residues, and the C-terminus S3 segment (29GAIIGLMVGGVVIA42, the last two residues are missing in Ab40) has no polar side chains and is thus very hydrophobic.
Transmembrane (TM) β strands were modeled to be components of radially symmetric TM β barrels. We typically require all polar backbone atoms of TM residues exposed primarily to lipid alkyl chain to form H-bonds. This criterion is best satisfied for isolated TM segments by making them helical. However, for models in which β-strands span the bilayer, this criterion can be satisfied well by β-barrel motifs with the central axis of the barrel oriented perpendicular to the membrane and the strands spanning the bilayer. In fact, all known structures with transmembrane β-strands are β-barrels. β-barrel strands are always tilted relative to the axis of the barrel and spiral about the axis in a right-handed manner. Thus, if a β-barrel is composed of parallel β-strands of identical segments from differing subunits, the tilt of the strands prevents them from all being in register, as occurs in β-fibrils 56. The magnitude of the tilt is typically stipulated by the shear number 44. Although the shear number is typically about the same as the number of strands, it can vary, and the diameter of the barrel increases as a function of the shear number.
In almost all known crystal structures of transmembrane proteins composed of identical subunits, all subunits have identical conformations and are symmetrically related about a central axis oriented orthogonal to the surface of the membrane 6, 12, 23, 67, 26, 35. The α-hemolysin channel, in which seven identical β-hairpins form a 14-stranded transmembrane β-barrel, is an example of this type of structure 67. We thus restrained our initial hexameric models of transmembrane oligomers to have 3-fold radial symmetry (see accompanying manuscript); all subunits have identical conformations in the antiparallel models, but two conformations are present in the parallel models. Our hexamer-of-hexamer models of ion channels were constrained to have 6-fold symmetry about the central axis of the pore. If one constrains barrels composed of identical strands of a fixed number of subunits to have radial symmetry, then the shear number is restricted to a multiple of two times the symmetry number; e.g., if the barrel has 3-fold symmetry around its axis, then the shear number is limited to the series (6,12,18,…). Unless noted otherwise, the shear number of our β-barrel models equals the number of strands. These symmetry criteria highly constrain backbone structure of the β-barrel models, making the number of possible backbone structures relatively small.
Initial models were developed using the PSSHOW program (Swanson, 1995) to manually position subunits and alter dihedral angles. Adjustments were made to reduce steric clashes and to improve energetically favorable interactions such as backbone hydrogen bonds of β-barrels and α-helices and salt bridges between side chains. Side chain torsion angles were selected that are observed frequently in known structures for that residue type. Energies of these structures were then minimized using CHARMM 10 with the symmetry constraints of the specific assembly design. This process was repeated with manual adjustments if minimization perturbed the structure substantially, if side chains adopted energetically unfavorable conformations, or if hydrogen bonds of the β-barrels or α-helices broke.
The molecular dynamics simulations were run using the program Gromacs (http://www.gromacs.org). Coordinates for the phosphatidylethanolamine (POPE) lipid bilayer used in these simulations were kindly provided by Dr. Peter Tieleman. The electrostatic calculations were done using the Particle Mesh Ewald (PME) method, and the Van-der-Waals cutoff was 1.0 nm. The time step was 2fs and the LINCS algorithm was used to constrain bond-lengths. The simulations were run under NPT conditions with the protein, lipid and water each coupled separately to a temperature bath at 310K with a coupling constant τT of 0.1ps, and at a constant pressure of 1 bar in all directions with a pressure constant of τp = 1.0 ps. The lipid parameters were based on Berger et al 7, the lipid-protein interactions were based on the GROMOS parameters. Each simulation was preceded by an energy minimization using the steepest descent method. This was followed by a short equilibration run of 200 ps with harmonic restraints on the backbone atoms of the protein to allow packing of the lipid molecules around the protein and relaxing of the water molecules. Each simulation was run for 6ns on the BioWulf computer system. The models were examined at the end of 6 ns simulations to determine if most of the subunits changed conformations in a similar, or systematic manner from the initial symmetrical model. If they did, a new symmetric model was constructed that incorporated these molecular shifts and changes, and the simulation was repeated. This process was stopped, usually after a few iterations, when systematic changes were no longer observed. Then, a 20ns simulation was performed to evaluate the stability of the final models. Thus, a final model represents a total simulation time of at least 40ns. Approximately 50 simulations on membrane-bound peptides were performed throughout the course of this study.
Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081) 57.
Experimental
Microscopy
Our models were designed to be consistent with results from microscopy. Atomic force 42 and freeze fracture studies (presented here) indicate that Aβ peptides form large assemblies ranging from 7 to 13 nm in diameter. Atomic force microscopy studies of Aβ assemblies bound to membranes have identified large (8–12 nm in diameter) assemblies in which four to six peaks surround a central cavity 42. These peaks are too large to be due to single Aβ monomers. It is more likely that each peak is an Aβ oligomer and that four to six of these oligomers aggregate to form assemblies with a central pore. Thus, the first stage of our modeling was to identify plausible structures for isolated oligomers. Here we have limited the oligomers to hexamers that have hydrophobic β-barrel structures formed by S3 segments, similar to those proposed in the accompanying manuscript for soluble hexamers. These membrane-bound hexamer models were then modified to develop numerous hexamer-of-hexamers models of channels consistent with dimensions of microscopically observed Aβ assemblies.
Freeze-fracture procedure
Aliquots of Aβ-incorporated proteoliposomes were attached to a glass surface, covered with a small piece of copper and rapidly frozen by immersion in liquid propane 74. The samples were fractured in a JOEL freeze-fracture apparatus and the cleaved surfaces were shadowed first with a layer of carbon and then with platinum-carbon 38, 75. The replicas were mounted on substrates with 1 μm diameter holes (“holey” grids), and the regions spanning the holes were used for imaging to eliminate the contribution of the support film to the thickness of the specimen.
Preparation of Proteoliposomes
Liposomes were prepared by hydration of air-dried palmitoyloleoyl phosphatidylserine (10 mg) with 1 M potassium aspartate,pH 7.0 (1 ml), followed by water sonication for 5 min. The liposomesuspension (50 μl) was mixed with a stock aqueous solution of Aβ (1http://www.jbc.org/cgi/inline?StressGen mg/ml, obtained from Bachem and from AnaSpec, California, USA), followed by sonication for an additional period of5 min.
Planar Lipid Bilayer Methodology
Planar lipid bilayers were made as described previously 5. Briefly, a suspension of palmitoyloleoyl phosphatidylserine and palmitoyloleoyl phosphatidylethanolamine, 1:1, in n-decane was applied to an orifice of about 100–120 μm in diameterwith a Teflon™ film separating two compartments. The ionic solutions in thecompartments contained asymmetrical concentrations of CsCl (200cis/50trans mM) and symmetrical 0.5 mM CaCl2 and 5 mM K-HEPES, pH 7. Aliquots (5 μl) of the proteoliposome suspension wasadded to the solution in the cis side of the planar lipid bilayerchamber andstirred. After successful ion channel incorporation was achieved, proteoliposomes were perfused from the chamber and the ionic current was recorded and stored on computer disk memory. Off-line analysis of the channel activity was carriedout using the software package Pclamp (Axon Instruments, Foster City, CA).
Results
Freeze Fracture Studies
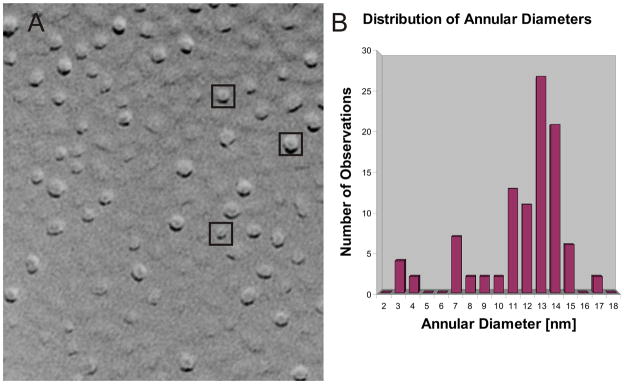
Atomic force microscopy studies of membrane-bound Aβ assemblies have revealed assemblies 8–12 nm in diameter in which four to six peaks surround a central indentation that has been postulated to correspond to the pore of an Aβ channel 42. These peaks extended ~ 1nm above 5nm-thick membranes. Here we report results of freeze fracture electron microscopy studies of Aβ assemblies inserted in phospholipid membranes. The Aβ-containing proteoliposomes were provided by N. Arispe and the freeze fracture experiments were performed by Dr. G. Zampighi (Departments of Neurobiology, Physiology, and Jules Stein Eye Research Institute, University of California at Los Angeles School of Medicine, Los Angeles, California, USA). In these studies, freshly prepared Aβ reconstituted in phospholipid bilayers was frozen and fractured down the middle, exposing complementary fracture faces to metal shadowing. The center of the phospholipid bilayer appeared as a flat, featureless surface and the membrane fractures between the two leaflets of the bilayer reveal cylindrically-shaped transmembrane Aβ assemblies of various sizes (Fig. 1A). The diameters of the three most frequently observed sizes of assemblies are ~13–14 nm, ~11nm, and ~7nm (Fig. 1B). Later we present a series of models designed to be consistent with these data.
