Abstract
Prion diseases are fatal neurodegenerative disorders that involve the conversion of the normal cellular form of the prion protein (PrPC) to a misfolded pathogenic form (PrPSc). There are many genetic mutations of PrP associated with human prion diseases. Three of these point mutations are located at the first strand of the native β-sheet in human PrP: G131V, S132I and A133V. To understand the underlying structural and dynamic effects of these disease-causing mutations on the human protein, we performed molecular dynamics of wild-type and mutated human PrP. The results indicate that the mutations induced different effects but they were all related to misfolding of the native β-sheet: G131V caused the elongation of the native β-sheet, A133V disrupted the native β-sheet, and S132I converted the native β-sheet to an α-sheet. The observed changes were due to the reorientation of side chain-side chain interactions upon introducing the mutations. In addition, all mutations impaired a structurally conserved water site at the native β-sheet. Our work suggests various misfolding pathways for human PrP in response to mutation.
Keywords: prion disease, prion protein, genetic mutations, structural misfolding, molecular dynamics
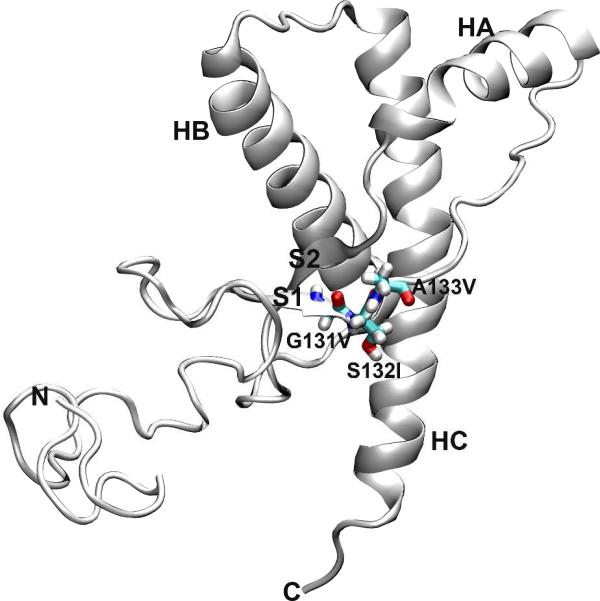
Prion diseases, or transmissible spongiform encephalopathies (TSEs)1, are fatal neurodegenerative disorders, including Creutzfeldt-Jakob disease (CJD), Gerstmann-Straüssler-Scheinker syndrom (GSS), fatal familial insomnia (FFI), and kuru in humans, scrapie in sheep, bovine spongiform encephalopathy (BSE) in cattle, and chronic wasting disease (CWD) in cervids (1, 2). Prion diseases are attributed to the conformational conversion of the normal cellular prion protein (PrPC) to its misfolded ‘scrapie’ form, PrPSc (2, 3). PrPC is a glycoprotein anchored to the cell membrane via a glycophosphatidylinositol (GPI) moiety (2, 4). PrPC contains a flexible and largely unstructured N-terminal region and a C-terminal globular domain (Figure 1). The C-terminal domain consists of three α-helices (HA, HB, and HC) and a short two-stranded antiparallel β-sheet (S1 and S2). While PrPC is mainly α-helical and is sensitive to protease digestion, PrPSc is enriched in β-sheet structure and is proteinase K-resistant (5, 6). The precise structure of PrPSc, however, is still unknown, and in fact it appears to be quite heterogeneous.
Figure 1.
Structure of wt human PrPC. The starting structure used for MD simulations, residues 90-230. Secondary structure elements in the C-terminal globular domain are labeled. The three residues involved in the point mutations studied in this work are shown as sticks.
Human prion diseases can be infectious, sporadic, or genetic in origin (1, 2). Many single nucleotide mutations in the human prion protein gene (PRNP) have been linked with human prion diseases: familial CJD, GSS, and FFI (7, 8). Three mutations that are related to GSS, G131V (9), S132I (10), and A133V (11), are located at the first strand of the native β-sheet, S1 (Figure 1). The pathological mechanism as to how these mutations cause GSS is unknown. To reveal the underlying structural ramifications upon mutation, one crucial step is to study how these mutations affect and convert PrPC to PrPSc.
To this end, we have performed molecular dynamics (MD) simulations of human PrPC structures containing residues 90-230 with the three mutations mentioned above at neutral pH and physiological temperature (310 K). Our simulations suggest that these mutations induced different conformational changes in human PrPC, although they are all located at S1. With G131V, two additional hydrogen bonds formed between the main chains of strands S1 and S2, resulting in the extension of the native β-sheet. In contrast, A133V caused the loss of hydrogen bonds and disrupted the native β-sheet. By introducing S132V, hydrogen bonds of the native β-sheet were reordered. As a result, the native β-sheet was converted to an α-sheet (12). Reorganization of side chain-side chain interactions at the native β-sheet and the neighboring part of the N-terminal region was responsible for the observed conformational changes. Furthermore, the three mutations impaired a structurally conserved water site connecting S1, S2, and HC. In particular, the G131V mutation fully abolished the water site. These results suggest that there are various pathways for the early misfolding of PrPC due to mutations.
METHODS
Structure Preparation
The starting structure of wild-type (wt) human PrPC for MD simulations was modeled based on a NMR structure of human PrPC (PDB code 1QLX) (13), which contains residues 125-228. Because the major proteinase K-resistant fragment of PrPSc (i.e., PrP 27-30) consists of residues ~90-230 (14), we modeled the N-terminal residues 90-127 and the C-terminal residues 229-230 as described previously (15). Consequently, the starting structure for the MD simulations of wt human PrPC contained residues 90-230. As before, His140 was protonated on Nδ1 and other histidines were protonated on Nε2 (15).
