Abstract
Bioluminescence, due to its high sensitivity, has been exploited in various analytical and imaging applications. In this work, we report a highly stable, cell-transductable and wavelength-tunable bioluminescence system achieved with an elegant and simple design. Using aqueous in-situ polymerization on bioluminescent enzyme anchored with polymerizable vinyl groups, we obtained nano-sized core-shell nanocapsules with enzyme as the core and crosslinked thin polymer net as the shell. These nanocapusles possess greatly enhanced stability, retained bioactivity, and readily engineered surface. In particular, by incorporating polymerizable amines in the polymerization, we endowed the nanocapsules with efficient cell-transduction and sufficient conjugation sites for follow-up modification. Following in-situ polymerization, decorating the polymer shell with fluorescent quantum dots allowed us to access continuous tunable wavelength, which extends the application of such bioluminescent nanocapsules, especially in deep tissue. In addition, the unique core-shell structure and adequate conjugation sites on surface enabled us to maximize the BRET efficiency by adjusting the QD/enzyme conjugation ratio.
Bioluminescence1, the light emission resulted from enzymatic reactions within living organisms, is commonly used for various applications2-3, such as whole-cell biosensors, immunoassays, nucleic-acid hybridization assays and in-vivo imaging. Many of these applications, however, require transfection of bioluminescent reporter genes, accompanied with many other limitations, such as safety. Moreover, the wavelengths of bioluminescence are still limited to blue/green light, which hinders its use in deep-tissue applications 4-5.
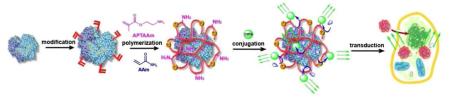
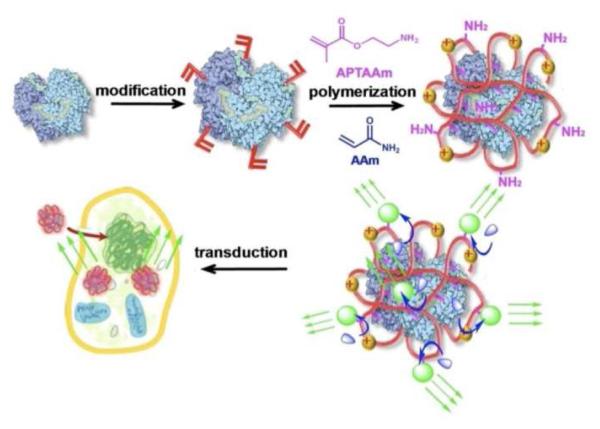
Herein, we report a novel class of bioluminescent nanocapsules (BNs) that are robust, cell-permeable, and tunable in wavelength. Scheme 1 illustrates our synthesis strategy. Starting with a bioluminescent protein, mild chemical modification attaches the protein with polymerizable vinyl groups; subsequent polymerization in an aqueous solution containing acrylamide (AAm) and N-(3-aminopropyl)methacrylamide (APMAAm) wraps the protein molecule with a thin polymer layer. Finally, linking the polymer-protein conjugate with fluorophores enables the conversion of short-wavelength bioluminescence to long-wavelength emission through bioluminescence resonance energy transfer (BRET)6.
Scheme 1.
Scheme of forming robust, cell-permeable bioluminescent nanocapsules
This unique architecture endows the BNs with many advantages: 1) the thin polymer layer stabilizes the protein and enables rapid transport of the substrate to the encased protein, ensuring bioactivity and stability of the BNs7; 2) fluorophores can be attached to the protein conjugates with controlled density, ensuring an efficient BRET. Particularly, quantum dots (QD), a class of fluorophores with high photo-stability and quantum efficiency, wide-range excitation and tunable emission, can be readily used as BRET receptors8-13; 3) as formed BNs are nano-sized and their surface charge can be readily tuned by controlling the ratio of neutral monomer (AAm) and cationic monomer (APMAAm) used, allowing their effective intracellular delivery; 4) targeting components, such as antibody, may be linked to the BNs for targeting purposes.