Figure 1.
Freeze fracture studies of transmembrane Aβ assemblies. (A) Image of a fractured liposome reconstituted with Aβ and shadowed with platinum–carbon. The small particles represent the external domains of the transmembrane Aβ assemblies and the smoother regions, the center of the phospholipid bilayer. The enclosed images are those illustrated in Fig. 3G. (B) The histogram show the distribution of Aβ assemblies according to their diameter (nm, n = 102). The freeze fracture figure was generated by Dr. G. Zampighi (University of California at Los Angeles School of Medicine, Los Angeles, California, USA).
Membrane surface hexamers
Experiments using Fourier transform infrared-polarized attenuated total reflection (FTIR-PATR) spectroscopy to study ganglioside-containing membranes at high concentrations of Aβ40 have been interpreted to indicate that the peptides form antiparallel β-sheets that lie parallel to the membrane surface where they dehydrate lipid interfacial groups and perturb alkyl chain orientation 48, 50. We have developed models of Aβ hexamers to be consistent with these findings (see Fig. 2). The axis of the antiparallel S3 barrel of our models of surface assemblies is proposed to be parallel to the membrane surface with only the lower side of the barrel interacting with lipid alkyl chains of the cis lipid leaflet. The asymmetric nature of the lipid interaction will likely make the assembly asymmetric. The dynamic nature of the S1/S2 segments may allow some to interact with lipids, while others shield the upper surface of the S3 barrel from water. Experimental studies indicate that S3 segments can adopt either α helical or β-strand secondary structures 1, 14, 46, depending upon factors such as time, environment, peptide concentration, peptide sequence, and lipid composition. We have developed models of surface hexamers in which the S2 segments are helices, β-strands, or a combination of both.
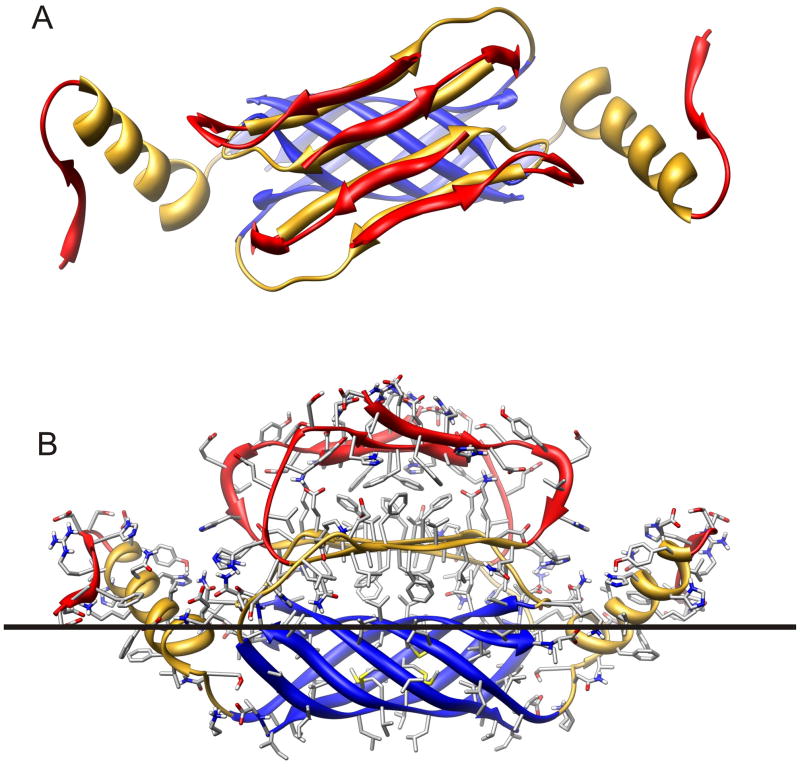
Figure 2.
Ribbon representation of Aβ42 hexamer model bound to the cis surface of the membrane. Segment S1 (residues 1–14) is red, S2 (15–28) is gold, and S3 (29–42) is blue. (A) Hexamer as viewed looking from the aqueous phase toward the membrane. The hexamer has 2-fold symmetry, with three monomeric subunit conformations. The S3 segments from six subunits form a six stranded antiparallel β-barrel with its axis parallel to the membrane surface. S1 and S2 segments of Sub1 and Sub2 comprise two antiparallel β-sheets. The red S1a sheet is on the outer surface were it is exposed to water, the gold S2 sheet is sandwiched between the S1a sheet and S3 β-barrel. S2 of the remaining two Sub3 subunits form α-helices that interact with both water and lipid. The red S1 segment of Sub3 is ill-defined and may have extended and/or coiled conformations. (B) Side view of the hexamer illustrating side chains, colored by element (red = O, blue = N, gray = C, white = H). The black line approximates the boundary between the lipid head group and alkyl chain portions of the membrane. Note that almost all hydrophobic side chains are either buried between sheets, inside the β-barrel, are exposed to lipid alkyl chains and almost all polar and charged groups are exposed to water and/or located where they can form H-bonds and salt bridges.
The model of surface hexamers we currently favor is similar to our favored model of soluble hexamers (accompanying manuscript) in that the S3 segments form a six-stranded antiparallel β-barrel and the assembly has two-fold symmetry from end to end. However, it differs in that there is no symmetry about the axis of the S3 barrel, due to the asymmetric environment of the lipid-water interface. Thus, there are two copies of three different subunit conformations in the assembly, which we will refer to as Sub1, Sub2, and Sub3. Each subunit is individually composed of S1, S2 and S3 segments. The three subunit types differ primarily in the S1–S2 region. S2 segments of Sub3 interact with lipids and are helical. S2 segments of Sub1 and Sub2 are β-strands and form a four-stranded antiparallel β-sheet that shields the upper portion of the S3 barrel from water (Fig. 2): S2 segments of Sub2 are on the edges of the sheet and those of the Sub1 are in the center. S1 segments of these assemblies are the most exposed segments and are likely to be relatively disordered. Thus, the basic model of the S2–S3 segments is not contingent upon a specific model for S1. For illustrative and computational purposes, the N-terminus end of the S1 segments (D1-H6 or S1a segment) was tentatively modeled as a β strand. In Sub1 and Sub2, these S1a strands form an antiparallel β sheet in which the A2 and F4 side chains of the S1a sheet interact with the V18 and F20 side chains of the S2 sheet (see Fig. 2B). Residues 10-13 of these subunits were modeled with extended structures with Y10 and V12 buried inside the assembly. Residues D7-S8-G9 have no regular secondary structure.
This model appear to be structurally sound; i.e., all of the hydrophobic side chains are either buried within the protein or exposed to lipid alkyl chains, all of the buried and most of the exposed charged groups form salt bridges, and backbone hydrogen bonding is extensive due to a high content of secondary structure. This model was relatively stable during molecular dynamic simulations; its root mean square deviation (RMSD) form the starting model at the end of a 20 ns simulation was ~ 0.42 nm, which is slightly less than observed for the most stable soluble hexamer but is ~ 0.10 nm greater than for the transmembrane hexamer (see Fig. Sup1 of supplement and Fig. Sup2 of the accompanying manuscript).
Hexamer-of hexamers Assemblies
The next stage of the Aβ modeling was to envision how these types of membrane-bound hexamers can interact to form assemblies consistent with experimental findings: specifically, structural images observed in atomic force microscopy and freeze fracture studies, as well as membrane currents that have been recorded in bilayers. In our antiparallel models of transmembrane hexamers, some of the S1/S2 segments of the hexamer must pass through the bilayer in order for the S3 β-barrel to span the membrane. We propose the following multistep insertion process by which these polar segments could pass through the hydrophobic membrane and form the lining of transmembrane pores without exposing many polar atoms to lipid alkyl chains.
Stage 1: Formation of hexamers on the cis membrane surface
In the accompanying manuscript we describe models of soluble hexamers and assemblies of hexamers. These pre-formed soluble hexamers or larger assemblies could bind to membranes to form the types of surface hexamers described above; however, it is also possible that monomers or smaller oligomers bind to the membrane surface, and then assemble into hexamers, or some monomers of membrane-bound assemblies may never leave the membrane after being cleaved from APP.
