Abstract
Stat5 proteins modulate gene transcription upon cytokine- and growth factor action. Stat5a and Stat5b proteins alone are weak activators of transcription. They can modify chromatin organization through oligomerization and they act predominantly in co-operation and interaction with other proteins. The conservative view of exclusively nuclear functions of Stat5 was challenged by the observation of additional Stat5 effects in the cytoplasm, resulting in activation of the PI3K-Akt pathway. We summarize biological consequences of mutations in conserved domains of Stat5 or of deletions in the N- or C-terminal domains with impact on target gene transcription. Formation of higher-order oligomers is dramatically changed upon amino- or carboxyterminal deletions in Stat5 proteins. Mutations in or deletion of the Stat5 N-terminus leads to diminished leukemogenic potential of oncogenic Stat5, probably due to the inability to form Stat5 tetramers. The Stat5 N-terminal domain prevents persistent activation and can act as a DNA-docking platform for the glucocorticoid receptor (GR). The corresponding protocols should facilitate follow-up studies on Stat5 proteins and their contribution to normal physiological versus pathological processes through differential chromatin binding.
Keywords: Stat5, Chromatin Alteration, Oligomerisation, Cytoplasmic Function, Review
2. STAT5 ISOFORMS: THEIR ACTIVATION AND INVOLVEMENT IN DISEASE
Stat5 proteins play crucial roles in controlling physiological processes like hematopoiesis or hepatocyte function. Yet, unrestricted Stat5 activation leads to pathological conditions such as cancer promotion and - progression, myelo-proliferative diseases, inflammation, or auto-immunity. Too little Stat5 activity, however, also can cause diseases such as myeloid hypoplasia (anemia, thrombocytopenia), dwarfism, infertility, immunodeficiency or metabolic syndromes (1-14). Thus, Stat5 activity needs to be tightly controlled, a demanding task given the broad spectrum of Stat5 functions in various parts of the organism. How and where Stat5 proteins exert their function and which post-translational modifications of Stat5 are actually necessary for proper function is still largely unknown, not least because other proteins interacting with Stat5 were not sufficiently taken into consideration until now.
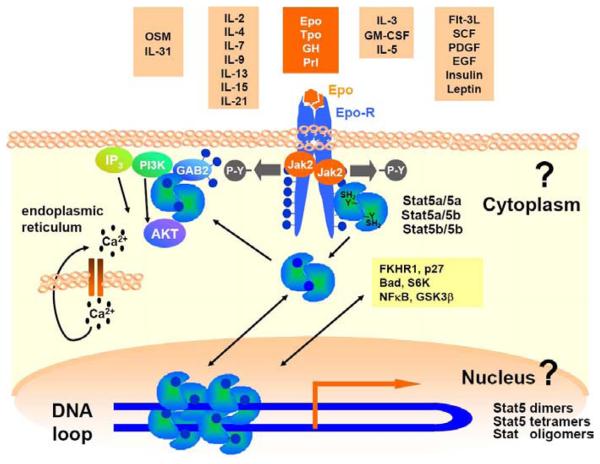
Stat5a and Stat5b transcription factors are rapidly activated after stimulation of cells with different cytokines or growth factors in conjunction with the corresponding receptors. Subsequently, signals are transduced mainly through Jak1, Jak2 and Jak3 tyrosine kinases, which leads to efficient nuclear translocation of Stat5 molecules (Figure 1). Stat5a and Stat5b proteins can form homo- or heterodimers. In addition, multiple shorter isoforms of Stat5a and Stat5b exist. Whether the latter arise from splicing, proteolytic processing or alternative start codons (Figure 1) is controversial (7, 15-20). Finally, Stat5 proteins are modified by post-translational modifications such as glycosylation, ubiquitinylation, serine/threonine phosphorylation and alternative tyrosine phosphorylation (18-22). Thus, Stat5 proteins can be found in many different homo- or heterodimeric complexes regulating differential gene expression. Overall, it is economical for different cell types to utilize these ancient transcription factors (homologues in dictyostelium or drosophila exist) to modulate the transcriptome in response to a variety of cytokine and growth factor challenges (Figure 1).
Figure 1.
Cytokines, hormones or growth factors that activate Stat5. Cytokines can be grouped based on structural homologies, similar signal transduction pathways and shared receptor chains. The specific example shown refers to definitive erythropoiesis. Self renewal and differentiation of erythroid cells requires functional Epo signaling through its receptor EpoR and Jak2, leading to tyrosine-phosphorylated, transcriptionally active Stat5a and Stat5b proteins. Stat5 molecules involved were found to be dimers and to be attached to the receptor via the N-terminus prior to tyrosine phosphorylation (38). Efficient nuclear translocation and transcriptional activation of Stat5 requires tyrosine phosphorylated Stat5 proteins. Unrestrained Stat5 activation is found in the presence of many different transforming tyrosine kinases, which can contribute to diseases such as cancer. In contrast, too little Stat5 activation causes diseases like anemia. Recently, we found that Stat5 molecules also have an important function in the cytoplasm to activate the PI3K-Akt pathway. Stat5 molecules can form dimers, homo- or hetero-tetramers on DNA (so called oligomers), which can induce the formation of DNA loop structures.
The observation that Stat5 proteins are prime targets of transforming tyrosine kinases in cancers was not unexpected, yet a recent surprise was that Stat5 proteins are essential to render malignant cells largely independent of external proliferation stimuli (Figure 1). The activation of Stat5 proteins by tyrosine kinases enables cancer cells to overcome cell cycle control and to survive within the cancer microenvironment. Many questions about the “the role of Stat5 in cancer” remain unanswered. It is unknown whether Stat5 activation in cancer contributes to immune escape, vascularization, metastasis or immortalization. Such roles were reported for activated Stat3, but in many cell types the functions of Stat3 and Stat5 proteins are distinct (23, 24). In general, it is well established that Stat5 plays an essential role in leukemogenesis. Whether it has a broader role in the emergence of solid tumors is not well studied. Data on the role of Stat5 in breast cancer are controversial (25-27), but lymphomas, liver cancer, prostate cancer, lung cancer, ovarian cancer or head and neck cancer were shown to require Stat5 proteins (28-33). Stat5 proteins are also implicated in infectious diseases, immune cell homeostasis or autoimmunity.
In general, proper function of Stat5 molecules is essential for all immune cells and controls e.g. lymphocyte development, NK cell activity, cytotoxic T cell function, T helper or suppressor/regulatory T cell function, mast cell-, platelet/megakaryocyte-, and macrophage responses or stress erythropoiesis (2, 5, 6, 14, 34-39). Most hematopoietic cytokines and growth factors signal through Stat5. In addition, cytokines and growth factors acting on epithelial and mesenchymal cell types as well as oncogenic tyrosine kinases can activate Stat5 (Figure 1). Thus, it is important to understand how Stat5 proteins are activated and how they modulate signal transduction, also under conditions when they are found to execute their function in the cytoplasm.
Recent work of several laboratories has demonstrated that activation of phosphoinositol-3-kinase (PI3K) and of the Akt pathway is induced by protein-protein interaction of Stat5 with the scaffold protein Gab2 (40, 41). Interestingly, Gab2- or Stat5-deficient fetal liver cells have both hematopoietic deficiencies due to diminished cytokine responses, although the phenotype of Gab2-deficiency is milder than in Stat5-deficiency (7, 42-44). This underlines the notion that Stat5 proteins have additional functions in the cytoplasm and are not mere nuclear transcription factors (40, 41).