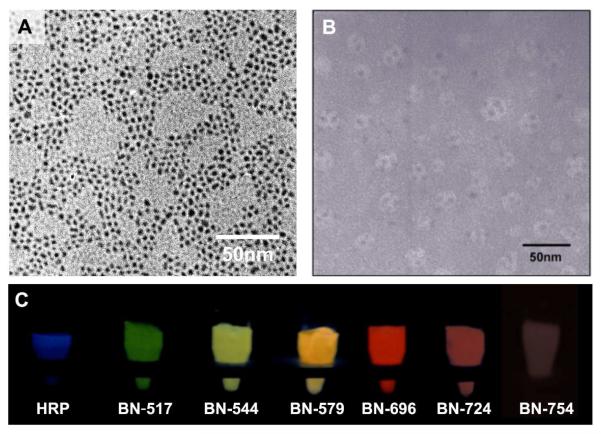
To demonstrate this concept, we used horseradish peroxidase (HRP) as a model bioluminescent protein and QDs of CdTe as the model fluorophores. Water-soluble QDs14 with emission wavelengths of 517, 544, 579, 696, 724 and 754 nm (Fig S1a) were respectively attached to the HRP-polymer conjugates (nHPR), creating a series of BNs with tunable emission (denoted as BN-wavelength, such as BN-517). Figure 1A shows a representative TEM image of the QDs with an emission wavelength of 544 nm (denoted as QD-544), demonstrating uniform size distribution with an average diameter of 2-3 nm. Figure 1B shows a TEM image of BN-544, suggesting that each BN has an average diameter of 17 nm and contains 3-4 QDs. The zeta potentials of the BNs were typically around 9 mV (Figure S2).
Figure 1.
TEM images of (A) QD-544 and (B) BN-544; (C) photographs of HRP and BNs in the presence of H2O2, luminol and p-iodophenol showing tunable-wavelength bioluminescence
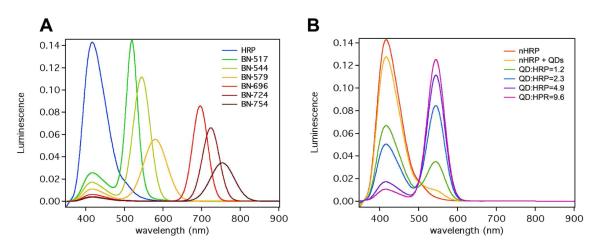
Note that the absorbance wavelengths of the QDs (Figure S1b) well overlap with that of the bioluminescence generated from HRP-mediated oxidation of luminol (425 nm), which allows their effective energy transfer. Figure 2A shows the fluorescent spectra of BNs with different emission wavelengths. As expected, besides the HRP bioluminescence at 425 nm, these BNs emits additional intense luminescence centered at 517, 544, 579, 696, 724 and 754 nm, respectively, which are in accordance with the fluorescence emissions of their conjugated QDs. Consistently, these BNs show bioluminescence color tunable from blue to red (Figure 1C). BNs with emission wavelength in the range of 700 to 800 nm are, of particular, importance for deep-tissue imaging owning to its deep-tissue penetrating capability4-5.
Figure 2.
(A) luminescent spectra of (A) native HRP, BN-517, BN-544, BN-579, BN-724 and BN-754, (B) HRP-polymer conjugates (nHRP), mixture of nHRP and QD-544, and BN-544 with various QD/HRP ratios
Besides tunable wavelength, these BNs also show tunable bioluminescent intensity, which was achieved by tuning the number of QDs attached to each BN (denoted as QD/HRP ratio). For example, Figure 2B demonstrates bioluminescence spectra of BN-544 with different QD/HRP ratios. Clearly, the luminescent intensity at 544 nm increases with increasing QD/HRP ratio. The BRET ratios determined are 0.48, 1.39, 5.06 and 8.92 for the QD/HRP ratio of 1.2, 2.3, 4.9 to 9.6, respectively. It has been demonstrated that directly linking luciferase (another bioluminescent protein) to QDs resulted in bioluminescence conjugates containing QD cores and luciferase shells. In this configuration, BRET occurred only between the core QDs and the excitation-state products diffused inwards the conjugates. Consequently, although the conjugates could emit bioluminescence with tunable wavelength, their BRET ratios were significantly lower (i.e., <0.5)10. In contrast, the BNs reported herein contain bioluminescent-protein cores and polymer shells containing tunable number of QDs. Such a unique architecture maximizes the harvest of bioluminescence, creating a series of BNs with significantly higher intensity.
It is also worth mentioning that BRET occurs only when the distance between donor and acceptor is less than 10 nm15. To create such a BRET response, the excitation-state products of the enzymatic reaction must be diffused to adjacent QDs before they decay to their stable species. Indeed, mixing the HRP-polymer conjugates with QD-544 resulted in much weaker emission intensity (544 nm), in comparison with BN-544 containing the same HRP to QD ratio (Figure 2B). This observation further confirms BRET is the main mechanism to illuminate the BNs.