Stage 2: Aggregation of hexamers on the membrane surface
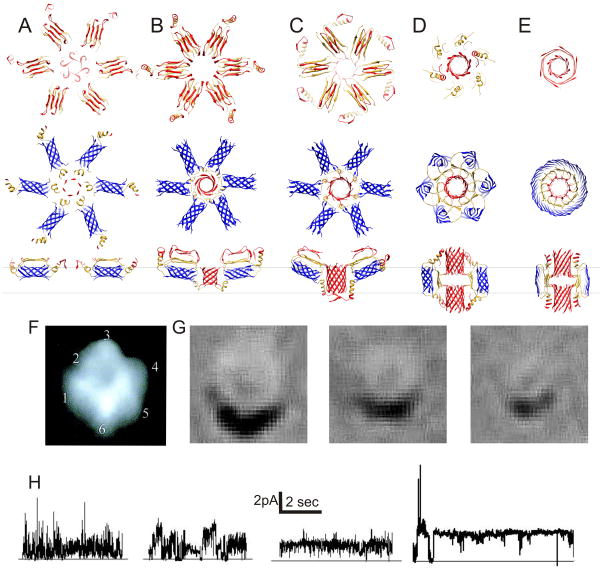
The surface hexamers may next form an assembly of several hexamers, such as the hexamer-of-hexamer models illustrated in Fig. 3A. Interactions among S1/S2 segments of different hexamers in the center of the complex may contribute to the aggregation. For example, in the model of Fig. 3A, S1–S2 segments of the centrally located Sub3 subunits form an αβ-barrel in which six S2 helices surround a six-stranded parallel S1a β-barrel with a shear number of 12. The outer diameter of this entire assembly is about 14 nm, consistent with the dimensions of the larger assemblies that have been observed microscopically. A substantial portion of each hexamer extends above the membrane, consistent with peaks observed with atomic force microscopy 42.
Figure 3.
Models of membrane bound assemblies of six hexamers and their proposed relationships to microscopy images and recorded currents. (A–E) Aβ assemblies at various stages of insertion colored as is Fig. 2. Top view of aqueous phase and of transmembrane regions are in the 1st and 2nd row; side views of two of the hexamers are in the 3rd row where black lines approximate boundries of the membrane’s alkyl phase. (F) Atomic force microscopy image of a large Aβ assembly 42 (reproduced with permission from Dr. R. Lal, Center for Nanomedicine University of Chicago, USA). (G) Freeze fracture images of Aβ assembly with diameter of ~13–14, 11 and 7 nm. (H) Different patterns of current activity from Aβ40 ion channels incorporated into planar lipid bilayers at zero bilayer potential. Currents are driven by a chemical potential gradient.
Stage 3. Penetration of the trans leaflet by Sub3 S1/S2 segments
In Fig. 3B, the central S1–S2 segments of Sub3 reorient to penetrate the trans leaflet of the bilayer. The central α-β-barrel model of the previous step is maintained, but it is oriented in the opposite direction. This arrangement places all of hydrophobic S1/S2 residues in apolar environments (lipid-exposed or buried) while allowing all of the charged side chains to be in contact with water and/or positioned where they can form salt bridges. The assembly may also be stabilized by interactions on the cis side of the bilayer. In these models, residues G9-H13 of centrally-located Sub1 and Sub2 S1 segments form β-strands that bind to their counterpart of adjacent hexamers.
Dimensions of the hexamer-of-hexamers models with S3 barrels on the membrane’s surface (Fig. 3A&B) are consistent with the larger assemblies observed microscopically. The atomic force microscopy studies reveal up to six peaks surrounding a central cavity 42 (Fig. 3F). These peaks extend ~ 1 nm above 5 nm thick membranes, and the diameters of the largest assemblies are ~12 nm; about the same as the larger freeze-fracture images. If one approximates the volume of the protein in each peak that extends above a 3 nm thick alkyl phase of the bilayer by a hemisphere with a diameter of 4 nm and assumes a protein density of ~7.5 residues/nm3, then ~120–130 residues/peak should reside above the alkyl phase. If each peak corresponds to a an Aβ hexamer, then about half of its residues should extend into the lipid head group and aqueous phase region and the other half should extend into the membrane’s alkyl phase. These values are consistent with the surface models.
Stage 4: Tilting of S3 barrels into TM region, passage of some S1–S2 segments to the trans side of the membrane, and formation of pores by other S1 segments
Both types of microscopy studies also identify smaller assemblies: in the freeze fracture studies most of these have diameters of ~ 11 and 7 nm (Fig. 1). These smaller structures could be surface assemblies composed of four or five hexamers. If so, it is possible that the S3 β-barrels remain on the surface and never span the bilayer. However, it is also possible some of the smaller diameter assemblies are hexamer-of-hexamers in which the S3 β-barrels do span the bilayer. In the mechanism that we envision, this reorientation of the S3 β-barrels can occur without exposing polar residues of S1 and S2 to the lipid alkyl chains. As the S3 barrels begin to tilt into the transmembrane region, the Sub3 S1/S2 segments may move into the trans aqueous phase. Simultaneously, some S1 segments of the Sub1 and Sub2 may move into the transmembrane region and form parallel β-barrels with hydrophilic linings through which ions may flow; e.g., Fig. 3C illustrates a parallel 12-stranded S1 barrel.
Stage 5: Reorientation of S3 barrels to span the bilayer
A complete reorientation of the S3 barrels to span the bilayer also places antiparallel S2 strands of Sub1 and Sub2 in the transmembrane region. The Sub2 S2 strands may bind to their counterparts of adjacent hexamers to form a 24-stranded S3 β-barrel in the center of the transmembrane region. The 12-stranded parallel S1 β-barrel of the preceding step may move to the trans side of the assembly while an identical S1 barrel forms on the cis side (see Fig. 3D). This model is now symmetric with respect to the central plain of the membrane. The S1–S2 segments of the Sub3 may surround these S1 barrels and have α, β, coiled, or mixtures of secondary structures.
Final stage 6: Merging of S3 barrels to form a 36-stranded S3 barrel
In the accompanying manuscript, we proposed a model for the transition from ‘beaded’ to ‘smooth’ annular protofibrils that involves the merging of six S3 barrels to form a single 36-stranded S3 barrel. Here we propose that a similar process occurs for transmembrane Aβ assemblies. The transition from Stage 5 to Stage 6 models involves a splitting apart of the 6-stranded S3 barrels between the Sub3’s followed by a binding of these Sub3 S3-strands to their counterparts of adjacent hexamers to form a 36-stranded antiparallel S3 β-barrel (Fig. 3E). The core S1–S2 segments remain relatively unaltered. This assembly has an outer diameter of ~7nm, consistent with that of the smallest major assemblies observed in the freeze fracture microscopy studies (Fig. 3G).
Relationship between models and Aβ-induced currents
Electrical recordings of Aβ-induced currents in lipid bilayers typically begin with very noisy currents immediately after exposure of the membrane to the peptides. With time more discrete single channel currents are observed and eventually relatively high conductance, stable, channels that seldom close appear (see Fig. 3H). The depiction of the insertion process described above and illustrated in Fig. 3A–E should be viewed as simplified illustrations of this highly dynamic and less ordered process for all but the Final Stage. The process is unlikely to have six discrete stages (a movie illustrating a more continuous process is in the supplement), initial assemblies are unlikely to be exactly symmetric, and the pores of the inserting assemblies are unlikely to be idealized β-barrels. Thus, we will not dwell on precise details of the inserting models. However, the Final Stage model (Fig. 3E) is intended to correspond to the highly stable, long-lived channels. The next section deals with its details and properties.
Details of the Final Stage model
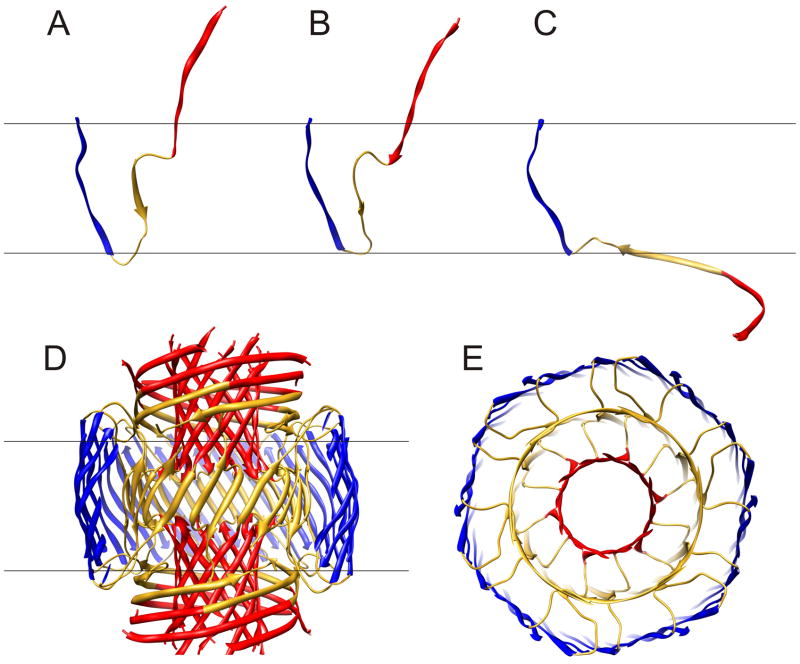
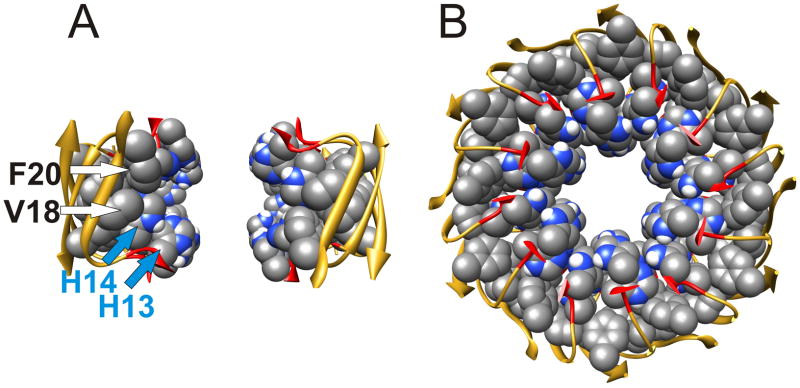
The three subunit types of the Final Stage model are illustrated in Fig. 4a. The S3 segments have almost identical conformations in all three subunits due to the approximate 18-fold symmetry of the 36-stranded S3 barrel (shear # of 36) about its axis and the 2-fold symmetry of the assembly relative to the central plain of the membrane. S1–S2 segments of Sub1 and Sub2 have similar conformations that span the transmembrane region without contacting the alkyl phase; however, they differ slightly due to staggering of adjacent strands in the 12-stranded S1 β-barrels, and due to transitions from a 24-stranded S2 β-barrel with approximate 12-fold symmetry to the 36-stranded S3 β-barrel with approximate 18-fold symmetry. S1–S2 segments of Sub3 differ drastically; instead of spanning the bilayer, they reside in the aqueous phases on each side of the membrane. These highly exposed segments are the most speculative, but also least essential, parts of the model; they may have substantial disorder in the actual assembly. We have tentatively modeled them as long curved β-strands that surround the S1 β-barrels of Sub1 and Sub2.