To model the starting structures of mutants G131V, S132I, and A133V, for which experimental structures don't exist, we mutated the corresponding residues in silico. First, the side chain atoms of the mutated residue, except for Cβ, were deleted. Then, different rotamers of the target residue type were obtained from the Dynameomics rotamer library (16, 17) and added to generate multiple structures; after 100 steps of steepest descent minimization on all side chains, the structure with the smallest interaction energy between the side chain of the mutated residue and the surrounding residues were selected as the starting structure for MD simulations.
MD simulations
Explicit solvent all-atom MD simulations were performed with in lucem molecular mechanics (ilmm) (18). The Levitt et al. force field (19) and the F3C model (20) were used for protein and water, respectively. Electrostatic interactions between atoms separated by 3 bonds or less were scaled by 0.4. The force-shifted non-bonded cutoff (21) was set to 10 Å and the non-bonded list was updated every 2 steps. A time step of 2 fs was used.
The starting structures were first energy-minimized for 1000 steps with the steepest descent method in vacuo. Then, they were solvated in pre-equilibrated water boxes at 310 K. The final water density of the system was adjusted to the experimentally-measured density of 0.993 g/mL (22). The minimal distance between the protein and the walls of the box was 10 Å. To preclude bad interactions during solvation, water molecule positions were adjusted by 1000 steps of steepest descent minimization, 500 steps of MD, and another 500 steps of minimization. After that, 500 steps of steepest descent minimization were performed for the proteins. The production MD simulation of the whole system were then performed in the microcanonical (NVE, constant number of particles, volume, and energy) ensemble for 50 ns. System coordinates were saved every 1 ps. Five and three independent MD simulations were performed for wt PrPC and each mutant, respectively, using different random number seeds to generate independent trajectories, for a total of 700 ns, or 0.7 μs.
Analysis
Analysis of MD trajectories was performed with ilmm (18) and VMD (23). Root-mean-square deviations (RMSDs) of the Cα atoms of the C-terminal globular domain (residues 128-228) and HA (residues 144-156) were calculated by aligning structures on the Cα atoms of the globular domain and a stable core region (residues 174-186 and 200-219) (15, 24), respectively. A hydrogen bond was defined if the hydrogen-acceptor distance was ≤ 2.6 Å and the donor-hydrogen-acceptor angle was > 135°. Hydrophobic contacts were identified when the C-C distance between CHn groups was ≤ 5.4 Å. The DSSP algorithm (25) with additional definitions (26) was used to assign secondary structures. Molecular graphics were generated using VMD (23).
RESULTS
HA Shifts Position in Both Wt and Mutant Simulations
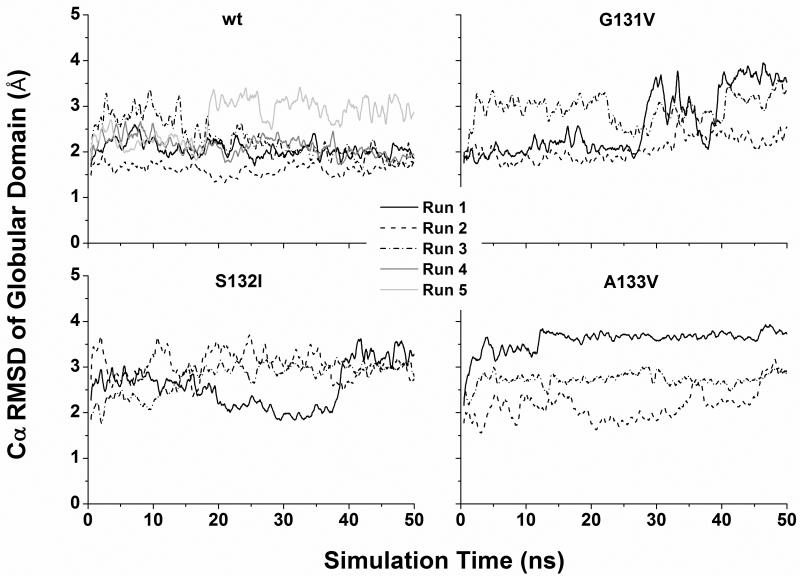
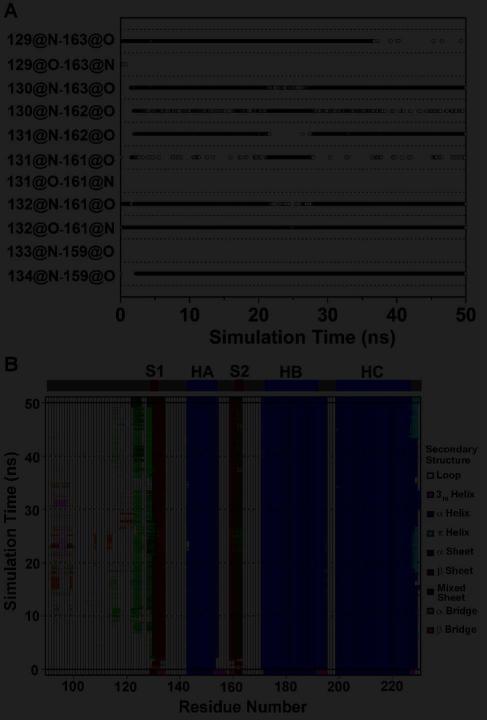
The three disease-related mutations (G131V, S132I, and A133V) are located at the S1 strand (Figure 1). Three independent simulations at neutral pH and 310 K were performed for each mutant. For comparison, five simulations at neutral pH and 310 K were performed for the wt PrP structure. In some simulations, the RMSD of all Cα atoms of the globular domain (residues 128-228) relative to the starting structure was ~2 Å through the simulations (Figure 2), indicating that the globular domain was stable. However, in other cases the Cα RMSD of the globular domain jumped to 3-4 Å, manifesting conformational changes. This is attributed to the movements of HA as indicated by the similar behavior of the RMSD of HA alone when structures were aligned on the stable core region (residues 174-186 & 200-219) (24) (Figure S1). Different types of HA movements were observed (Figure S2). HA moved away from the HB/HC plane (e.g., G131V run 1), moved downward (e.g., wt run 5), or moved toward the center (e.g., A133V run 1). By measuring the moving vectors of the HA N-terminus (residues 144-147) relative to its starting position as described previously (27), we quantified the HA movements along different directions (Figure S3). The biggest outward movements of HA occurred in G131V run 1 and S132I run 1 while the largest downward movements were observed in wt simulation 5 and G131V run 2. Because HA movements occurred in both wt and mutant simulations, this structural flexibility might be an intrinsic property of human PrP at 310 K.