Moreover, new data have shown that Stat5 regulates Akt activity in T cells, eventually involving also late or indirect transcriptional effects resulting in Akt activation (45, 46). It is currently unknown whether in leukemic cells Stat5 interacts directly with Gab2 (e.g. in primary CML cells, where endogenous Stat5 proteins are tyrosine phosphorylated and predominantly localized to the cytoplasm) to recruit the p85 subunit of PI3K, but tyrosine phosphorylation of Stat5 proteins is a prerequisite (41). Still, whether cytokine activated Stat5 or oncogenic Stat5 can activate Akt directly through a Gab2-p85-Stat5 interaction or indirectly through transcriptional effects remains controversial (40, 41, 46). It is possible that different mechanisms are involved in a cell type specific manner. In any case, Stat5 activation and tyrosine phosphorylation bridge to Akt activation. The two alternatives do not exclude each other. Direct protein-protein interaction or indirect transcriptional regulation might have the same result or target, both leading to enhanced Akt activity. In the case of IL-7-induced Glut1 induction through Stat5-Akt (46), IL-7 is very potent in activating Stat3 besides Stat5. Whether Stat3 also regulates Akt expression via a transcriptional mechanism was not tested and is questionable. In summary, oncogenic P-YStat5 (as in CML patients or in mice transplanted with hematopoietic cells carrying oncogenic Stat5) is clearly cytoplasmically localized and very efficient in activating Akt through direct protein-protein interaction with Gab2 and p85 (40).
The ratio of nuclear versus cytoplasmic Stat3 protein is influenced by transforming tyrosine kinases (47), but the mechanism of nuclear-cytoplasmic shuttling of Stat5 in different cell types remains enigmatic (48). No conclusion can currently be drawn from the observation that activated Stat5 (P-Y-Stat5) is often found in the cytoplasm of transformed myeloid cells from CML or AML samples (40). We have extended these studies with additional patient samples and found that survival of myeloid cancer cells depends on the Stat5-PI3K-Akt axis (Gouilleux, Moriggl et al., manuscript submitted).
One further hallmark of AML is the occurrence of an oncogenic variant of the FLT3 (FLTmut) growth factor receptor, which leads to nuclear activation of Stat5 (49). Recent studies underscored the importance of Stat5 in FLTmut-driven leukemogenesis via binding and activation of Rac1, an essential component of the NAPDH-oxidase. The latter was crucial for generation of endogenous reactive oxygen species (ROS), leading to genomic instability and increased DNA damage and thus might account for the observed poor prognosis of AML patients harboring FLT3mut receptors (50).
Cytokine and growth factor receptor signaling systems activate a large variety of downstream signaling molecules such as kinases, phosphatases, transcription factors or negative feedback regulators (e.g. SOCS/PIAS/STAM proteins). Stat5 transcription factors are highly and broadly expressed proteins found in these cellular signaling pathways. Expression levels of Stat5 proteins vary from cell type to cell type but can reach an abundance of 3,000-100,000 molecules per cell (51). Quantification of Stat5 proteins will be important in future studies and repression of Stat5 gene expression could be relevant in diseases.
In that context it is also important to mention that only a fraction of a Stat protein pool gets tyrosine phosphorylated (18, 52). 35S-labeled cell extracts and pulse chase experiments with Stat1 and Stat5b revealed approximately 10-25% of the total Stat1 or Stat5b pool to be tyrosine phosphorylated upon cytokine action (18, 52). How many Stat molecules are activated might be cytokine and cell type specific, but quantitative studies even from cell lines are limited. P-Y-Stat proteins are recycled through tyrosine dephosphorylation (52), in contrast to the Jak kinase or cytokine receptor chains which are degraded. Recently, ubiquitination and proteasome-dependent degradation was postulated as an additional mechanism for inactivating Stat5a. Interestingly, nuclear Stat5a was mainly inactivated via ubiquitination and protein degradation whereas cytoplasmic Stat5a was dephosphorylated by abundantly available tyrosine phosphatases. This differential inactivation mechanism might argue for a distinct function of unphosphorylated nuclear Stat5a, as phosphorylation of nuclear Stat5a was not required for ubiquitination (53). Recently, Stat5a was also shown to be epigenetically silenced in NPM-ALK lymphomas (54). Usually, the limiting component during cytokine and growth factor receptor response (and even in transformed tyrosine kinase signaling) is the moderately expressed tyrosine kinase. There are exceptions to this rule, however, since BCR-ABL was calculated to be in a range of up to 100.000 molecules in K562 cells, a cell line derived from a human CML patient.
It should be emphasized again that Stat5 transcription factors are not only at the end of signaling cascades (40, 45, 46). Stat5 proteins are highly expressed signaling molecules with docking functions even when not activated. Cytoplasmic functions were also recognized for Stat1 and, as already mentioned, for Stat3 (55-57).
3. PHENOTYPES OF MOUSE MODELS WITH FULL OR PARTIAL DELETION OF STAT5
In the following we will provide an overview on “old” and “new” Stat5 knock out mouse models that have either no Stat5 proteins expressed (new) or that have a deletion in the Stat5-N terminus (old) (Figure 2). It is well understood that Stat5 molecules control a multitude of functions. The current picture is incomplete due to the multitude of Stat5 activities and due to the lack of more advanced mouse models (e.g. knock-in models). A further level of complexity originates from the broad expression (16) and activation pattern (58) of Stat5 proteins and their different specific functions in mammals. Here, we will not elaborate on studies targeting one single Stat5 isoform for deletion (8, 13), and focus instead on reports describing biological consequences of deleting both isoforms.
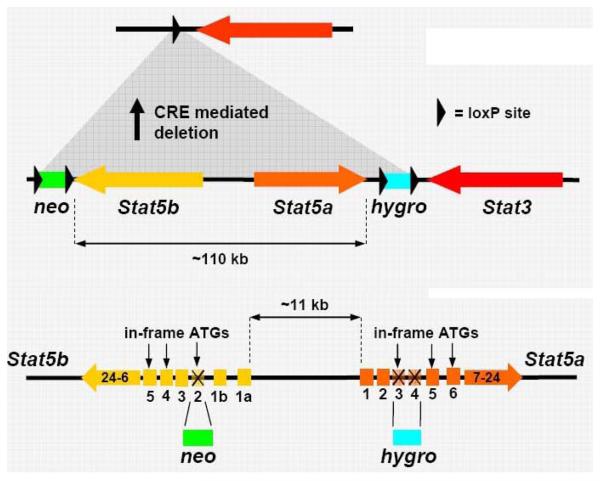
Figure 2.
Chromosomal organization of the Stat5 locus and strategies to generate knockout mouse models. Targeting strategies for complete or incomplete Stat5a/b deletion. The description of the different Stat5 mouse systems are originating from the following studies: (i) Complete gene deletion (s) of Stat5a, Stat5b or both: Gene deletion of Stat5a (8); Gene deletion of Stat5b (13); Gene deletion of Stat5a and Stat5b (1); or (ii) incomplete gene deletion (s): N-terminal gene deletion of Stat5a, N-terminal gene deletion of Stat5b and N-terminal gene deletions of Stat5a and Stat5b (12); Graphic representation of targeting strategies generate single deletions of Stat5a or Stat5b is omitted, since the focus here is on the consequences of deleting both isoforms. Top panel: reproduced with permission from 1. Bottom panel” reproduced with permission from 12.