Figure 4.
Ribbon representation of the final stage model in which blue S3 segments form a 36-stranded antiparallel β-barrel (same as model in Fig. 3E). (A–C) Conformations of the three types of subunits, Sub1, Sub2, and Sub3, that comprise the model; 12 copies of each are in the complete model. (D) Side view of the complete model. Some of the S3 strands nearest the viewer have been clipped off to reveal the structure of the core region. Red S1 segments form two 12-stranded parallel β-barrels, central hydrophobic residues (17–21) of gold S2 segments form a central 24-stranded anitparallel β-barrel. (E) Half of the transmembrane region viewed from the aqueous phase along the central axis of the pore.
The S3 β-barrel shown in Fig. 4B and C has near ideal properties for the portion of a transmembrane structure exposed to the alkyl phase of the membrane. It is composed exclusively of hydrophobic residues and glycines, and thus there are no polar side chain atoms or unpaired backbone polar atoms exposed to lipid alkyl chains. The ~30 Ǻ length of the barrel corresponds closely to the thickness of the alkyl chain region of biological membranes 71. Its relatively smooth surface should not perturb the normal structure of membrane lipid alkyl chains substantially. The positively charged K28 side chain, which immediately precedes S3, can interact favorably with negatively charged moieties of lipid head groups.
The five consecutive hydrophobic residues of the S2 segments of Sub1 and Sub2 comprise a 24-stranded antiparallel β-barrel with a shear # of 24 (gold barrel of Fig. 4B & C). The side chains (not shown) of these residues are relatively buried either between the S2 and S3 barrels (for L17, F19, and A21) or behind H14 side chains of the C-termini of S1 segments (for V18 and F20). The positively charged amine group of K16 forms salt bridges to carboxyl groups of E22 and/or D23 from adjacent subunits at the ends of the S2 barrel. Side chains of E22 and D23 can interact with each other but are not part of the Ca2+ permeation pathway, consistent with the finding that neutralization and covalently linking of side chains at these positions increases the toxicity of Aβ peptide 47. The segments linking A21 of the S2 barrel (gold) to G29 of the S3 barrel (blue) have a coiled conformation, consistent with secondary structure predictions. These segments differs somewhat between Sub1 and Sub2.
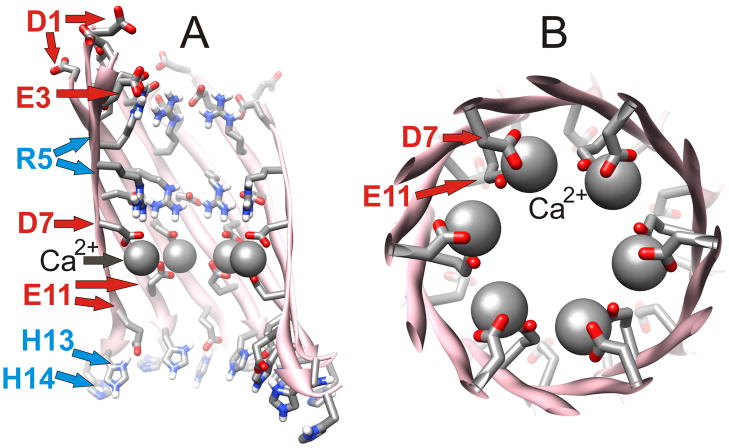
The transmembrane pore is formed by S1 segments of Sub1 and Sub2, which comprise two 12-stranded parallel β-barrels in series with one another. The C-termini ends of these two barrels approach one another near the central plain of the membrane. The lining of the central pore is formed almost exclusively by hydrophilic residues (D1, E3, R5, D7, G9, E11, H13) located at every other position in S1 (see Fig. 5A). The exterior of the barrels is composed of less polar residues (A2, F4, H6, S8, Y10, and V12) that are relatively buried between the S1 barrels and the S1–S2 segments of Sub3. Side chains of H14 are positioned at the interface between the pore and hydrophobic residues of S2; they interact with their counterparts of the opposing S1 barrel and shield hydrophobic residues of the S2 barrel (V18 and F20) from water that fills the pore. Adjacent S1 strands from Sub1 and Sub2 are staggered with respect to one another due to the tilt of the strands required to give the barrel a shear # of 12; i.e., on one side the strands are in register and bind to their counterpart of the adjacent subunit while on the other side they bind to residues that are shifted by two residue positions. This staggering reduces electrostatic repulsion due to interactions of charged side chains with their counterparts. For example, the positively charged R5 side chains of Sub1 extend toward the aqueous phases, where they salt bridge to negatively charged side chains of E3 and D1, whereas R5 side chains of Sub2 extend toward the center of the membrane where they salt bridge to D7 (Fig. 5A).
Figure 5.
Detailed representation of one of the12-stranded parallel S1 barrels that comprise the lining of the pore. Side chains of residues extending into the pore have been added and colored by element. (A) Side view in which five strands nearest the viewer have been removed to reveal the lining of the pore. Negatively and positively charged residues inside the barrel are labeled red and blue respectively. (B) View down the axis of a portion of the pore from D7 through E11. Space-filled spheres illustrate positions of Ca2+ ions prior to MD simulations. This model has six-fold symmetry about the axis of the pore/barrel.
There are several experimental findings supporting models in which S1 segments form the lining of Aβ channels. First, these channels are selective for cations and permeant to Ca2+ ions. This selectivity suggests that the lining is electronegative. Most of the negatively charged residues (and all of the negatively charged residues in highly toxic mutants in which E22 and D23 have been neutralized) reside in the S1 segments. Fig. 5B illustrates a crystalline-like symmetric model in which six Ca2+ ions bind to negatively charged carboxyl groups of D7 and Ell. Although these ion positions become more randomized during MD simulations, it is apparent that the putative pores formed by the S1 barrels are sufficiently electronegative to accommodate numerous Ca2+ ions.
Another reason for proposing that S1 segments are involved in pore formation is that Aβ channels are inhibited by Zn2+ 33 and Al3+ 5. These ions tend to bind with high affinity to sites that include imidazole rings (from histidine side chains) and carboxyl groups (from glutamate and aspartate side chains). Experimental and theoretical studies have found that side chains of His6, His13, His14, and Glu11 are involved in forming Aβ binding sites for Zn2+ 53 in solublized Aβ monomers. All of the histidine residues of Aβ are in the S1 segment. Thus, if these compounds act by blocking the pore, then S1 segments are likely to be involved in forming the pore. This hypothesis has been strengthened by the finding that peptides with sequences similar to the portion of Aβ that includes H13 and H14 inhibit Aβ-induced currents 3, 21. Histidine residues are an essential component of these inhibitory peptides: altering the histidines of the inhibitors to other residues reduces and/or eliminates the inhibition, and peptides composed exclusively of histidines are potent inhibitors. The typical explanation for inhibition of Aβ assembly processes by Aβ-like peptide fragments is that the peptide inhibitors act by binding to their sequential counterparts in the complete Aβ peptides, thus inhibiting self association of the peptides. This explanation is based on findings that β-strands of Aβ fibrils form in-register parallel β-sheets in which residues bind to their counterparts of adjacent subunits on each side 55. Although this type of mechanism may be responsible in this case, it is also possible that these inhibitors block ion permeation by binding within the pore. In either case, the results suggest that histidines of the S1 segments are important in the formation of channels.