Figure 2.
Cα RMSD of the globular domain in MD simulations. Structures were aligned on the Cα atoms of the globular domain (residues 128-228) before RMSD measurements. The results of both wt and mutants are shown. For clarity, a running average over 500 ps is shown.
The G131V Mutation Induces the Elongation of the Native β-sheet
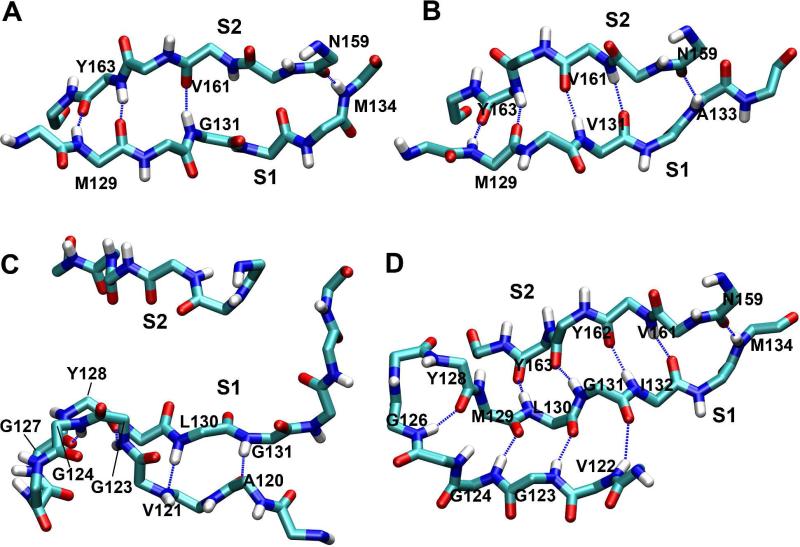
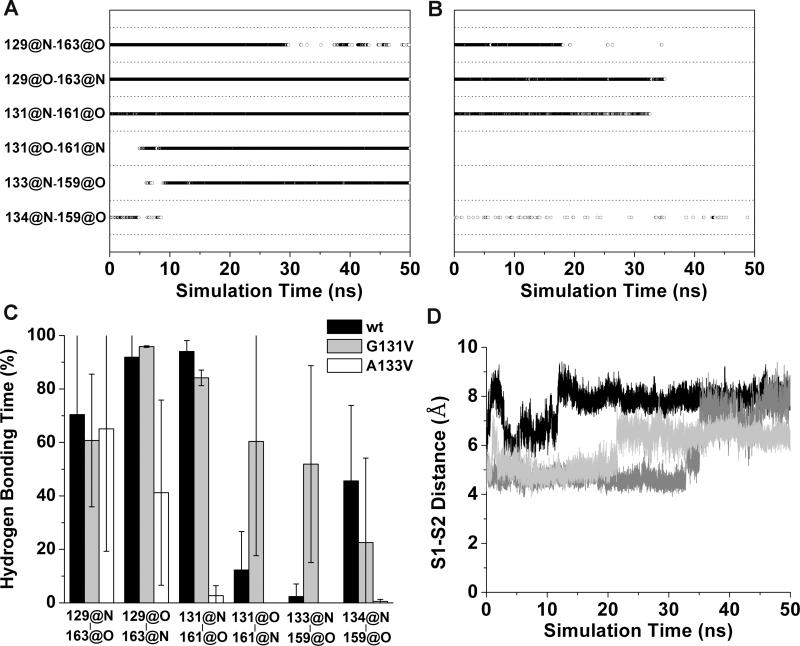
In the starting PrP structure, the short antiparallel native β-sheet contains a β-bulge (28) and consists of four main-chain hydrogen bonds: M129:N-Y163:O, M129:O-Y163:N, G131:N-V161O, and M134:N-N159:O (Figure 3A). Upon mutation to G131V, two new hydrogen bonds (V131:O-V161:N and A133:N-N159:O) formed while one of the original hydrogen bonds (M134:N-N159:O) broke, which resulted in the elongation of the native β-sheet and the elimination of the β-bulge (Figures 3B and 4A). In two out of three simulations of G131V, we observed the stable elongation of the native β-sheet (Figure S4). By comparison, in the wt simulations, the two new hydrogen bonds formed sporadically, but they were unstable (Figure S4). The hydrogen bond occupancies over the last 25 ns of the simulations were calculated for the six hydrogen bonds in the native β-sheet (Figure 4C). The two new hydrogen bonds were intact significantly longer in G131V than wt.
Figure 3.
Structures of the native β-sheet. A. The starting native sheet. B. The elongated native sheet at 49 ns of run 1 of PrP mutant G131V. C. The disrupted native sheet at 50 ns of run 2 of PrP mutant A133V. D. The converted native sheet to an α-sheet at 50 ns of run 2 of PrP mutant S132I. Only backbone atoms are shown. Dashed lines represent hydrogen bonds. Residues participating in hydrogen bonds are labeled.
Figure 4.
Changes in the native sheet upon G131V and A133V mutations. Main-chain hydrogen bonds of the native sheet are plotted vs. simulation time for (A) run 1 of PrP mutant G131V and (B) run 2 of PrP mutant A133V. A symbol represents that the hydrogen bond indicated by a y label exists at the time indicated by a x value. C. Percentage of hydrogen bonding times in the period of 25-50 ns averaged over all runs. x labels indicate the hydrogen bonds. Error bars show the standard deviation. D. The distances between the centers of masses of the backbone atoms of the S1 and S2 strands during MD simulations of PrP mutant A133V. The results of run 1 (black), run 2 (dark gray), and run 3 (light gray) are shown.