Old and new mouse models for Stat5a and Stat5b deletion display overlapping as well as distinct phenotypes. The generation of murine knock-out models for Stat5 proteins was complicated by the fact that the Stat5a-Stat5b locus spans only approximately 110 kb on chromosome 11 (in humans, the Stat5 genes are localized in identical configuration on chromosome 17; Figure 2). The original Stat5 knock out mouse, now referred to as Stat5ΔN, was generated via double targeting ES cells at two different loci on chromosome 11. Neomycin- and hygromycin-resistance cassettes were inserted into the first coding exon of the Stat5a and Stat5b gene, respectively (Figure 2) (12). The resulting Stat5ΔN mice were viable and were believed to represent true knock outs although they expressed N-terminally truncated Stat5 proteins at high levels. Surprisingly, N-terminally truncated Stat5a and Stat5b molecules were also found in vivo in wild type animals (7). Many studies were based on the Stat5ΔN mouse model (2-6, 9-12, 14). Today, these mice are regarded and described as a model to study functions of the N-terminal domain, particularly in the liver and in lymphocytes (3, 5, 14, 59). Apparently, the Stat5 N-terminus constitutes an important protein-protein interaction domain (Figure 3 to 4). Stat5ΔN mRNA harbors intact Kozak sequences and in-frame start codons at amino acid position 103 or 137 of murine Stat5a or Stat5b (Figure 3). According to observed migration patterns in Western blots or in DNA binding assays, most likely the ATG at position 137 is predominantly used (3, 5, 7, 59). Amino acid M137 is conserved in all mammalian Stat5 proteins, in contrast to M103, which is lacking in human Stat5a (Figure 4). Stat5ΔN mice express both N-terminally truncated Stat5a and Stat5b molecules (2, 3, 5, 7, 14, 59). Interestingly and in contrast to endogenous Stat5, Stat5ΔN proteins have lost the ability to physically interact with the glucocorticoid receptor (3, 60). It is currently questionable which parts of the phenotypes described in Stat5ΔN mice are due to loss of Stat5-GR protein-protein interaction. Moreover, N-terminal truncation of Stat5a, but not of Stat5b renders the molecule persistently active (3, 61). This underlines a regulatory role of the N-terminus of Stat5a to prevent persistent tyrosine phosphorylation.
Figure 3.
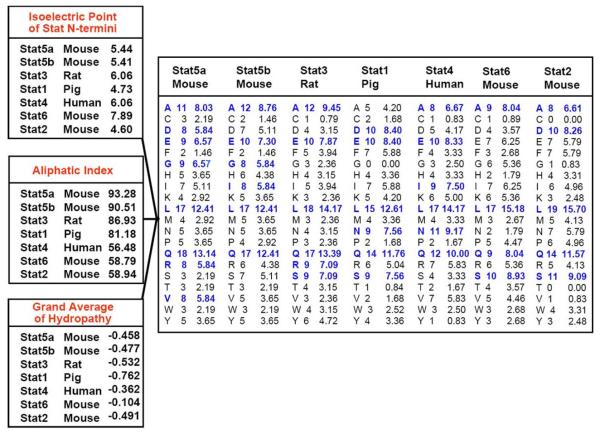
Identity and semi conservative amino acid changes. The N-termini of different Stat family member were compared. NCBI protein blast was used for similarity searches. Conserved amino acids in Stat5 N-termini are highlighted in bold. Mammalian species with closest homology to mouse Stat5a and Stat5b were used for comparison. Rodent, pig, bovine and human sequences were chosen, while ovine Stat5a was excluded from analysis. Ovine Stat5b was so far not completely sequenced and ovine Stat5a displayed significant differences to other mammalian Stat5 sequences in a stretch of 17 amino acids in the N-terminus. The amino acid motif containing the O-Glc-NAc modification at T92 is underlined. Moreover, the two alternative start codons M103 and M137 are underlined.
Figure 4.
Amino acid composition and biochemical protein parameters. Protein characterization tools were used from Swiss-Prot/TrEMBL public databases to highlight distinction criteria of the N-termini of different Stat proteins. Specifically, the ProtParam program was used as a protein identification and analysis tool for the calculation of isoelectric points, aliphatic indices, grand averages of hydropathy and amino acid composition (103). The aliphatic index of a protein is defined as the relative volume occupied by aliphatic side chains (alanine, valine, isoleucine, and leucine). The GRAVY value for a peptide or protein is calculated as the sum of hydropathy values of all the amino acids, divided by the number of residues in the entire sequence. The amino acid composition is distinct among the different Stat N-termini. Amino acids with higher appearance of n=7 residues within the N-terminal domain are highlighted in bold and underlined.
The complete knock out of Stat5a and Stat5b (Stat5Null), was created in the laboratory of Lothar Henninghausen, who already had designed the complete knock-out of Stat5a (1, 62). The Cre-loxP technology used allowed deletion of the entire 110kb locus either in the germ line or via conditional deletion (Figure 3) (1, 59). Genetic complementation with a retrovirus encoding wild type Stat5a into the Stat5Null background restored critical functions, indicating that genome integrity was preserved despite the large deletion on chromosome 11 (Figure 2). Stat5 deletion during embryonic development was lethal, particularly on a pure C57Bl/6 or Balb/c genetic background. Surprisingly, mixed Sv129 × C57Bl/6 F1 mice developed to term, but over 97% died perinatally (1, 5, 59). Recent data suggest that lethality in Stat5-deficient animals is due to anemia resulting from defects in iron metabolism of fetal erythroid cells and the ensuing elevated levels of apoptosis (Kerenyi et al., manuscript submitted). Complete deletion of both Stat5 genes can result in activation of other Stat family members, a general phenomenon in Jak-Stat-SOCS gene deletion studies, complicating the interpretation of phenotypes (63).
Numerous conditional Stat5 knock out mouse models have already been created. To start with, Stat5 was deleted specifically in mammary epithelial cells (WAPCre) or ablated in a broader range of epithelial cell types (MMTV-Cre) (1), B-cells (CD19-Cre) (64), T lymphocytes (efficient recombination from CD4/CD8 double-positive cells via Lck-Cre; (5) less efficient in the CD4+ compartment with CD4-Cre; (59)). In addition, Stat5 was deleted from hepatocytes plus biliary epithelial cells using albumin-alpha-fetoprotein-Cre (3), while liver-restricted deletion was achieved with Albumin-Cre (65), although less efficiently. Moreover, pancreatic beta-cells and hypothalamus (Rip-Cre) (66), endocrine and exocrine pancreas cells (Pdx1-Cre) (66), as well as skeletal-muscle (Myf5-Cre) (67) were targeted. Finally, transplant models using Stat5Null cells and the analysis of Stat5Null survivors unraveled new phenotypes in B- and T lymphocytes (5, 14, 59).
From all studies listed above, it is clear that various cell types critically depend on Stat5 proteins for proliferation, differentiation or survival. Important functions in hematopoietic lineages were appreciated already for a long time but also mammary gland, ovary, prostate, testis, liver, bone or fat tissues depend on Stat5 proteins for full specification. Referring to hematopoiesis, already the repopulation capacity of hematopoietic stem cells depends to a large extent on Stat5 (7, 43, 44, 59). In more differentiated hematopoietic compartments, Stat5 is important for proper development of T-, NK- or B cells: The generation of CD8+ T cells, gamma/delta T cells, regulatory T cells, but also T helper cells depends on Stat5 proteins (5, 14, 34, 59). Similarly, NK cell number and biological activity depend on Stat5 (35, 36, 39, 68), also B lymphocyte development and transformation are closely connected to Stat5 (5, 64). Moreover, Stat5 functions obviously are involved in definitive and stress erythropoiesis (2, 69), monocyte maturation and granulopoiesis (70), as well as activities of macrophages, eosinophilic granulocytes or mast cells (6, 71, 72). It is therefore not surprising that expression of oncogenic Stat5 variants or overexpression of Stat5 can lead to the development of leukemias, lymphomas or myeloproliferative diseases (9, 28, 33, 61, 104).