The pore structure of the Final Stage model is consistent with results of inhibitor studies. The central region where the C-termini of the two S1 barrels meet contains 48 imidazole groups from H13 and H14 side chains positioned near the axis of the pore (Fig. 6A). This region is flanked on each side by rings of 12 carboxyl groups from E11 side chains. Thus, one would expect this region to bind numerous Zn2+ and/or Al3+ ions with high affinity, and that binding of these cations would inhibit permeation of Ca2+ ions through the pore. Steep dose-response curves have been observed for inhibition of Aβ-induced Ca2+ influx into neurons and Aβ-induced apoptosis by tetrahistidine peptides (N. Arispe, to be published elsewhere), which suggest cooperative processes that may involve multiple binding sites. Based on these findings, we have developed models in which up to four tetrahistidine peptides bind in the central region of the final stage model where their imidazole groups interact with side chains of E11, H13, and H14 (to be published elsewhere). These models are consistent with findings that histidine residues within proteins interact most frequently with negatively charged and other histidine residues 54.
Figure 6.
Central region of the pore formed by residues 13–21. Space-filled representations of side chains that line the pore are colored by element. (A) Side view showing only four subunits on each side of the pore. (B) Top view through the pore of all subunits. Note that V18 and F20 side chains are buried behind H14 side chains, and that H13 side chains are relatively exposed near the axis of the pore.
These models are also consistent with blockade of Aβ-induced currents and cell apoptosis by two small drug enantiomers: MRS2481 and MRS2485 22. These drugs have a positively charged nitrogen moiety connected to an phenyl ring by an alkyl chain and ester group. Although both of these drugs inhibit in the micromolar range, inhibition by MRS2481 is reversible whereas that by MRS2485 is irreversible. This difference suggests that they bind to a specific site22. Magnitudes of single channel current are typically reduced before they are completely blocked. This suggests that more than one drug may bind to the channel assembly, and that binding of only one reduces, but does not eliminate, the single channel conductance. We have developed models in which up to six MRS2485 molecules may bind in the central region of the Final Stage model (see Fig. 7B & C). Their positively charged region binds to carboxyl groups of E11 residues and their aromatic ring and oxygen atoms interact with imidazole rings of H13 and H14 residues.
Figure 7.
Model of the binding of MRS2485 in the central region of the pore. (A) Side view of portion or the pore formed by residues 11–21 with one MRS2485 molecule (space-filled colored by element) bound. Subunits nearest the viewer have been removed to reveal the lining of the central pore. (B) Same as (A) but with six bound MRS2485 drugs. These are related by 3-fold symmetry about the axis of the pore and 2-fold symmetry about the central plain. (C) Top view of the C-termini ends (residues 11–14) of S1 segments with three bound MRS2485 drugs.
We analyzed the stabilities of these models and refined their structures (both with and without blocking peptides and drugs) by performing MD simulations of the assemblies embedded in a lipid bilayer with water and ions on each side and within the pore. The all atom root-mean-squared-deviation (RMSD) of the entire structure averaged over the last 3 ns of a 20 ns simulation from the initial symmetrical model was only 0.33 nm (supplement Fig. S2A), and 1366 non-aqueous H-bonds were formed. These values are substantially better than for the Stage 5 model (0.45 nm RMSD and 1138 H-bonds) or for the models of the smooth annular protofibrils that have similar 36-stranded S3 barrels ((0.40 nm RMSD and 1296 H-bonds) (see accompanying manuscript). The RMSD values are exceptionally low considering that the assembly has 36 water-exposed N-termini that have been too disordered for their structures to be determined in all NMR studies of different types of Aβ assemblies. This stability is likely due to the near optimal physiochemical properties of the assembly (see discussion). Calculations of root-mean-squared-fluctuations (RMSF) about their mean position during the last 3 ns indicate that residues of the putative ion selectivity and drug binding regions are exceptionally static (residues 4–23 of Sub1 and Sub2 have average RMSF value of 0.06–0.07 nm) (supplement Fig. Sup 2D). Residues that surround this core (10–23 of Sub3, and residues 31–39 of all subunits) are only slightly more dynamic (RMSF values of 0.07–0.08 nm). The most dynamic residues those exposed to the aqueous phases (residues 1–3, 25–30, and 40–42 plus residues 4–9 of Sub3).
Other Models
In our long-term iterative approach (see discussion) it is useful to distinguish the following general features of our models from precise details that are less likely to be correct. 1) Hexamers may form in which S3 segments construct a six-stranded β-barrel. Although we currently prefer antiparallel structures because they are more consistent with some interpretations of experimental results and are slightly more stable, we cannot exclude parallel structures (see supplement). 2) Parallel S1 β-barrels may comprise some portion of the permeation pore and the E11-H14 segment may form multivalent cation, drug, and peptide binding sites; however, the number of strands in these barrels may vary and these motifs are compatible with numerous specific models of other segments (see Fig 3 and supplement). 3) TM six-stranded S3 β-barrels may merge to form more stable, much larger single transmembrane S3 β-barrels that surround other TM segments. Although we currently favor antiparallel 36-stranded S3 barrels, we cannot exclude the parallel β-barrels, which are almost a stable in our simulations (see supplement), nor can we be certain that such barrels always form from six hexamers and will always have 36 strands. 4) The TM region may be composed of concentric β-barrels with the outer barrel having more stands than the barrel it surrounds. We have found that when the number of strands differs by twelve and the barrels have a shear number equal to the number of strands, then the gap between barrels is ~ 1.0 nm, which is reasonable to accommodate side chains; e.g, in our final preferred model a 36 stranded S3 barrel with a shear number of 36 surrounds a 24-stranded S2 barrel with a shear number of 24. However, this concept is not limited to this model alone; e.g., we have made numerous models of channels and annular protofibrils (see supplement and accompanying manuscript) and we cannot exclude the possibility that a channel of this type will not have six-fold symmetry about the axis of the pore.
In our search for the ‘best’ models, we developed models not presented in the text in which 1) S2 and S3 segments are helical, 2) S2 and S3 segments form β-hairpins, 3) S3 segments form parallel β barrels, 4) S1 segments form antiparallel beta barrels that line the pores, either as extended strands or as β-hairpins, 5) S2 segments form β-barrels that line the pore, and 6) S3 strands are the only transmembrane segments, and lipid head groups line the pore. The supplement describes some of the more plausible alternatives, and explains why we currently consider them to be inferior. In general these alternative models were less stable than our favored models and/or were less consistent with the microscopy and other data.
Discussion and Conclusions
Here we have proposed six-stranded β-barrel models for the hydrophobic portions of Aβ proteins that interact with membranes. We have used the same motif to develop models of soluble Aβ (accompanying manuscript) and Prion Protein (Guy & Durell, Biophysical Journal, 2007, 2667-PosB32a) assemblies. If these β-barrel models are correct, they could be key to understanding the molecular basis of Alzheimer’s and prion-related spongiform encephalopathies. We have also developed models of larger assemblies formed from these hexamers. Our proposed multi-step membrane insertion process for Aβ oligomers involves formation of several types of pores. Although the description of the insertion pathway is overly simplified, it was included for several reasons: 1) to explain the AFM results with peaks surrounding a central region (the dimensions in the AFM images are more consistent with the surface hexamer-of-hexamers than with the transmembrane models), 2) to explain why some groups have found that Aβ assemblies form antiparallel beta sheets on the surface of the membrane with the strands oriented parallel to the membrane’s surface50, 3) to explain how an antiparallel structure can span the bilayer since polar residues must cross the membrane to form the antiparallel structure, 4) to explain the sequence and variety of conductances observed in bilayer experiments in which highly noise current records are frequently followed by more stable, higher conductance channels, 5) to explain why some of the channels appear to be assymetric relative to channel blockers3, and 6) because the proposed insertion process was an integral part of our rationale for developing the channel models. In the last stage of the channel-forming process, the 6-stranded S3 β-barrels split apart and then merge to form a 36-stranded S3 β-barrel. We proposed a similar process for the transition from ‘beaded’ to ‘smooth’ annular protofibrils in the accompanying manuscript. We and others30 have developed a variety of alternative models; however, the models presented here are those more consistent with our modeling criteria and experimental results. The Final Stage models are physiochemically sound; i.e., the outer hydrophobic wall corresponds closely to the thickness of the membrane’s alkyl phase, positively charged residues interact with negatively charged lipid head group moieties, virtually all polar atoms located in apolar environments (transmembrane) form hydrogen bonds, almost all charged groups form salt bridges or bind to counterions in the pore, almost all hydrophobic groups are in hydrophobic environments (exposed to lipid alkyl chains or buried within the protein), the β-barrels have geometries and shear numbers commonly observed for transmembane β-barrels, and the structure is exceptionally stable during MD simulations. The stabilities of our models contrasts with the instabilities of Aβ channel models of Jang et al. 30. We thus reject their conclusion that hydrogen-bonded β-strands tend to break in fluidic bilayers, and refer to the stabilities of porins and other transmembrane β-barrel channel proteins of known structure such as α-hemolysin as evidence that this is not the case for correct structures. Our models are also consistent with numerous experimental results: Their dimensions correspond closely to both atomic force and freeze fracture microscopy images. The proposed insertion process explains observed transitions from very noisy short-lived channels to very stable long-lived channels. The modeled pore has a net negative charge, consistent with the cation selectivity and Ca2+ permeability of the pore. It also can explain how Zn2+ 33 and Al3+ 5, histidine-containing peptides 3, 21, and the MRS2481 and MRS2485 drugs 22 inhibit Aβ-induced channels. The most compelling evidence that Ca2+ permeation through Aβ channels is responsible for Aβ’s toxicity is the observation that all of these putative channel-blocking agents also inhibit Aβ-induced apoptosis 3, 22. The histidine-containing peptides have also been shown to inhibit Aβ-induced entry of Ca2+ into neurons; whereas blockers of voltage-gated Ca2+ channels do not (Arispe, et al, Biophysical Journal, Volume 98, Issue 3, Supplement 1, January 2010, Page 293a).