The A133V Mutation Disrupts the Native β-sheet
In contrast to the G131V mutation, introduction of A133V resulted in partial or full disruption of the native β-sheet hydrogen bonding and a shift with S1 interacting with the neighboring part of the N-terminal region (Figure 3C). In all three simulations at least two of the original hydrogen bonds broke (Figure S4), and all of them broke in run 2 (Figure 4B). Except for the M129:N-Y163:O hydrogen bond, the hydrogen bonding times of the other three original hydrogen bonds were much shorter for A133V than wt, indicating the disruptions of these hydrogen bonds due to the A133V mutation (Figure 4C). In conjunction with the loss of hydrogen bonds, S1 and S2 separated (Figure 4D). In different simulations, the separation of S1 and S2 occurred at different times and to different degrees. In run 1, the separation occurred at the start of the simulation and the distance between the centers of mass of the backbone atoms of S1 and S2 increased to ~8 Å. In run 2, the separation occurred at ~33 ns. The distance between S1and S2 first increased to ~5 Å for a short period of time and then to ~8 Å. In run 3, S1 was separated from S2 at ~22 ns and stayed at a position ~6.5 Å away from S2.
The S132I Mutation Causes α-sheet Formation
In the first simulation of mutation S132I, the M129/L130 and Y162/Y163 peptide planes □flipped□ at ~41 ns, allowing for formation of the L130:N-Y162:O hydrogen bond while the original M129:O-Y163:N hydrogen bond was lost (Figure S4). The resulting sheet is now in an α-sheet conformation (12, 29). In run 2, even more significant changes were observed (Figure 3D). Besides the flipping of the peptide planes, hydrogen bonding between S1 and S2 shifted by one residue: G131:N now formed an hydrogen bond with Y162:O (instead of V161:O) (Figure 5A). Two new hydrogen bonds, I132:N-V161:O and I132:O-V161:N, also formed. Together, these changes resulted in a mixed α/β-sheet, with the α-sheet on one side (residues 130-132 and 161-163) and the β-sheet on the other (residues 132-134 and 159-161). This conformational change occurred at the very beginning of the simulation (Figure 5). Around 10 ns, an additional α-strand (residues 122-126) formed in the N-terminal region (Figures 3D and 5), yielding a three-stranded mixed α/β-sheet. However, in run 3 such changes were not observed (Figure S4).
Figure 5.
Changes in the native sheet upon S132I mutation. A. Time course of main chain hydrogen bonds of the native sheet in run 2 of PrP mutant S132I. B. Secondary structure analysis of PrP in run 2 of PrP mutant S132I. The secondary structure elements present in the starting structure are shown on the top.
Reorientation of Side Chain-Side Chain Interactions is Responsible for Changes in the Native β-sheet
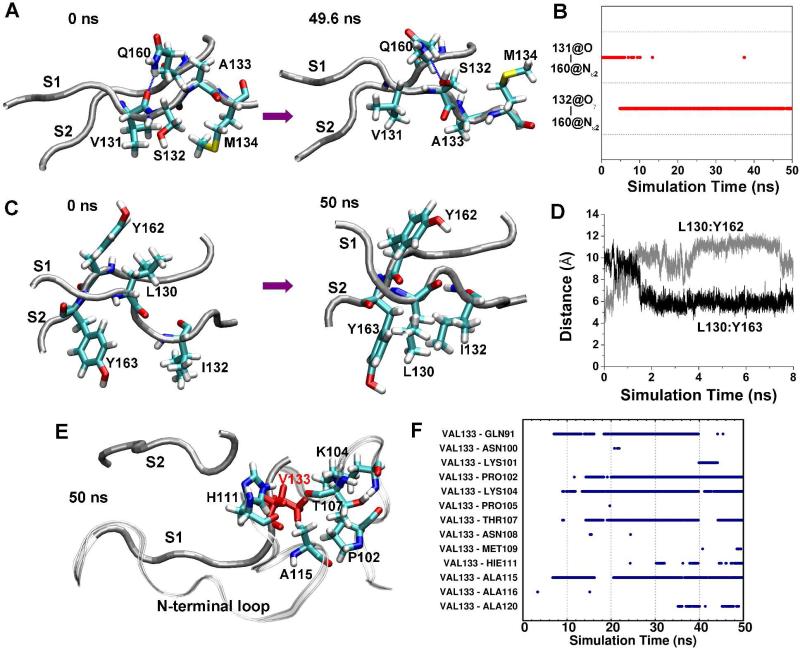
To understand why the mutations had different effects on the native β-sheet, we investigated the side chain-side chain interactions in detail. With the G131V mutation, the bulky side chain of V131 tended to push the side chain of S132 away to the other side of the β-sheet (Figure 6A). This repositioning allowed the side chain oxygen (Oγ) of S132 to form an hydrogen bond with the side chain nitrogen (Nε2) of Q160 (Figure 6B). As a result, the original hydrogen bond between Q160:Nε2 and V131:O was disrupted, which in turn allowed V131:O to hydrogen bond with V161:N. The local conformational changes further propagated down to A133 to allow the formation of the A133:N-N159:O hydrogen bond. Ultimately, the native β-sheet was elongated by 2 more hydrogen bonds. In run 1 of G131V, we even saw the reorientation of the side chains of A133 and M134 (Figure 6A). Initially, the A133 side chain pointed up while the M134 side chain pointed down. During the simulation, the side chains changed their orientations so that the A133 side chain pointed down while the M134 side chain pointed up. This is reasonable because the favored configuration of a β-sheet is that side chains are alternatively oriented to the both sides.
Figure 6.