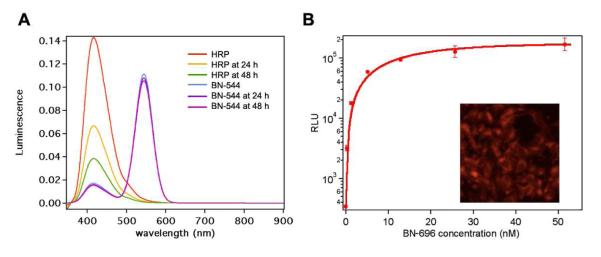
Furthermore, such BNs are extremely robust and cell transductable. For example, native HRP lost 52% and 73% of its original luminescence intensity after incubation at 37 °C for 24 hrs and 48 hrs, respectively (Figure 3A). On the contrary, BNs retained 99% and 98% of its luminescence intensity under the same conditions. This result validates our hypothesis that polymer network around HRP could protect the core protein from denaturation. Figure 3B shows a fluorescent image (Inset) of HeLa cells after 3-hour incubation with BN-696, clearly suggesting cellular uptake of the BNs. The bioluminescent intensity increases with increasing BN concentration (Figure 3B), indicating a concentration-dependent cellular uptake and retained intracellular activity. Such robust, cell transductable BNs are of particular interest for in vivo imaging and other applications.
Figure 3.
(A) Luminescence spectra of HRP and BN-544 after incubation at 37 °C for 0 hr, 24 hr and 48 hr. HRP and BN-544 were exposed to same amounts of H2O2, luminol and p-iodophenol during acquisition of spectra. (B) Relative luminescence unit (RLU) of HeLa cells pretreated with BN-696 with different concentrations. Inset: Fluorescent microscope images of HeLa cells pretreated with BN-696, showing cellular uptake of BNs
In summary, we have demonstrated a novel class of robust, cell-transductable bioluminescent nanocapsules with tunable emission wavelengths, providing new potentials for bioluminescence imaging, therapeutic and other applications.
Supplementary Material
ACKNOWLEDGMENT
This work was partially supported by Defense Threat Reducing Agency (DTRA), NSF-CAREER, Sandia National Laboratories, and NIH.
Footnotes
Supporting Information Available: Full experimental details for preparation of bioluminescent nanocapsules; TEM images; fluorescence and bioluminescence spectra; cell internalization protocols. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Wilson T, Hastings JW. Annu. Rev. Cell Dev. Bio. 1998;14:197. doi: 10.1146/annurev.cellbio.14.1.197. [DOI] [PubMed] [Google Scholar]
- (2).Roda A, Pasini P, Mirasoli M, Michelini E, Guardigli M. Trends Biotechnol. 2004;22:295. doi: 10.1016/j.tibtech.2004.03.011. [DOI] [PubMed] [Google Scholar]
- (3).Roda A, Guardigli M, Michelini E, Mirasoli M. Trends Analyt. Chem. 2009;28:307. [Google Scholar]
- (4).Weissleder R. Nature Biotech. 2001;19:316. doi: 10.1038/86684. [DOI] [PubMed] [Google Scholar]
- (5).Rice BW, Contag CH. Nature Biotech. 2009;27:624. doi: 10.1038/nbt0709-624. [DOI] [PubMed] [Google Scholar]
- (6).Xia ZY, Rao JH. Curr. Opin. in Biotech. 2009;20:37. doi: 10.1016/j.copbio.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Yan M, Du J, Gu Z, Liang M, Hu Y, Zhang W, Priceman S, Wu L, Zhou ZH, Liu Z, Segura T, Tang Y, Lu Y. Nature Nanotech. 2009;5:48. doi: 10.1038/nnano.2009.341. [DOI] [PubMed] [Google Scholar]
- (8).Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Nature Mat. 2005;4:435. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- (10).So MK, Xu CJ, Loening AM, Gambhir SS, Rao JH. Nature Biotech. 2006;24:339. doi: 10.1038/nbt1188. [DOI] [PubMed] [Google Scholar]
- (11).Yao HQ, Zhang Y, Xiao F, Xia ZY, Rao JH. Angewandte Chemie-International Edition. 2007;46:4346. doi: 10.1002/anie.200700280. [DOI] [PubMed] [Google Scholar]
- (12).Xia ZY, Xing Y, So MK, Koh AL, Sinclair R, Rao JH. Analytical Chemistry. 2008;80:8649. doi: 10.1021/ac801562f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Huang XY, Li L, Qian HF, Dong CQ, Ren JC. Angew. Chem. Int. Ed. 2006;45:5140. doi: 10.1002/anie.200601196. [DOI] [PubMed] [Google Scholar]
- (14).Qian H, Dong C, Peng J, Qiu X, Xu Y, Ren J. J. Phys. Chem. C. 2007;111:16852. [Google Scholar]
- (15).Pfleger KDG, Eidne KA. Nature Meth. 2006;3:165. doi: 10.1038/nmeth841. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.