The most thorough analyses of the relationships among Aβ40 concentration in membranes, its secondary structure, its insertion into membranes, affects of lipid compensation, the size of oligomer, and Aβ-induced permeation have been performed in the Gafni laboratory60, 72. These studies were published after development of the models presented here, so it is important to relate our models to these results. This group finds that Aβ40 binds to all types of membranes studied, but inserts (as measured by tryptophan fluorescence in a Y10W mutant) only in membranes with a substantial component of negatively charged head groups72. Interactions between these head groups and the K28 side chain are the most likely explanation for this dependency in our models. At low peptide to lipid (P:L) ratios most Aβ40 peptides are monomers distributed evenly over the membrane surface, have substantial α-helical secondary structure, and do not cause permeation72. As P:L increases to moderate levels, large oligomers (tetramers to octamers, but primarily hexamers) form, the secondary structure shifts to predominantly β, and small permeability increases occur, but only if the membranes have a liquid crystal alkyl phase72. This finding that increases in permeability coincide with a transition from helical to β structure appears to be inconsistent with our previous predominately helical hexameric models of channels24 (see supplement), but would be consistent with our hexameric β-barrel models if the hexamers can induce low conductance channels. Binding and insertion are not sufficient to cause permeation; e.g., these processes are not dependent upon the nature of the alkyl phase, are not affected by cholesterol, and occur quickly; whereas permeation requires a relatively dynamic liquid crystal phase, is inhibited by cholesterol, and increases relatively slowly72. Our explanation for these findings is that their binding and insertion mechanisms correspond to formations of the surface hexamers of Fig. 2; whereas, permeation requires penetration of the bilayer by a portion of the peptide, which occurs slowly and only if the alkyl phase is relatively fluid. At still higher P:L they observe formation of much larger assemblies (N>20), and dramatic increases in permeation due to formation of large pores, but only if the membranes have a liquid crystal alkyl phase60. Unlike the smaller hexameric oligomers, these larger assemblies change slowly with time. These larger assemblies may correspond to assemblies similar to the hexamer-of-hexamer models of Fig. 3, and the slow changes may involve types of transmembrane translocations similar to those of Fig. 3. Thus, differing experimental conditions may produce different results because they favor and/or inhibit different stages of the relatively complex membrane insertion process proposed here. This may explain why various groups have obtained so many different results in studying effects of Aβ peptides on membranes. For example, numerous groups have observed formation of relatively stable channels with multiple conductances that are often accompanied or preceded by more noisy currents 2, 4, 5, 31, 32, 34, 36, 43, 65, some have observed only noisy currents that were interpreted to be indicative of membrane perturbations 19, some have observed no channels and little perturbations but have observed changes in membrane conductance and capacitance 66, and others have observed membrane disruptions in which the membranes break 39, 51. Which effects are observed appears to depend upon numerous factors such as the lipid composition of the membrane, the state of the Aβ assemblies prior to incorporation into the membranes, the methods used to place them in the membrane, the concentration of the peptides, which buffer is present in the solution, and whether all solvent used to prepare the peptides has been removed 11. For example, negatively charged head groups are required to produce stable cation-selective channels 2, 4, 5, 72, zwitterion lipids (phosphatidylcholine) favor β-structures on the membrane surfaces and relatively unstable currents 19 or no permeation72, oxidized cholesterol is required to produce anionic selective channels 52, Tris buffer used in some studies blocks Aβ channels 4, 5, and the presence of hexafluoroisopropanol solvent, often used to prepare Aβ peptides, produces capacitance changes and inhibits channel formation 11. The freeze fracture studies presented here clearly indicate that under some conditions, Aβ assemblies can penetrate the lipid bilayer. These transmembrane assemblies likely form rather classic types of channels that may have numerous conductance states, dependent upon the number of peptides or oligomers in the assembly, the structure of the portion of the assembly that forms the pore, and whether the assemblies substantially perturb the structure of the lipids with which they interact.
Our models raise numerous issues that should be analyzed experimentally. For example, higher resolution microscopy studies that also include side views of the membrane assemblies should help determine whether some assemblies are asymmetric, with most of the protein on the cis side of the bilayer (Fig. 3A–C), or span the bilayer symmetrically (Fig. 3D–E); or whether the variation of sizes of Aβ assemblies observed in the freeze fracture studies is due to variability in the number of Aβ peptides that comprise the different assemblies, or due to the stage of membrane insertion, as depicted in Fig. 3. Effects on ion selectivity of mutations that change the charges of S1 and/or S2 segments should help determine which, if either, of these segments lines the pores; e.g. in our models D7 and E11 are important in determining the Ca2+ selectivity of the channel, whereas in some alternative models 30 E22 is crucial. The models also predict that E11, H13, and H14 are involved in binding of putative channel blocking agents, and thus mutating any of these residues to glutamine should have substantial effects on the binding of these agents. Antibodies could be used to determine whether S1 and/or S2 segments cross the membrane, as predicted in the antiparallel models of Fig. 3D–F. Experiments similar to those describe above from the Gafni lab 72 should be repeated with Aβ42 to determine whether the results are affected by the peptide length. The models could also be useful in developing strategies to produce homogeneous populations of membrane-bound Aβ assemblies required for more detailed solid-state NMR studies.
A major goal of our research is to assist in the developments of improved methods to prevent and/or treat Alzheimer’s disease. Our proposal for how some small drug molecules may inhibit neurotoxicity by blocking Aβ channels 22 is an initial step in this direction. This suggests that the models could be useful in structure-based drug design if Aβ channels are an integral component of the disease. However, potential therapeutics may not to be limited to this site or mechanism. For example, if the toxic moieties are composed of, or derived from, Aβ hexamers, then any mechanism that inhibited formation of these hexamers might prove effective.
Supplementary Material
Acknowledgments
We thank Dr. G. Zampighi (University of California at Los Angeles School of Medicine, Los Angeles, California, USA) for performing the freeze fracture microscopy of the Aβ assemblies, Adina Milac and Sijung Yun for comments, and Sijung Yun for making the Movie in the supplement. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (NIH), Bethesda, MD. This work was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Abbreviations
- Aβ
amyloid beta
- AD
Alzheimer’s disease
- MD
molecular dynamics
- TM
transmembrane
Contributor Information
Yinon Shafrir, Email: yinon.shafrir@nih.gov.
Stewart Durell, Email: stewart.durell@nih.gov.
Nelson Arispe, Email: narispe@usuhs.mil.