4. STAT5 GAIN-OF-FUNCTION APPROACHES
Persistent Stat5 activation is known to be crucial for the persistent proliferation of various cancer cell types. Indeed, activation of Stat5 proteins is associated with the most frequently encountered transforming tyrosine kinases. The list of such persistently active kinase mutants or variants includes prominent examples like v-Abl, Jak2 (V617F), Jak3 (A572V), c-Kit (D816V), Flt-3-ITD, v-ErbB or persistently activating Stat5 is generated by chromosomal translocation events, exemplified by BCR-ABL, p185 and p210, TELABL, TEL-PDGF, TEL-JAK2, TEL-Jak3, EML1-ABL or ZNF198-FGFR1. Accordingly, the first successful attempt to create a persistently active Stat5 gain-of-function mutant was the generation of a Stat5-tyrosine kinase fusion chimera, in which the Jak2 kinase domain was fused to the C-terminus of Stat5a, together with a strong transactivation domain in between (73). Transgenic mice expressing this construct developed breast cancer (25).
So far, only a limited number of studies addressed the important question whether Stat5 proteins are essential for transformation. So far, the requirement for the presence of Stat5 to induce transformation could be genetically proven only for BCR-ABL (5). Recently, we were able to show that a gain-of-function mutant of Stat5 was able to largely complement Jak2- or erythropoietin receptor deficiency during erythro- and myelopoiesis (69). The current view that too much Stat5 activity can indeed result in transformation of hematopoietic progenitors is best illustrated by the ability of one persistently activated Stat5 molecule to substitute for exogenous cytokine signaling and to cause multi-lineage leukemia. This well studied oncogenic variant is a point mutant of Stat5a, harboring a serine-to-phenylalanine mutation in the C-terminal domain, termed cS5F (or S710F; Figure 3). Upon transduction of cS5F into wild type hematopoietic stem cells and subsequent transplantation into lethally irradiated wild type mice, leukemia develops within 4 weeks (40, 61).
The predecessor of cS5F had been another Stat5 mutant, designated caStat5a1*6, containing two independent mutations, S710F plus H299R in the DNA binding domain (74). This variant had originally been found in a screen for factor-independent proliferation of Ba/F3 cells, with which a PCR mutagenesis on Stat5 had been performed. CaStat5a1*6 conferred IL-3 independent growth to Ba/F3 cells. Both, the double mutant and the single point mutant cS5F used in our studies do not differ significantly in their biochemical properties: (i) They are constitutively activated through persistent tyrosinephosphorylation, even after cytokine starvation. (ii) Both display hyper-activation upon cytokine stimulation and (iii) enhanced DNA binding (61). Thus, CaStat5a1*6 and cS5F could be termed “constitutively active” as well as “hyper-activatable”. There is, however, one important difference: In contrast to cS5F, caStat5a1*6 was unable to genetically complement Stat5null- or Stat5ΔN cells (61) (and data not shown), requiring full length, endogenous Stat5 proteins to exert its transforming function. In contrast, cS5F transformed Stat5ΔN cells (61) as well as Stat5null hematopoietic cells, causing multi-lineage leukaemia upon transplantation of cS5F-expressing Stat5null or Stat5ΔN fetal liver-derived hematopoietic cells into irradiated mice (unpublished data). Moreover, we observed that cS5F was capable of transforming primary mast cells from Stat5-deficient animals. The mutant was also able to activate Stat5-reporter gene constructs in Cos7 cells which do not express any endogenous Stat5 (unpublished data). Together, these observations argue that cS5F does not require endogenous Stat5 protein and is structurally and functionally related to endogenous Stat5 since it genetically complements Stat5-deficiencies.
Mutations equivalent to caStat5a1*6 were also introduced into Stat5b to generate gain-of-function variants for the study of lymphopoiesis in corresponding transgenic mice (34, 39). Another screen for persistently active Stat5 mutants identified a point mutant in the SH2 domain of Stat5a (N642H), which is also a residue conserved in Stat5b. Since Stat5b is the only isoform expressed at significant levels in the liver, Stat5b (N642H) was used for in vivo studies with hepatocytes (75).
In summary, it is recommendable to use the cS5F gain-of-function mutant rather than the double mutant caStat5a1*6, since cS5F was able to complement Stat5-, Jak2- or EpoR-deficiencies (61, 69). Contrarily, caStat5a1*6 required endogenous Stat5 for gain-of-function effects. Whether Stat5a (N642H), Stat5b (N642H) or the Stat5-tyrosine kinase fusion chimera can complement Stat5 deficiency is currently unknown. One transgenic mouse models with cS5F was reported, which resulted in B-cell lymphomas in absence of p53 (104).
Tyrosine phosphorylation, DNA binding activity and the C-terminal transactivation domain were essential for the ability of cS5F to substitute for myeloid growth factors or cytokines (69) (and Moriggl, unpublished data). We worked extensively with the cS5F oncogene to study transforming characteristics in immature and mature hematopoietic cell types (40, 69). Extracts of cS5F expressing cells displayed enhanced Stat5 chromatin binding activity. In addition, cytoplasmic cS5F efficiently activated the PI3K-Akt pathway. This led to the speculation that both, nuclear and cytoplasmic functions of cS5F, might be required for full transforming ability. Moreover, cS5F relieved cytokine dependence and prolonged the duration of Stat5 signaling in response to growth factors or cytokines (40, 61, 69). Interestingly, the constitutively active caStat5a1*6 mutant promoted senescence in fibroblasts, similar to oncogenic Ras (76, 77). It is questionable whether the more physiologic cS5F mutant can promote senescence similarly. Knock in and inducible transgenic mouse models harboring the cS5F mutation are currently established to gain further insights into persistent cytokine and growth factor signaling through oncogenic Stat5 variants. These studies in immuno-competent adult transgenic mice will be important to illuminate how persistent Stat5 protein activation can lead to development and progression of cancers.
5. STAT5 PROTEIN-PROTEIN INTERACTIONS WITH FOCUS ON THE STAT5 N-TERMINUS
The changes in chromatin structure upon binding of Stat5 proteins are induced by the N-terminal tetramerization domain, whereas the C-terminal region is responsible for transactivation and also regulates P-Y-Stat5 turnover (Figure 5) (17, 18). For protein-protein interactions, both, the N- and C-terminal domains are of particular importance. These interactions influence signal transduction and transcription (Figure 5). Putative protein binding sites with NCoA/SRC, the TUDOR domain coactivators p100 and CBP/p300 cofactors were both mapped to the N- and/or C-terminal domains of Stat5 (78, 79). The N-terminal region contains a secondary modification at threonine 92 (O-Glc-NAc glycosylation), which is believed to be crucial for high affinity interaction with CBP/p300 (Figures 3 and 5) (80). The C-terminus of Stat5a and Stat5b is serine/threonine phosphorylated, most likely increasing the acidic blob of the amphiphatic alpha-helical transactivation domain core (17, 18). This core can interact with octamer transcription factors through a short amino acid motif similar to the one found in the octamer cofactors OBF-1 (BOB) and SNAP190 (81).
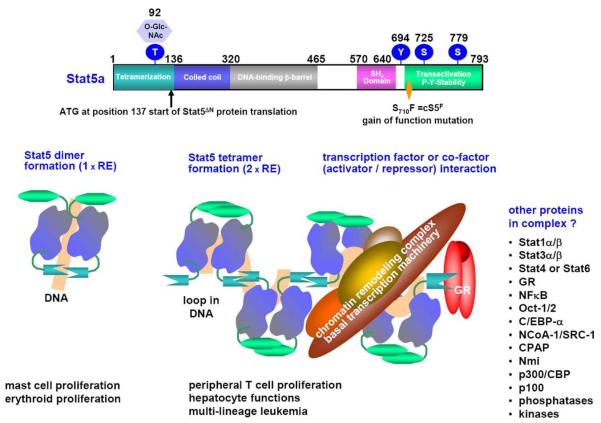
Figure 5.