Reconfiguration of side chain interactions of the native sheet due to mutations. A. Conformations of strands S1 and S2 at the indicated times in run 1 of PrP mutant G131V. The backbones of the strands are shown as tubes and relevant residues are represented as sticks and labeled, as is the case in panels C and E. B. Time course of hydrogen bonds between S131:O and Q160:Nε2 and between S132:Oγ and Q160:Nε2 in run 1 of PrP mutant G131V. C. Conformations of strands S1 and S2 at indicated times in run 2 of PrP mutant S132I. D. The distance between the centers of mass of the L130 side chain and the Y162 side chain in run 2 of PrP mutant S132I. E. The structure of the S1, S2 strands, and N-terminal region at 50 ns in run 2 of PrP mutant A133V. V133 is in red. The backbone of the N-terminal region is shown as a ribbon. F. Time course of hydrophobic side chain-side chain contacts between V133 and residues of the N-terminal region in run 2 of PrP mutant A133V.
Changes in side chain-side chain interactions were also observed in the S132I mutant. Because the I132 side chain is bulky and hydrophobic, it interacted with the hydrophobic side chain of Y163, bringing them closer together. This movement caused a shift of hydrogen bonding between S1 and S2 by one residue, as mentioned above. This shift brought the L130 side chain closer to Y162. As a result, the L130 side chain turned downward (Figure 6C). These changes are illustrated by the increase in distance between the L130 and Y162 side chains and the decrease in distance between the L130 and Y163 side chains (Figure 6D). The shift between S1 and S2 also eliminated the β-bulge at S132 and A133, eventually resulting in the elongation of the native β-sheet.
A different mechanism was observed for the A133V mutation. V133 formed hydrophobic interactions with residues 91-122 in the flexible N-terminal region for significant periods during the simulations (Figures 6E, 6F, and S5), presumably due to the solvent-exposed starting position of its hydrophobic side chain. In comparison, A133 in the wt simulations seldom interacted with the residues of the N-terminal region. The new hydrophobic interactions brought the neighboring N-terminal sequence (residues 117-125) closer to S1, which led to formation of both hydrogen bonds and hydrophobic interactions between S1 and neighboring N-terminal residues (Figure 3C). Altogether, these interactions disrupted the hydrogen bonds between the backbones of S1 and S2, ultimately leading to their separation.
Mutations Affect a Conserved Water Site in PrPC
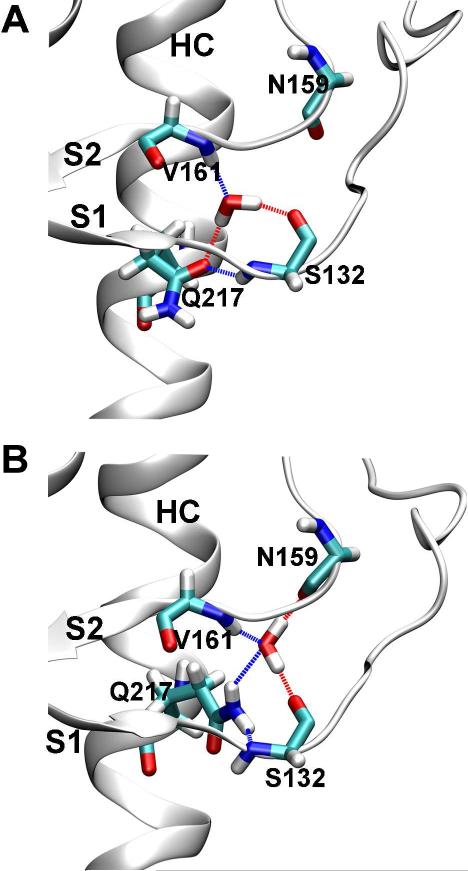
A tightly bound, structurally conserved water in the native β-sheet of PrPC was identified by the crystal structures of the sheep PrPC (pdb 1UW3) (30) and the human PrPC (pdb 1I4M and 3HAK) (31, 32) and MD simulations (33). The water forms hydrogen bonds with the carbonyl oxygen of S132, the amide nitrogen of V161, and the side chain oxygen of Q217 (Oε1), which connects three PrPC regions (S1, S2, and HC) (Figure 7A). In our simulations of wt human PrPC, we also observed that water molecules entered the water site and stayed for long or short periods of time (Figure S6). Besides the hydrogen bonding pattern predicted from the crystal structures, a different pattern also occurred in the simulations: the water molecule interacted with the carbonyl oxygen of S132, the amide nitrogen of V161, the carbonyl oxygen of N159, and the side chain nitrogen of Q217 (Nε2) (Figure 7B).
Figure 7.
The conserved water site at the native β-sheet. A. One mode of water coordination at the water site as observed in both the crystal structures of PrP and MD simulations of wt human PrP. B. Another mode of water coordination at the water site as observed in MD simulations of wt human PrP. Coordinating residues and water are shown as sticks. Dashed lines indicate hydrogen bonds.
To investigate how the mutations would affect the conserved water site, we analyzed water occupancy in the site over the simulations. We defined a water molecule as a coordinating water if it simultaneously connected S1, S2, and HC, i.e. it formed hydrogen bonds with S132:O, N159:O or V161:N, and Q217:Oε1 or Q217:Nε2 at the same time. In all simulations of wt PrP, coordinating waters were observed (Figure S6). Particularly in run 4, a water molecule was locked in the site throughout the simulation. However, waters were in exchange with bulk water in runs 1 and 3. In contrast to the wt simulations, no coordinating waters were observed in the site in the G131V simulations, indicating that the water site was abolished due to the elongation of the native β-sheet. For the S132I mutant, several water molecules moved in and out of the site sequentially in run 1. But no coordinating waters were found in run 2 because of the shift and elongation of the native β-sheet, as described above. In run 3, only at the very beginning was coordination water present because the bulky side chain of I132 pushed the side chain of Q217 aside. For the A133V mutant, coordinating waters were present in the first halves of runs 2 and 3 but was displaced half way through the simulation and almost no coordinating waters existed in run 1. The disappearance of the coordinating waters were well correlated with the separation of S1 and S2 (Figure 4D). In run 1, the two strands separated from the beginning, so no coordinating waters were possible. In runs 2 and 3, the separation occurred at ~22 ns and ~33 ns, respectively. As a result, no coordinating waters were found in the second halves of the simulations. Altogether, the three mutations at S1 impaired the water site although to different degrees.