Reference List
- 1.Abedini A, Raleigh DP. A role for helical intermediates in amyloid formation by natively unfolded polypeptides? Physical Biology. 2009:6. doi: 10.1088/1478-3975/6/1/015005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alarcon JM, Brito JA, Hermosilla T, Atwater I, Mears D, Rojas E. Ion channel formation by Alzheimer’s disease amyloid beta-peptide (Abeta40) in unilamellar liposomes is determined by anionic phospholipids. Peptides. 2006;27:95–104. doi: 10.1016/j.peptides.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Arispe N, Diaz JC, Simakova O. Abeta ion channels. Prospects for treating Alzheimer’s disease with Abeta channel blockers. Biochim Biophys Acta. 2007;1768:1952–1965. doi: 10.1016/j.bbamem.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 4.Arispe N, Pollard HB, Rojas E. Giant multilevel cation channels formed by Alzheimer disease amyloid beta-protein [A beta P-(1–40)] in bilayer membranes. Proc Natl Acad Sci U S A. 1993;90:10573–10577. doi: 10.1073/pnas.90.22.10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arispe N, Rojas E, Pollard HB. Alzheimer disease amyloid beta protein forms calcium channels in bilayer membranes: blockade by tromethamine and aluminum. Proc Natl Acad Sci U S A. 1993;90:567–571. doi: 10.1073/pnas.90.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bass RB, Strop P, Barclay M, Rees DC. Crystal structure of Escherichia coli MscS, a voltage-modulated and mechanosensitive channel. Science. 2002;298:1582–1587. doi: 10.1126/science.1077945. [DOI] [PubMed] [Google Scholar]
- 7.Berger O, Edholm O, Jahnig F. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys J. 1997;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bitan G, Kirkitadze MD, Lomakin A, Vollers SS, Benedek GB, Teplow DB. Amyloid beta-protein (Abeta) assembly: Abeta 40 and Abeta 42 oligomerize through distinct pathways. Proc Natl Acad Sci U S A. 2003;100:330–335. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bokvist M, Lindstrom F, Watts A, Grobner G. Two types of Alzheimer’s beta-amyloid (1–40) peptide membrane interactions: aggregation preventing transmembrane anchoring versus accelerated surface fibril formation. J Mol Biol. 2004;335:1039–1049. doi: 10.1016/j.jmb.2003.11.046. [DOI] [PubMed] [Google Scholar]
- 10.Brooks BR. In: CHARMM: a program for macromolecular energy minimization and dynamic calculations. Bruccoleri REOBDSDJSSaKM., editor. 1983. p. 217. [Google Scholar]
- 11.Capone R, Quiroz FG, Prangkio P, Saluja I, Sauer AM, Bautista MR, Turner RS, Yang J, Mayer M. Amyloid-beta-induced ion flux in artificial lipid bilayers and neuronal cells: resolving a controversy. Neurotox Res. 2009;16:1–13. doi: 10.1007/s12640-009-9033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang G, Spencer RH, Lee AT, Barclay MT, Rees DC. Structure of the MscL homolog from Mycobacterium tuberculosis: a gated mechanosensitive ion channel. Science. 1998;282:2220–2226. doi: 10.1126/science.282.5397.2220. [DOI] [PubMed] [Google Scholar]
- 13.Chi EY, Ege C, Winans A, Majewski J, Wu G, Kjaer K, Lee KY. Lipid membrane templates the ordering and induces the fibrillogenesis of Alzheimer’s disease amyloid-beta peptide. Proteins. 2008;72:1–24. doi: 10.1002/prot.21887. [DOI] [PubMed] [Google Scholar]
- 14.Chimon S, Ishii Y. Capturing intermediate structures of Alzheimer’s beta-amyloid, Abeta(1–40), by solid-state NMR spectroscopy. J Am Chem Soc. 2005;127:13472–13473. doi: 10.1021/ja054039l. [DOI] [PubMed] [Google Scholar]
- 15.Coles M, Bicknell W, Watson AA, Fairlie DP, Craik DJ. Solution structure of amyloid beta-peptide(1–40) in a water-micelle environment. Is the membrane-spanning domain where we think it is? Biochemistry. 1998;37:11064–11077. doi: 10.1021/bi972979f. [DOI] [PubMed] [Google Scholar]
- 16.Crescenzi O, Tomaselli S, Guerrini R, Salvadori S, D’Ursi AM, Temussi PA, Picone D. Solution structure of the Alzheimer amyloid beta-peptide (1–42) in an apolar microenvironment. Similarity with a virus fusion domain. Eur J Biochem. 2002;269:5642–5648. doi: 10.1046/j.1432-1033.2002.03271.x. [DOI] [PubMed] [Google Scholar]
- 17.Curtain CC, Ali F, Volitakis I, Cherny RA, Norton RS, Beyreuther K, Barrow CJ, Masters CL, Bush AI, Barnham KJ. Alzheimer’s disease amyloid-beta binds copper and zinc to generate an allosterically ordered membrane-penetrating structure containing superoxide dismutase-like subunits. J Biol Chem. 2001;276:20466–20473. doi: 10.1074/jbc.M100175200. [DOI] [PubMed] [Google Scholar]
- 18.Dahlgren KN, Manelli AM, Stine WB, Jr, Baker LK, Krafft GA, LaDu MJ. Oligomeric and fibrillar species of amyloid-beta peptides differentially affect neuronal viability. J Biol Chem. 2002;277:32046–32053. doi: 10.1074/jbc.M201750200. [DOI] [PubMed] [Google Scholar]
- 19.de Planque MR, Raussens V, Contera SA, Rijkers DT, Liskamp RM, Ruysschaert JM, Ryan JF, Separovic F, Watts A. beta-Sheet Structured beta-Amyloid(1–40) Perturbs Phosphatidylcholine Model Membranes. J Mol Biol. 2007 doi: 10.1016/j.jmb.2007.02.063. [DOI] [PubMed] [Google Scholar]
- 20.Deshpande A, Mina E, Glabe C, Busciglio J. Different conformations of amyloid beta induce neurotoxicity by distinct mechanisms in human cortical neurons. J Neurosci. 2006;26:6011–6018. doi: 10.1523/JNEUROSCI.1189-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diaz JC, Linnehan J, Pollard H, Arispe N. Histidines 13 and 14 in the Abeta sequence are targets for inhibition of Alzheimer’s disease Abeta ion channel and cytotoxicity. Biol Res. 2006;39:447–460. doi: 10.4067/s0716-97602006000300007. [DOI] [PubMed] [Google Scholar]
- 22.Diaz JC, Simakova O, Jacobson KA, Arispe N, Pollard HB. Small molecule blockers of the Alzheimer Abeta calcium channel potently protect neurons from Abeta cytotoxicity. Proc Natl Acad Sci U S A. 2009;106:3348–3353. doi: 10.1073/pnas.0813355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyle DA, Morais CJ, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 24.Durell SR, Guy HR, Arispe N, Rojas E, Pollard HB. Theoretical models of the ion channel structure of amyloid beta-protein. Biophys J. 1994;67:2137–2145. doi: 10.1016/S0006-3495(94)80717-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esler WP, Wolfe MS. A portrait of Alzheimer secretases--new features and familiar faces. Science. 2001;293:1449–1454. doi: 10.1126/science.1064638. [DOI] [PubMed] [Google Scholar]
- 26.Gonzales EB, Kawate T, Gouaux E. Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature. 2009;460:599–604. doi: 10.1038/nature08218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guy HR, Seetharamulu P. Molecular model of the action potential sodium channel. Proc Natl Acad Sci U S A. 1986;83:508–512. doi: 10.1073/pnas.83.2.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirakura Y, Carreras I, Sipe JD, Kagan BL. Channel formation by serum amyloid A: a potential mechanism for amyloid pathogenesis and host defense. Amyloid. 2002;9:13–23. doi: 10.3109/13506120209072440. [DOI] [PubMed] [Google Scholar]
- 29.Hirakura Y, Lin MC, Kagan BL. Alzheimer amyloid abeta1–42 channels: effects of solvent, pH, and Congo Red. J Neurosci Res. 1999;57:458–466. [PubMed] [Google Scholar]
- 30.Jang H, Arce FT, Capone R, Ramachandran S, Lal R, Nussinov R. Misfolded Amyloid Ion Channels Present Mobile beta-Sheet Subunits in Contrast to Conventional Ion Channels. Biophys J. 2009;97:3029–3037. doi: 10.1016/j.bpj.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kagan BL, Azimov R, Azimova R. Amyloid peptide channels. J Membr Biol. 2004;202:1–10. doi: 10.1007/s00232-004-0709-4. [DOI] [PubMed] [Google Scholar]
- 32.Kagan BL, Hirakura Y, Azimov R, Azimova R, Lin MC. The channel hypothesis of Alzheimer’s disease: current status. Peptides. 2002;23:1311–1315. doi: 10.1016/s0196-9781(02)00067-0. [DOI] [PubMed] [Google Scholar]
- 33.Kawahara M, Arispe N, Kuroda Y, Rojas E. Alzheimer’s disease amyloid beta-protein forms Zn(2+)-sensitive, cation-selective channels across excised membrane patches from hypothalamic neurons. Biophys J. 1997;73:67–75. doi: 10.1016/S0006-3495(97)78048-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawahara M, Kuroda Y. Molecular mechanism of neurodegeneration induced by Alzheimer’s beta-amyloid protein: channel formation and disruption of calcium homeostasis. Brain Res Bull. 2000;53:389–397. doi: 10.1016/s0361-9230(00)00370-1. [DOI] [PubMed] [Google Scholar]
- 35.Kawate T, Michel JC, Birdsong WT, Gouaux E. Crystal structure of the ATP-gated P2X(4) ion channel in the closed state. Nature. 2009;460:592–598. doi: 10.1038/nature08198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kourie JI, Henry CL, Farrelly P. Diversity of amyloid beta protein fragment [1–40]-formed channels. Cell Mol Neurobiol. 2001;21:255–284. doi: 10.1023/A:1010995121153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LaFerla FM, Green KN, Oddo S. Intracellular amyloid-beta in Alzheimer’s disease. Nat Rev Neurosci. 2007;8:499–509. doi: 10.1038/nrn2168. [DOI] [PubMed] [Google Scholar]