Stat5 domains and the cS5F gain-of-function mutant with a model for oligomerization. Diagram based on solved Stat structures (upper part) and models of oligomer formation (lower half). Structural data are available for Stat1 (87, 89, 92), Stat3 (93), Stat4 (88, 94, 95) and Stat5a (86). DNA loop formation by Stat5 tetramers is catalyzed by the N-terminal domains of Stat5 molecules. Stat5 tetramer formation is involved in regulation of Stat5 target gene expression in normal, cytokine and/or growth factor responsive cells, but also in transformation processes (61). The association and formation of Stat5 complexes with cofactors, co-repressors or other transcription factors is critical in the decision which genes are specifically activated or repressed. Multiple other proteins interacting with Stat5a and/or Stat5b on DNA were described as illustrated to the lower right.
Additional interaction sites were mapped within the coiled-coil domain, the region regulating Stat5 nuclear versus cytoplasmic localization (48). Yeast-Two-Hybrid screens underlined the importance of the coiled-coil domain in interacting with the corepressor molecule SMRT (82) and the transactivator molecule Nmi (83). The SH2 domain of Stat5 and its critical tyrosine residue are essential for tyrosine kinase contact, dimer specificity, nuclear translocation and attack by tyrosine phosphatases in conjunction with the carboxyl terminal domain.
In summary, the very N- and C-terminal domains of Stat5 proteins have multiple functions with respect to interaction with other proteins. Both domains are exposed on the surface of Stat5 dimers or tetramers as recognized by native DNA binding assays (EMSA) or antibody supershift experiments (Figure 6) (61, 84). Only antibodies directed against N- or C-terminal epitopes recognize Stat5 in native polyacrylamide gels or are able to immunoprecipitate Stat5 proteins efficiently, irrespective of their activation status (3, 36, 85). The postulated protein surface expression of the N- and C-terminus is in line with protein biology, as the N-terminus usually folds first and the C-terminus folds at the end of protein synthesis.
Figure 6.
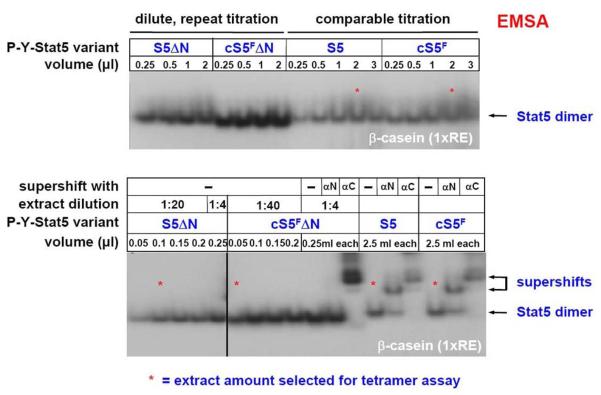
Comparison of Stat5a extracts to test them prior to tetramer stability assays. Dilutions of extracts have to be subjected to Stat5 EMSAs prior to the use of extracts in tetramer assays. After titration, four different volumes of protein extract for the indicated Stat5 variants (61) were used in tetramer assays (*). Equal activities have to be re-checked and loaded in an EMSA using the beta-casein site and Stat5 specific antisera for supershifting against the extreme N- and C-terminus to verify integrity of extracts.
Compared to other Stat family members, Stat5a and Stat5b N-terminal domains show differences in amino acid composition, limited conservation, different isoelectric points, highest aliphatic indexes and distinct grand average of hydropathy (Figure 3 and 4). Only between Stat5a and Stat5b themselves, there is 93% amino acid conservation at the N-termini (Figure 4), while those of other Stat protein family members show less than 50% conservation (Figure 3). Nevertheless, important conserved residues can be delineated throughout the entire N-terminus, in addition to unique amino acid motifs. Homologies among Stat family members with Stat5a and Stat5b N-terminal domains can be found particularly in the sequence of Stat4, followed by Stat1 and Stat3 (Figure 3). Stat1 and Stat4 N-termini were shown to assist in efficient oligomer formation, in contrast to activated Stat3, which we found to be a weak tetramer former when compared to the ability of Stat5 to tetramerize (R. Moriggl and A. Ecker, data not shown). Oligomers for Stat6 and Stat2 were not reported so far and the N terminal domains show only weak homology to the other family members.
Thus, extrapolations from these results onto other Stat family members have to be taken with caution. The Stat5 N-terminus was so far only modeled and not crystallized. So far, only unphosphorylated Stat5 dimers lacking the N- or C-terminus were crystallized (86). The structure of these unphosphorylated Stat5ΔNΔC molecules reveals a high degree of similarity to non-phosphorylated Stat1ΔNΔC dimers (87); In addition, Stat5ΔNΔC also can form antiparallel dimers. This occurs through an interface consisting of the four alpha helix bundles (coiled-coil domain) and the beta-barrel (DNA binding domain) of the dimerization partners (Figure 5) (86). Structures of isolated Stat5a and Stat5b N-termini and their interaction with other proteins in complexes (e.g. with the GR or p300/CBP) are unresolved as yet. Further elucidation of these protein-protein interactions might become especially useful as a way to alter or inhibit Stat5 functions.
The N-terminal oligomerization domain of Stat is also involved in receptor docking and is important for pre-formation of dimers in absence of tyrosine phosphorylation (88, 89) Kinetics of nuclear shuttling of mutants devoid of the Stat5 N-terminus was not studied so far but efficient chromatin binding persisted upon deletion of the Stat5b N-terminus in hepatocytes as measured by CHIP assays in vivo (3). Moreover, mutants of Stat5 deficient in tetramerization (mutation of W37A or deletion of the first 136 amino acids) could translocate to the nucleus when cytoplasmic versus nuclear extracts were analyzed (40). Interestingly, deletion of the Stat5a N-terminus led to a constitutively active Stat5aΔN dimer, whereas deletion of the Stat5b N-terminus did not (3, 85). Thus, at least the Stat5a N-terminus inhibits auto-activation in the absence of cytokines or growth factors.
Importantly, we recently discovered that the Stat5 N-terminus serves as docking platform for binding of the GR (3), a known cofactor of Stat5 proteins (90, 91). The GR could provide the AF-1 and AF-2 transactivation domains, even when C-terminally truncated Stat5 molecules (lacking their transactivation domain) are bound to DNA (91). The result could be a stable DNA-bound complex of C-terminally truncated Stat5 and GR proteins that can moderately but not fully activate target gene transcription. DNA binding or dimerization of GR proteins was not needed for GR-Stat5 protein interaction. This was shown through the use of GRdim mice (knock in mice expressing a mutated GR). The corresponding mutation resides in the second zinc finger domain, resulting in a loss of DNA binding capacity of the GR. GRdim mice have an intact GR-Stat5 interaction and do not suffer from dwarfism, in contrast to Stat5ΔHep, GRΔHep or Stat5ΔN mice that display dwarfism due to growth hormone defects (3, 85). From these results we concluded that the GR interacts with Stat5 through the N-terminus in a classic cofactor fashion.
It is evident from previous work and our current analyses of GR- and Stat5-transgenic mouse models that the molecular details of the Stat5-GR interaction are not restricted to a simple linear transcription factor model. A rather complex scenario appears to integrate positive versus negative transcriptional regulation, short- or long-term cytokine and hormone responses, dependent and interdependent signaling pathways in the cytoplasm or the nucleus (Kornfeld, Tuckermann, Friedbichler and Moriggl, unpublished data). Overall, the Stat5-GR axis is more important than previously anticipated and similar surprises might be revealed from closer studies on other Stat5-interacting proteins.