DISCUSSION
Many single nucleotide mutations in PRNP have been identified to be associated with human prion diseases (7, 8), but the underlying mechanism of pathogenesis is still unclear. Here we describe MD simulations of human PrP with point mutations located on or near the S1 strand that are implicated in the development of GSS: G131V (9), S132I (10), and A133V (11). The simulations showed that these three mutations induced different effects on the native β-sheet: G131V elongated the native β-sheet from 3 to 5 hydrogen bonds, A133V disrupted the native β-sheet, and S132I converted the native β-sheet to an α-sheet. The various effects caused by these mutations suggest that PrP mutants can undergo diverse misfolding pathways and eventually may form different structures of PrPSc that manifest as different prion strains.
Previously, Santini et al. also simulated the G131V mutant although their simulations are only 4 ns long, much shorter than ours (34). However, Santini et al. did not observe the elongation of the native β-sheet from 3 to 5 hydrogen bonds for the G131V mutant. The elongation of the native β-sheet eliminated the β-bulge at S132 and A133 (Figure 3B), which may facilitate interactions of S1 with the N-terminal region and thus the growth of the native β-sheet from 2 strands to 3 or more strands, as observed in simulations under acidic conditions (35, 36).
In contrast to the G131V mutation, A133V caused separation of the S1 and S2 strands. We have not seen any reports of the effects of this mutation on the PrP structures. However, another mutation, T183A, has been shown by coarse grained MD simulations to disrupt the native β-sheet (37). Therefore, other than the growth of the native sheet, the disruption of the native β-sheet is probably another early misfolding event.
Interestingly, the S132I mutation converted the native β-sheet to an α-sheet and further extended to 3 strands with participation of the N-terminal region (Figure 3D). In the α-sheet, the main chain amide groups are on one side and carbonyl groups are on the other side, forming a polar sheet. This alignment has been suggested to give rise to attractive forces for self-assembly and thus facilitate formation of amyloid protofibrils (29). In the early stage of the unfolding of bovine PrPC and simulations of human and hamster PrPC, an α-sheet was also observed (29, 36, 38). The fact that the formation of α-sheets occurs in the simulations of numerous amyloidogenic proteins under amyloidogenic conditions (low pH or upon mutation, typically) and also in our simulations of S132I suggest that it is a common mechanism for amyloidogenesis (12, 29, 36, 38-43).
A structurally conserved water site was identified in the β-bugle of the native β-sheet in several crystal structures (Figure 7) (30-33). Water in this site connects S1, S2, and HC and has been suggested to stabilize the structure (33). Indeed, in the simulations of sheep PrP by De Simone et al., a water molecule was locked in the site once it entered (33). By comparison, in our wt simulations, which are much longer than De Simone et al.'s simulations (50 vs. 10 ns), we observed exchange of waters in the site with bulk water, except that in run 4 a water was locked in the site throughout the simulation. The three mutations in the native β-sheet all impaired the water site, either by eliminating the β-bulge at S132 and A133 or by separation of the S1 and S2 strands.
The predictions of the structural changes of the native β-sheet based on the MD simulations could be tested by experiments. One possible way is to measure hydrogen bond scalar couplings (h3JNC') for the native β-sheet by nuclear magnetic resonance (NMR) (44). h3JNC' depends strongly on the geometry of hydrogen bonds (45, 46). We calculated h3JNC' based on structures from the last 25 ns of the MD trajectories. As expected, the changes of h3JNC' due to mutations were observed corresponding to the formation or breakage of hydrogen bonds in the native β-sheet (Figure S7). Compared with wt, the G131V mutant increases the magnitude of h3JNC' of the V131:O-V161:N and A133:N-N159:O hydrogen bonds while the A133V mutant reduces h3JNC' for several hydrogen bonds: M129:O-Y163:N, G131:N-V161:O, and M134:NN159:O. On the other hand, the S132I mutant should yield new couplings. In addition, mutagenesis experiments could be used to test our predictions. A hydrogen bond between Q160:Nε2 and G131:O exists in the wt PrP structure. In G131V this hydrogen bond was broken when the V131:O formed a hydrogen bond with V161:N (Figure 6A), suggesting that these two hydrogen bonds compete with each other. Therefore, if Q160 is mutated to another residue such as Alanine, G131:O should be able to form a hydrogen bond with V161:N, resulting in the elongation of the native β-sheet.
The simulations described here suggest that multiple early misfolding pathways for PrP exist in response to mutation: one is the growth of a β-sheet via the elongation of the native β-sheet; the second is the disruption of the native β-sheet; the third is the growth of an α-sheet via the conversion of the native β-sheet. In contrast we have found more consensus among misfolding events in the wild-type protein at low pH even among different species (15, 26, 35, 36, 38). The simulations presented here shed further light on the mechanisms of the conversion of PrPC to PrPSc, particularly variations due to mutation.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for computational resources provided by Microsoft. We thank Alex D. Scouras for technical assistance.
Footnotes
We are grateful for support provided by the National Institutes of Health (GM81407 to V.D.).
Abbreviations: PrP, prion protein; PrPC, cellular prion protein; PrPSc, □scrapie□ prion protein; TSE, transmissible spongiform encephalopathy; CJD, Creutzfeldt-Jakob disease; GSS, Gerstmann-Straüssler-Scheinker syndrom; FFI, fatal familial insomnia; BSE, bovine spongiform encephalopathy; CWD, chronic wasting disease; GPI, glycophosphatidylinositol; MD, molecular dynamics; wt, wild-type; ilmm, in lucem molecular mechanics; RMSD, root-mean-square deviation; NMR, nuclear magnetic resonance.