- 38.Lanzavecchia S, Cantele F, Bellon PL, Zampighi L, Kreman M, Wright E, Zampighi GA. Conical tomography of freeze-fracture replicas: a method for the study of integral membrane proteins inserted in phospholipid bilayers. J Struct Biol. 2005;149:87–98. doi: 10.1016/j.jsb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 39.Lau TL, Ambroggio EE, Tew DJ, Cappai R, Masters CL, Fidelio GD, Barnham KJ, Separovic F. Amyloid-beta peptide disruption of lipid membranes and the effect of metal ions. J Mol Biol. 2006;356:759–770. doi: 10.1016/j.jmb.2005.11.091. [DOI] [PubMed] [Google Scholar]
- 40.Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. A specific amyloid-beta protein assembly in the brain impairs memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 41.Lim KH, Nguyen TN, Damo SM, Mazur T, Ball HL, Prusiner SB, Pines A, Wemmer DE. Solid-state NMR structural studies of the fibril form of a mutant mouse prion peptide PrP89–143(P101L) Solid State Nucl Magn Reson. 2006;29:183–190. doi: 10.1016/j.ssnmr.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 42.Lin H, Bhatia R, Lal R. Amyloid beta protein forms ion channels: implications for Alzheimer’s disease pathophysiology. FASEB J. 2001;15:2433–2444. doi: 10.1096/fj.01-0377com. [DOI] [PubMed] [Google Scholar]
- 43.Lin MC, Kagan BL. Electrophysiologic properties of channels induced by Abeta25–35 in planar lipid bilayers. Peptides. 2002;23:1215–1228. doi: 10.1016/s0196-9781(02)00057-8. [DOI] [PubMed] [Google Scholar]
- 44.Liu WM. Shear numbers of protein beta-barrels: Definition refinements and statistics. Journal of Molecular Biology. 1998;275:541–545. doi: 10.1006/jmbi.1997.1501. [DOI] [PubMed] [Google Scholar]
- 45.Luhrs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Dobeli H, Schubert D, Riek R. 3D structure of Alzheimer’s amyloid-beta(1–42) fibrils. Proc Natl Acad Sci U S A. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mandal PK, Pettegrew JW. Alzheimer’s disease: soluble oligomeric Abeta(1–40) peptide in membrane mimic environment from solution NMR and circular dichroism studies. Neurochem Res. 2004;29:2267–2272. doi: 10.1007/s11064-004-7035-1. [DOI] [PubMed] [Google Scholar]
- 47.Masuda Y, Uemura S, Ohashi R, Nakanishi A, Takegoshi K, Shimizu T, Shirasawa T, Irie K. Identification of Physiological and Toxic Conformations in A beta 42 Aggregates. Chembiochem. 2009;10:287–295. doi: 10.1002/cbic.200800411. [DOI] [PubMed] [Google Scholar]
- 48.Matsuzaki K. Physicochemical interactions of amyloid beta-peptide with lipid bilayers. Biochim Biophys Acta. 2007 doi: 10.1016/j.bbamem.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 49.Matsuzaki K. Physicochemical interactions of amyloid beta-peptide with lipid bilayers. Biochim Biophys Acta. 2007 doi: 10.1016/j.bbamem.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 50.Matsuzaki K, Horikiri C. Interactions of amyloid beta-peptide (1–40) with ganglioside-containing membranes. Biochemistry. 1999;38:4137–4142. doi: 10.1021/bi982345o. [DOI] [PubMed] [Google Scholar]
- 51.McLaurin J, Chakrabartty A. Membrane disruption by Alzheimer beta-amyloid peptides mediated through specific binding to either phospholipids or gangliosides. Implications for neurotoxicity. J Biol Chem. 1996;271:26482–26489. doi: 10.1074/jbc.271.43.26482. [DOI] [PubMed] [Google Scholar]
- 52.Micelli S, Meleleo D, Picciarelli V, Gallucci E. Effect of sterols on beta-amyloid peptide (AbetaP 1–40) channel formation and their properties in planar lipid membranes. Biophys J. 2004;86:2231–2237. doi: 10.1016/S0006-3495(04)74281-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Minicozzi V, Stellato F, Comai M, Serra MD, Potrich C, Meyer-Klaucke W, Morante S. Identifying the minimal copper- and zinc-binding site sequence in amyloid-beta peptides. J Biol Chem. 2008;283:10784–10792. doi: 10.1074/jbc.M707109200. [DOI] [PubMed] [Google Scholar]
- 54.Miyazawa S, Jernigan RL. Residue-residue potentials with a favorable contact pair term and an unfavorable high packing density term, for simulation and threading. J Mol Biol. 1996;256:623–644. doi: 10.1006/jmbi.1996.0114. [DOI] [PubMed] [Google Scholar]
- 55.Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer’s beta-amyloid fibrils. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s beta-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 58.Quist A, Doudevski I, Lin H, Azimova R, Ng D, Frangione B, Kagan B, Ghiso J, Lal R. Amyloid ion channels: a common structural link for protein-misfolding disease. Proc Natl Acad Sci U S A. 2005;102:10427–10432. doi: 10.1073/pnas.0502066102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sato T, Kienlen-Campard P, Ahmed M, Liu W, Li H, Elliott JI, Aimoto S, Constantinescu SN, Octave JN, Smith SO. Inhibitors of amyloid toxicity based on beta-sheet packing of Abeta40 and Abeta42. Biochemistry. 2006;45:5503–5516. doi: 10.1021/bi052485f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schauerte JA, Wong PT, Wisser KC, Ding H, Steel DG, Gafni A. Simultaneous single-molecule fluorescence and conductivity studies reveal distinct classes of Abeta species on lipid bilayers. Biochemistry. 2010;49:3031–3039. doi: 10.1021/bi901444w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Selkoe DJ. Translating cell biology into therapeutic advances in Alzheimer’s disease. Nature. 1999;399:A23–A31. doi: 10.1038/399a023. [DOI] [PubMed] [Google Scholar]
- 62.Shafrir Y, Durell SR, Guy HR. Models of the structure and gating mechanisms of the pore domain of the NaChBac ion channel. Biophys J. 2008;95:3650–3662. doi: 10.1529/biophysj.108.135327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shafrir Y, Durell SR, Guy HR. Models of voltage-dependent conformational changes in NaChBac channels. Biophys J. 2008;95:3663–3676. doi: 10.1529/biophysj.108.135335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shao H, Jao S, Ma K, Zagorski MG. Solution structures of micelle-bound amyloid beta-(1–40) and beta-(1–42) peptides of Alzheimer’s disease. J Mol Biol. 1999;285:755–773. doi: 10.1006/jmbi.1998.2348. [DOI] [PubMed] [Google Scholar]
- 65.Singer SJ, Dewji NN. Evidence that Perutz’s double-beta-stranded subunit structure for beta-amyloids also applies to their channel-forming structures in membranes. Proc Natl Acad Sci U S A. 2006;103:1546–1550. doi: 10.1073/pnas.0509892103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sokolov Y, Kozak JA, Kayed R, Chanturiya A, Glabe C, Hall JE. Soluble amyloid oligomers increase bilayer conductance by altering dielectric structure. J Gen Physiol. 2006;128:637–647. doi: 10.1085/jgp.200609533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 68.Takuma K, Fang F, Zhang W, Yan S, Fukuzaki E, Du H, Sosunov A, McKhann G, Funatsu Y, Nakamichi N, Nagai T, Mizoguchi H, Ibi D, Hori O, Ogawa S, Stern DM, Yamada K, Yan SS. RAGE-mediated signaling contributes to intraneuronal transport of amyloid-{beta} and neuronal dysfunction. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0905686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tashima Y, Oe R, Lee S, Sugihara G, Chambers EJ, Takahashi M, Yamada T. The effect of cholesterol and monosialoganglioside (GM1) on the release and aggregation of amyloid beta-peptide from liposomes prepared from brain membrane-like lipids. J Biol Chem. 2004;279:17587–17595. doi: 10.1074/jbc.M308622200. [DOI] [PubMed] [Google Scholar]
- 70.Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 71.White SH. A study of lipid bilayer membrane stability using precise measurements of specific capacitance. Biophys J. 1970;10:1127–1148. doi: 10.1016/S0006-3495(70)86360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wong PT, Schauerte JA, Wisser KC, Ding H, Lee EL, Steel DG, Gafni A. Amyloid-beta membrane binding and permeabilization are distinct processes influenced separately by membrane charge and fluidity. J Mol Biol. 2009;386:81–96. doi: 10.1016/j.jmb.2008.11.060. [DOI] [PubMed] [Google Scholar]
- 73.Younkin SG. Evidence that A beta 42 is the real culprit in Alzheimer’s disease. Ann Neurol. 1995;37:287–288. doi: 10.1002/ana.410370303. [DOI] [PubMed] [Google Scholar]
- 74.Zampighi GA, Hall JE, Ehring GR, Simon SA. The structural organization and protein composition of lens fiber junctions. J Cell Biol. 1989;108:2255–2275. doi: 10.1083/jcb.108.6.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zampighi GA, Kreman M, Lanzavecchia S, Turk E, Eskandari S, Zampighi L, Wright EM. Structure of functional single AQP0 channels in phospholipid membranes. J Mol Biol. 2003;325:201–210. doi: 10.1016/s0022-2836(02)01200-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.