6. SIGNIFICANCE OF STAT5 TETRAMER FORMATION/OLIGOMERIZATION FOR ACTIVATION OF TARGET GENES
Further oligomeric protein-protein interactions can occur through the N-terminal domain of Stat5-containing homo-or hetero-oligomers, also called Stat5 tetramers (Figures 3 to 5). A simplified scheme of Stat5 tetramer formation is depicted in Figure 5, based on structural data of Stat family members. Detailed structural information is available for several Stat isoforms including Stat1 (87, 89, 92), Stat3 (93), Stat4 (88, 94, 95) and Stat5a (86). These 3D structure models have contributed to clarification of the molecular action and complex formation of DNA-bound Stat proteins and the mechanism for oligomerization through their N termini. These studies were either based on protein crystal structures or 3D modeling, combined with mutational analyses to verify the structural predictions. We would like to mention that the high resolution structure of the crystallized Stat4 N-terminal domain was reinterpreted. Two alternate organizations were suggested but irrespective of this reinterpretation the N-termini apparently form stable protein-protein aggregates, like two interconnecting hooks (88, 94, 95).
Up to now, Stat5 proteins were mainly analyzed as transcription factors bound to a single high affinity DNA binding element, but this does most probably not reflect the situation in vivo (Figure 5). Stat5 proteins need to find their target sequences in open chromatin within minutes, where upon binding they modify chromatin structure. As already outlined above, Stat5 proteins do not only function in the nucleus, they also contribute to PI3K-Akt activation in the cytoplasm (Figure 1). Already several years after its discovery, it was recognized that Stat5 can influence the level of target gene expression through oligomers, so called Stat5 tetramers. In this case, two weak or high affinity Stat5 response elements in the promoter allow formation/recruitment of Stat5 tetramers onto DNA. These can even be two very weak Stat5 response elements (unpublished data). Multiple DNA/protein complexes were found upon analyses of patient samples using DNA probes with two Stat5 response elements, so called tetramer sites (61). These complexes contained Stat5 proteins and probably other unknown factors displaying high chromatin-binding affinity of their own.
One reason for the appearance of such diverse Stat5 protein complexes could be that Stat5 proteins from leukemic patient samples often show N- and/or C-terminal deletions. This can be explained by the observation that, in contrast to Stat3, Stat5 proteins are highly susceptible to proteolysis (unpublished data). The complexes from patients migrated differently and distinctly on native gels, as compared to controls. For analysis, patient extracts were first adjusted to equal Stat5 dimer DNA-binding activity and then employed in tetramer assays. The result was a strong enhancement of oligomer formation in leukemic extracts when compared to controls. Thus, it is likely that also other proteins or post-translational processing of Stat5 proteins might play a role in Stat5 oligomer complex formation (61). In summary, leukemic patient samples displayed an increased abundance of Stat5 tetramer complexes, which might not only be attributed to the mere presence of sufficient amounts of Stat5 protein.
A rare phenomenon in the world of transcription factors is the fact that Stat5 proteins can alter chromatin structure through oligomerization to redirect gene transcription upon cytokine action. Oligomerization is driven through the Stat5 N-terminal domain, where the GR and p300/CBP molecules can bind as cofactors. Both, p300/CBP and the GR are known to be strong chromatin modifiers. A significant part of the changes in chromatin structure is likely to be due to Stat5 proteins themselves. Unfortunately, currently no data are available on measurements or visualizations of Stat5 in complexes with these chromatin modifying proteins.
Until some time ago, the DNA binding activity of Stat5 proteins was mainly assessed by in vitro DNA binding electromobility shift assays (EMSA), frequently using the beta-casein milk protein gene response element (Figure 6) (17, 96). More recently, chromatin immunoprecipitation (ChIP) experiments using Stat5 proteins were performed in a limited set of cell lines or cell types in vivo (3, 58, 85). No global ChIP analysis was reported for Stat5 proteins based on their DNA binding preference so far, which might facilitate the analysis of binding site selection to understand if and where Stat5 proteins bind as dimers or oligomers. Such experiments could be elegantly performed using the different available genetic mouse models, since wild type Stat5, Stat5ΔN (no tetramers) or conditional Stat5NullCreX mice would be available to study dimers and oligomers versus, as control, the results in the absence of Stat5.
Stat5 proteins were also shown to repress genes in a promoter- and (co)-repressor-dependent manner. Since not many studies have been conducted on such mechanisms of gene repression by Stat5, we will rather mention a number of the most relevant Stat5 target genes with known and significant functions in the organism. Many Stat5-induced genes have two to four high-affinity binding sites in their promoter.
Prominent direct transcriptional targets of Stat5 (97) include (i) proteins important for cell cycle progression and cellular growth, together with cytokineand growth factor-receptor signaling components (IL-2R-alpha, all three D-type cyclins, c-Myc, OSM, ALS, IGF-1, Pim kinases, epidermal growth factor receptor, prolactin receptor), (ii) tissue invasion (MMP-1, MMP-3, Spi-2.1), (iii) survival (A1, Mcl-1, Bcl-2, Bcl-xL, survivin), (iv) negative feedback inhibition in tyrosine kinase signaling pathways (CIS, Socs-1, Socs-2, Socs-3), (v) lymphocyte function (FoxP3, CD25, TCR-gamma/delta rearrangement region, perforin, lymphotoxin-alpha, Pax5, EBF, Glut1), (vi) cofactor regulation (Cited2), (vii) liver function (sexual dimorphic proteins like p450 cytochrome genes), (viii) major urinary proteins, (ix) ribosomal proteins, (x) acute phase response genes (such as alpha2-macroglobulin, HNF-6), but also (xi) genes involved in DNA damage repair (Gadd45-gamma, Rad51), (xii) reproduction or (xiii) mammary gland function and differentiation (3beta-HSD, 20alpha-HSD, alpha-, beta- and gamma-casein genes, beta-lactoglobulin, whey acidic protein). We would like to close this necessarily rudimentary list of Stat5 target genes in the hope that it can be complemented by the characterization of additional prominent members in the near future.
7. METHODS TO STUDY DNA- AND CHROMATIN-BINDING PROPERTIES OF STAT5
The first two descriptions of higher order complex formation involving Stat proteins were published in 1996, describing the capability of Stat1 and Stat4 to form tetramers via their N-termini. The same was true for higher order oligomers while bound to DNA (92, 98). The first, a technically detailed biochemical study performed in the lab of James Darnell, revealed, how purified Stat1 would bind to DNA (92). Here, for the first time in Jak-Stat research, the migration behavior of Stat tetramer complexes was shown in EMSAs. The second paper, originating in the group of Tim Hoey, recognized differential binding site occupancies through oligomerization of heterologous expression of Stat1, Stat4 or Stat5 DNA-bound complexes, using the interferon-gamma promoter and DNA-footprinting as experimental read-out (98).
Subsequent studies performed with Stat5 proteins were pioneered by the laboratory of Warren Leonard (99-101). Here, complex formation of activated Stat5a proteins and its consequences for regulation of a 3′enhancer region of the IL-2Ralpha chain gene locus were delineated. The high affinity IL-2R-alpha chain (or CD25) constitutes an essential molecule of activated or regulatory T cells, whose functions are indeed controlled through Stat5 (5, 14, 36, 59, 100). A further approach focused on the CIS promoter and the multiple high and low affinity Stat5 elements therein (102).