SUPPORTING INFORMATION AVAILABLE
Cα RMSD of HA in MD simulations (Figure S1), HA movements in MD simulations (Figure S2), movements of the HA N-terminus (Figure S3), time course of main chain hydrogen bonds between the S1 and S2 strands in MD simulations (Figure S4), time course of hydrophobic side chain-side chain contacts between V133 and residues of the N-terminal region (Figure S5), and time course of water molecules residing in the conserved water site at the native β-sheet (Figure S6). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Aguzzi A, Baumann F, Bremer J. The prion's elusive reason for being. Annu. Rev. Neurosci. 2008;31:439–477. doi: 10.1146/annurev.neuro.31.060407.125620. [DOI] [PubMed] [Google Scholar]
- 2.Aguzzi A, Calella AM. Prions: protein aggregation and infectious diseases. Physiol. Rev. 2009;89:1105–1152. doi: 10.1152/physrev.00006.2009. [DOI] [PubMed] [Google Scholar]
- 3.Caughey B, Baron GS, Chesebro B, Jeffrey M. Getting a grip on prions: oligomers, amyloids, and pathological membrane interactions. Annu. Rev. Biochem. 2009;78:177–204. doi: 10.1146/annurev.biochem.78.082907.145410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linden R, Martins VR, Prado MA, Cammarota M, Izquierdo I, Brentani RR. Physiology of the prion protein. Physiol. Rev. 2008;88:673–728. doi: 10.1152/physrev.00007.2007. [DOI] [PubMed] [Google Scholar]
- 5.Caughey BW, Dong A, Bhat KS, Ernst D, Hayes SF, Caughey WS. Secondary structure analysis of the scrapie-associated protein PrP 27-30 in water by infrared spectroscopy. Biochemistry. 1991;30:7672–7680. doi: 10.1021/bi00245a003. [DOI] [PubMed] [Google Scholar]
- 6.Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick RJ, Cohen FE, et al. Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. U. S. A. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mead S. Prion disease genetics. Eur. J. Hum. Genet. 2006;14:273–281. doi: 10.1038/sj.ejhg.5201544. [DOI] [PubMed] [Google Scholar]
- 8.Van der Kamp MW, Daggett V. The consequences of pathogenic mutations to the human prion protein. Protein Eng., Des. Sel. 2009;22:461–468. doi: 10.1093/protein/gzp039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panegyres PK, Toufexis K, Kakulas BA, Cernevakova L, Brown P, Ghetti B, Piccardo P, Dlouhy SR. A new PRNP mutation (G131V) associated with Gerstmann-Straussler-Scheinker disease. Arch. Neurol. 2001;58:1899–1902. doi: 10.1001/archneur.58.11.1899. [DOI] [PubMed] [Google Scholar]
- 10.Hilton DA, Head MW, Singh VK, Bishop M, Ironside JW. Familial prion disease with a novel serine to isoleucine mutation at codon 132 of prion protein gene (PRNP) Neuropathol. Appl. Neurobiol. 2009;35:111–115. doi: 10.1111/j.1365-2990.2008.00964.x. [DOI] [PubMed] [Google Scholar]
- 11.Rowe DB, Lewis V, Needham M, Rodriguez M, Boyd A, McLean C, Roberts H, Masters CL, Collins SJ. Novel prion protein gene mutation presenting with subacute PSP-like syndrome. Neurology. 2007;68:868–870. doi: 10.1212/01.wnl.0000256819.61531.98. [DOI] [PubMed] [Google Scholar]
- 12.Daggett V. α-sheet: The toxic conformer in amyloid diseases? Acc. Chem. Res. 2006;39:594–602. doi: 10.1021/ar0500719. [DOI] [PubMed] [Google Scholar]
- 13.Zahn R, Liu A, Luhrs T, Riek R, von Schroetter C, Lopez Garcia F, Billeter M, Calzolai L, Wider G, Wuthrich K. NMR solution structure of the human prion protein. Proc. Natl. Acad. Sci. U. S. A. 2000;97:145–150. doi: 10.1073/pnas.97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prusiner SB, Groth DF, Bolton DC, Kent SB, Hood LE. Purification and structural studies of a major scrapie prion protein. Cell. 1984;38:127–134. doi: 10.1016/0092-8674(84)90533-6. [DOI] [PubMed] [Google Scholar]
- 15.Van der Kamp MW, Daggett V. The influence of pH on the human prion protein: Insights into the early steps of misfolding. Biophys. J. 2010;99:2289–2298. doi: 10.1016/j.bpj.2010.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van der Kamp MW, Schaeffer RD, Jonsson AL, Scouras AD, Simms AM, Toofanny RD, Benson NC, Anderson PC, Merkley ED, Rysavy S, Bromley D, Beck DA, Daggett V. Dynameomics: a comprehensive database of protein dynamics. Structure. 2010;18:423–435. doi: 10.1016/j.str.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scouras AD, Daggett V. The dynameomics rotamer library: amino acid side chain conformations and dynamics from comprehensive molecular dynamics simulations in water. Protein Sci. 2010 doi: 10.1002/pro.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beck DAC, Alonso DOV, Daggett V. lucem Molecular Mechanics (ilmm) University of Washington; Seattle, WA: 2000-2010. [Google Scholar]
- 19.Levitt M, Hirshberg M, Sharon R, Daggett V. Potential-Energy Function and Parameters for Simulations of the Molecular-Dynamics of Proteins and Nucleic-Acids in Solution. Comput. Phys. Commun. 1995;91:215–231. [Google Scholar]
- 20.Levitt M, Hirshberg M, Sharon R, Laidig KE, Daggett V. Calibration and testing of a water model for simulation of the molecular dynamics of proteins and nucleic acids in solution. J. Phys. Chem. B. 1997;101:5051–5061. [Google Scholar]
- 21.Beck DAC, Daggett V. Methods for molecular dynamics simulations of protein folding/unfolding in solution. Methods. 2004;34:112–120. doi: 10.1016/j.ymeth.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 22.Kell GS. Precise Representation of Volume Properties of Water at 1 Atmosphere. J. Chem. Eng. Data. 1967;12:66–69. [Google Scholar]