Important mechanistic insights resulted from studies with the Stat4 N-terminus and subsequent extrapolation to other Stat family members. For instance, it became clear that the conserved W37A mutation resulted in unstable Stat proteins and that deletion of the N-terminus resulted in absence of tyrosine phosphorylation. As already mentioned, there are significant differences in the N termini of Stat5a and Stat5b. Tyrosine phosphorylation of Stat5aΔN was persistent while that of Stat5bΔN is still inducible by cytokine (3, 61). In addition, the mutation W37A in Stat5a did not cause a chaotropic molecule, since Stat5a-W37A or Stat5aΔN were able to complement the phenotype of Stat5Null mast cells (K. Bunting, K. Friedbichler and R. Moriggl, unpublished data). Therefore, apparently, over-simplification by extrapolation of experiments performed with one particular Stat protein variant to all Stat family members can be drastically misleading. Each Stat protein, splice isoform, secondary protein modification, or proteolytically processed Stat protein might have individual chromatin binding behavior. This hypothesis is open for experimental extension and verification since additional studies on Stat5 and chromatin functions will be required in the future.
The differential tetramer formation of Stat proteins and their distinct ability to interact with specific sets of proteins leads to a great variety of transcriptional responses. Tetramer formation capability of distinct Stat family members might alter chromatin structures differentially. So far, only gene transcription rather then gene repression by Stat tetramers was reported, but the ability of Stat5 proteins to bind onto low affinity Stat5 sites should have consequences for differential cytokine responses via fine-tuning chromatin alterations.
In the following we present in detail a typical experiment using Stat5a variants with altered chromatin binding characteristic. A description of the corresponding mutants can be found in (61), and the extensive “Material and Methods” section below enables eventual follow up studies for questions regarding the “N-terminus of Stat5 and chromatin binding”:
The first experiment before conducting the actual Stat5 tetramer assay is to normalize dimer-DNA binding activity of different Stat5 extracts on an EMSA gel. A typical example is shown for dilutions of extracts from cells expressing either wt Stat5a, cS5F, or the tetramer-deficient mutants Stat5aΔN nd cS5FΔN, using the beta-casein single Stat5 response element (RE; Figure 6, upper part). This procedure has to be repeated until equal amounts of loaded Stat5 dimer activity can be observed (Figure 6, lower part). In addition, specificity of DNA binding activity in each extract needs to be assessed to verify integrity of protein samples. To this purpose, supershifts are performed using antibodies specific for epitopes in the Stat5 N- or C-terminal region (here obtained from Santa Cruz) (Figure 6, lower part). The interpretation of tetramer assays with different extracts is impossible without this titration and adjustment of DNA binding activity on a single RE.
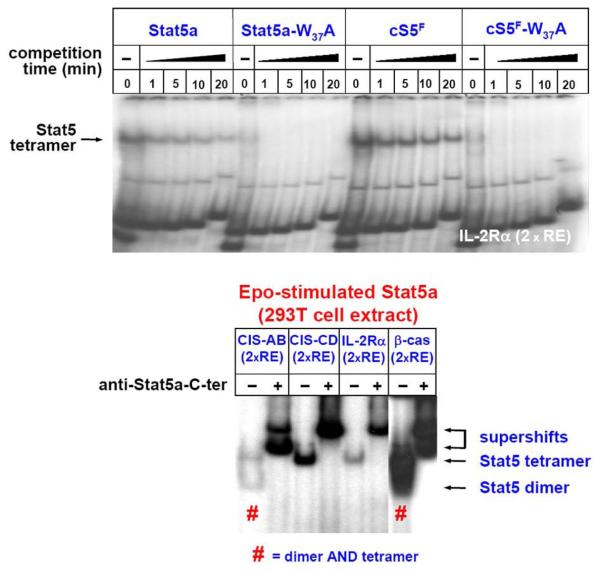
Tetramer assays with wt Stat5a, cS5F, or different tetramer-deficient mutants of Stat5a have been reported previously (61). Here, an unpublished experiment on the IL-2Ralpha chain tetramer element as a model is depicted (Figure 7, upper part). After mixing cell extracts and radioactive DNA oligonucleotide followed by brief centrifugation, equilibrium of the DNA-binding reaction is reached within two minutes at RT. The absence of unlabeled competitor DNA (0) represents the binding reaction under saturation conditions. The complexes of Stat5 proteins with labeled double stranded oligonucleotides are then competed with unlabelled double stranded oligonucleotides (2xRE) at 100fold molar excess and DNA off-on rates and oligomer complex stability are measured in EMSAs. Again, all reaction tubes containing extracts plus DNA are mixed vigorously to ensure homogenous distributions of protein, DNA and binding reaction components. Usually, a master mix for six reactions in one tube was prepared, and at specific time points equal amounts of extracts (here: 20 microliter) were loaded onto a continuously running native polyacrylamide gel. No pre-run of EMSA gels was required, but for better quality flushing of gel pockets with running buffer is recommended since chemicals remaining after acrylamide polymerization disturb the integrity of native complexes.
Figure 7.
Tetramerization of Stat5 protein variants. Tetramer formation of the indicated Stat5a variants (61) was analyzed using 293T cell extracts stimulated with Epo (50 U/ml) using the IL-2R-alpha chain tetramer element (upper part). Four different tetramer DNA elements (CIS-AB, 2xRE; CIS-CD, 2xRE; IL-2R-alpha, 2xRE beta-casein, 2xRE) were used for incubation with a Stat5a extract from Epo-activated cells to demonstrate dimer and/or tetramer formation (lower part). “#” indicates formation of both dimers and tetramers.
Quality of the extract is the crucial factor for complex formation and reduction of background. Samples with high Stat activity facilitate the analysis of oligomers and therefore, we frequently used transient transfection of Stat5 variants into 293T cells to enhance signal strength. We want to stress that also cytokine-stimulated cell lines or mouse tissues obtained after in vivo cytokine injection (growth hormone injected liver; IL-3 injected bone marrow cells; Epo stimulated erythroblasts, etc.) or samples from cancer patients work well provided sufficiently strong Stat5 activity can be detected on preliminary beta-casein bandshift assays.
In general, extracts from lymphoid cell types favor the formation of Stat5a/b higher order chromatin complexes of high affinity over dimer complexes. Until now, we did not observe tetramer complexes in myeloid cell types such as erythroid progenitors or Ba/F3 cells (R. Moriggl, unpublished). There is no straightforward explanation why samples from different hematopoietic cell types with Stat5 activity normalized on dimer sites are behaving so different in terms of biochemistry when employed in oligomerization assays. This is indeed surprising since extracts of fibroblasts or epithelial cell lines ectopically expressing Stat5a can form tetramers and also Stat5b proteins activated by growth hormone injection into mice in vivo do form efficient Stat5 oligomers.
We previously reported on the characterization of four different Stat5 tetramer elements (61) (Figure 7, lower part). Stat5a protein extracts isolated from Epo-stimulated 293T cells were incubated with these four tetramer sites, each of which contains two Stat5 REs either with high or weak affinity. Apparently, there must be alternatives in molecular composition, since three distinct DNA-binding complexes were observed with the same amount of extract and Stat5a activity (Figure 6). Two DNA elements displayed binding of Stat5 dimer- plus tetramer complexes (STD and CIS-AB), the two other elements displayed Stat5 tetramer complexes only (CD25 and CIS-CD) with extracts from Epo-stimulated 293T cells. Currently it is not understood how these differences in Stat5 oligomers bound to different Stat5 tetramer sites arise, but DNA structure conformation might be responsible (Figure 7).