- 23.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 24.Viles JH, Donne D, Kroon G, Prusiner SB, Cohen FE, Dyson HJ, Wright PE. Local structural plasticity of the prion protein. Analysis of NMR relaxation dynamics. Biochemistry. 2001;40:2743–2753. doi: 10.1021/bi002898a. [DOI] [PubMed] [Google Scholar]
- 25.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 26.Scouras AD, Daggett V. Species variation in PrPSc protofibril models. J. Mater. Sci. 2008;43:3625–3637. [Google Scholar]
- 27.Van der Kamp MW, Daggett V. Pathogenic mutations in the hydrophobic core of the human prion protein can promote structural instability and misfolding. J. Mol. Biol. 2010 doi: 10.1016/j.jmb.2010.09.060. DOI:10.1016/j.jmb.2010.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richardson JS, Getzoff ED, Richardson DC. The β bulge: a common small unit of nonrepetitive protein structure. Proc. Natl. Acad. Sci. U. S. A. 1978;75:2574–2578. doi: 10.1073/pnas.75.6.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armen RS, DeMarco ML, Alonso DO, Daggett V. Pauling and Corey's α-pleated sheet structure may define the prefibrillar amyloidogenic intermediate in amyloid disease. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11622–11627. doi: 10.1073/pnas.0401781101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haire LF, Whyte SM, Vasisht N, Gill AC, Verma C, Dodson EJ, Dodson GG, Bayley PM. The crystal structure of the globular domain of sheep prion protein. J. Mol. Biol. 2004;336:1175–1183. doi: 10.1016/j.jmb.2003.12.059. [DOI] [PubMed] [Google Scholar]
- 31.Knaus KJ, Morillas M, Swietnicki W, Malone M, Surewicz WK, Yee VC. Crystal structure of the human prion protein reveals a mechanism for oligomerization. Nat. Struct. Biol. 2001;8:770–774. doi: 10.1038/nsb0901-770. [DOI] [PubMed] [Google Scholar]
- 32.Lee S, Antony L, Hartmann R, Knaus KJ, Surewicz K, Surewicz WK, Yee VC. Conformational diversity in prion protein variants influences intermolecular β-sheet formation. EMBO J. 2010;29:251–262. doi: 10.1038/emboj.2009.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Simone A, Dodson GG, Verma CS, Zagari A, Fraternali F. Prion and water: tight and dynamical hydration sites have a key role in structural stability. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7535–7540. doi: 10.1073/pnas.0501748102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santini S, Claude JB, Audic S, Derreumaux P. Impact of the tail and mutations G131V and M129V on prion protein flexibility. Proteins. 2003;51:258–265. doi: 10.1002/prot.10348. [DOI] [PubMed] [Google Scholar]
- 35.Alonso DOV, DeArmond SJ, Cohen FE, Daggett V. Mapping the early steps in the pH-induced conformational conversion of the prion protein. Proc. Natl. Acad. Sci. U. S. A. 2001;98:2985–2989. doi: 10.1073/pnas.061555898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeMarco ML, Daggett V. From conversion to aggregation: protofibril formation of the prion protein. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2293–2298. doi: 10.1073/pnas.0307178101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chebaro Y, Derreumaux P. The conversion of helix H2 to β-sheet is accelerated in the monomer and dimer of the prion protein upon T183A mutation. J. Phys. Chem. B. 2009;113:6942–6948. doi: 10.1021/jp900334s. [DOI] [PubMed] [Google Scholar]
- 38.DeMarco ML, Daggett V. Molecular mechanism for low pH triggered misfolding of the human prion protein. Biochemistry. 2007;46:3045–3054. doi: 10.1021/bi0619066. [DOI] [PubMed] [Google Scholar]
- 39.Armen RS, Alonso DOV, Daggett V. Anatomy of an amyloidogenic intermediate: Conversion of β-sheet to α-sheet structure in transthyretin at acidic pH. Structure. 2004;12:1847–1863. doi: 10.1016/j.str.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 40.Armen RS, Bernard BM, Day R, Alonso DOV, Daggett V. Characterization of a possible amyloidogenic precursor in glutamine-repeat neurodegenerative diseases. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13433–13438. doi: 10.1073/pnas.0502068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Armen RS, Daggett V. Characterization of two distinct β2-microglobulin unfolding intermediates that may lead to amyloid fibrils of different morphology. Biochemistry. 2005;44:16098–16107. doi: 10.1021/bi050731h. [DOI] [PubMed] [Google Scholar]
- 42.Schmidlin T, Kennedy BK, Daggett V. Structural Changes to Monomeric CuZn Superoxide Dismutase Caused by the Familial Amyotrophic Lateral Sclerosis-Associated Mutation A4V. Biophys. J. 2009;97:1709–1718. doi: 10.1016/j.bpj.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steward RE, Armen RS, Daggett V. Different disease-causing mutations in transthyretin trigger the same conformational conversion. Protein Eng., Des. Sel. 2008;21:187–195. doi: 10.1093/protein/gzm086. [DOI] [PubMed] [Google Scholar]
- 44.Cordier F, Nisius L, Dingley AJ, Grzesiek S. Direct detection of N-H···O=C hydrogen bonds in biomolecules by NMR spectroscopy. Nature Protocols. 2008;3:235–241. doi: 10.1038/nprot.2007.498. [DOI] [PubMed] [Google Scholar]
- 45.Barfield M. Structural dependencies of interresidue scalar coupling h3JNC' and donor 1H chemical shifts in the hydrogen bonding regions of proteins. J. Am. Chem. Soc. 2002;124:4158–4168. doi: 10.1021/ja012674v. [DOI] [PubMed] [Google Scholar]
- 46.Sass HJ, Schmid FFF, Grzesiek S. Correlation of protein structure and dynamics to scalar couplings across hydrogen bonds. J. Am. Chem. Soc. 2007;129:5898–5903. doi: 10.1021/ja068336h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.