8. SUMMARY AND PERSPECTIVE
The way how Stat5 proteins exert their distinct and specific functions in different cell types under various physiological and pathological conditions is still unclear to a large extent. It is possible that additional proteins functionally interacting with Stat5 still await detection or were at least not taken into sufficient consideration so far. The next years of research on Stat5 will have to address the question of composition of different protein complexes in which Stat5 proteins reside and through which they execute their specific functions. It is also open whether still other proteins interact with DNA-bound Stat5 oligomers. Unfortunately, no further reports on the chromatin binding properties of Stat5 were published.
So far the Stat5 N-terminus is known to influence three distinct signaling properties of Stat5:
The N-terminus of Stat5a blocks auto-activation by intrinsic tyrosine kinases within the cell. N-terminal deletion mutants of Stat5a are persistently tyrosine phosphorylated, possess constitutive DNA binding activity and exhibit robust transactivation potential.
The N-terminus of Stat5 is important for interaction with transcriptional cofactors like CBP and p300 and transcription factor interaction with nuclear hormone receptors like the GR.
The Stat5 N-terminus is required for tetramer formation (or oligomerization) through protein-protein interactions, which subsequently lead to higher order complex formation on chromatin. The N-termini of Stat5 stabilize two dimer interactions bound to Stat5 responsive elements in the vicinity of transcriptional regulatory regions.
Several important questions regarding the chromatin altering capability of Stat5 proteins are largely unanswered:
How efficiently and on which DNA loci can Stat5 proteins modify chromatin structure and thus transcriptional competence?
Can these structural changes be visualized?
Do other proteins interact with Stat5 in higher order DNA complexes and are these potential additional proteins cell type specific?
Are post-translational modifications of Stat5 proteins involved in altered chromatin binding or transcriptional capacity?
We currently do not understand the secondary protein modifications of Stat5 and how they influence DNA binding and complex formations on different natural DNA elements. Future work around the truly multi-facetted transcription factors and signaling mediators Stat5a and Stat5b will address their functions in a broader context.
9. MATERIALS AND METHODS
9.1. Transient transfection and preparation of whole cell extracts
Extracts of cells expressing different Stat5 variants were generated by transient transfection of 293T cells using the calcium phosphate precipitation method (4 microgram of Stat5 cDNA in a pMSCV expression plasmid plus 2 microgram EpoR cDNA in pMSCV per 10 cm dish of 293T cells, 20-50% confluency). 48 hours post transfection, cells were harvested after a 30-min stimulation with 50 units/ml rhEpo. Culture dishes were rinsed twice with ice-cold PBS and cells scraped off and pelleted by centrifugation (1 min, 6,000 g). Cells were lysed by the addition of two pellet volumes of whole cell extract buffer (20 mM Hepes, pH 7.9; 20% glycerol; 50 mM KCl; 1 mM EDTA; 1 mM DTT (Sigma); 400 mM NaCl; 5 microgram/ml leupeptin (Roche); 0.2 units/ml aprotinin (Bayer); 1 mM PMSF (Roche); 5 mM Na3VO4; 10 mM NaF; 5 mM beta-glycerophosphate) and vigorous vortexing. Samples were frozen in liquid nitrogen, followed by thawing on ice. Freeze-thaw cycles were repeated 4 times. Subsequently, extracts were centrifuged at 15.000 g at 4°C for 20 minutes, and the supernatants used for EMSAs or Western blot assays or stored at −80°C until use.
9.2. Electrophoretic mobility shift assays (EMSAs)
DNA-Oligonucleotides were annealed at equimolar concentrations (>40 microM for each oligo) in 200 microliter using annealing buffer (10x = 0.625x PCR Buffer II (Roche); 9.4 mM MgCl2) by heating up to 95°C for 10 min, followed by slowly cooling down of samples to RT and an additional incubation on ice for another 10 min. Annealed oligos were then diluted to 2.5 microM for radioactive labeling. The labeling reaction (5 picomol of annealed ds-DNA in 20 microliter, containing 10 units of poly-nucleotide-kinase (Roche) and 5 microliter of gamma-ATP 32P (Amersham; 8,000 cpm/microliter) was incubated at 37°C for 1 hour. Labeled oligo-nucleotides were purified on size exclusion columns (Micro Bio-Spin 6 chromatography columns; BioRad) and stored at −20°C until use.
For in vitro Stat5-DNA-binding assays, a 20 microliter reaction was prepared for each sample, containing 2 microliter BSA (10 mg/ml in 20 mM KPO4, 50 mM NaCl, 0.1 mM EDTA, 5% glycerol), 2 microliter poly-dI-dC (1 mg/ml; Roche), 4 microliter 5x binding buffer (50 mM Tris; 5 mM DTT; 1 mM PMSF; 0.5 mM EDTA; 25% Glycerol; 250 mM NaCl; 0.5% NPO4) and 0.5-1 microliter labeled oligo-nucleotide probe (8,000 cpm/fM). Usually, 20-30 microgram of protein extract was used per reaction.
In the analysis of patient samples, low Stat5 activities are frequently encountered. In this case, up to 60 microgram of protein extracts can be loaded. In general, a maximum of 4 microliter of whole cell extract per 20 microliter of total EMSA sample volume should not be exceeded to ensure maintenance of proper salt concentrations. Extracts were incubated for approximately 5 min (equilibrium is reached within 2 min) at RT before gel loading. For supershift analyses, protein samples were pre-incubated for >2 min on ice with antibodies / antisera.
Complexes were separated on non-denaturating 4% acrylamide gels (BioRad; acrylamide / bis-acrylamide = 29 / 1) in 0.25x TBE and run in 0.25x TBE running buffer at 200 V for about 3 hours.
In the bandshift experiments the following high affinity Stat binding sites were used (Each of the Stat5 response elements (RE) listed contains an inverted repeat highlighted in bold). 1x response element for dimer assays: beta-casein 5′-3′ AGATTTCTAGGAATTCAATCC (binds Stat5/6 with high affinity, and Stat1/3/4 with very low affinity); 2x response elements for tetramer assays: CIS-AB (2xRE):5′-3′ GAGTTTTCCTGGAAAAGTTCTTGGAA CIS-CD (2xRE):5′-3′ CGCGGTTCTAGGAAGATGAGGCTTCCGGGAAGGGCT; IL-2R-alpha (2xRE): 5′-3′ GTTTCTTCTGAGAAGTACCAGACATTTCTGATAAGAGAG; beta-casein (2xRE): 5′-3′ AGATTTCTAGGAATTCAATCTTCTAGGAATTCAATC
9.3. Tetramer assays
For Stat5 oligomer assays, a classical approach (adapted from (92)) was used, relying on the stability of pre-formed DNA-Stat5 protein complexes on gamma-ATP 32P-labeled Stat5 tetramer elements (see 2xRE above).
For Stat5 tetramer assays, the volume was scaled up to 120 microliter. The binding reaction was finished 2 min after gentle mixing, and the time point 0 was loaded (i.e. 20 microliter of the labeled tetramer assay mixture loaded onto the gel before applying voltage). Subsequently, 10 microliter of unlabeled competitor oligo-nucleotide were added, the reaction mixed, centrifuged and 20 microliter loaded after 1, 5, 10 and 20 minute (s) onto the gel continuously running at a constant 200 V. The cold competitor DNA was used in 100x molar excess over the radioactive labeled oligo. The use of large gels (15 cm × 20 cm, 2 mm thickness) facilitates handling significantly. Electrophoresis was continued until the orange-G dye front, usually loaded in an empty lane together with free probe, reached the lowest 5 cm of the gel. Orange-G migrates comparable to an oligo of tRNA size, which is close to the length of double stranded annealed Stat REs. After electrophoresis, the gel was transferred to chromatography paper (Whatman), dried on a vacuum gel drier at 80°C for two hours and subjected to autoradiography. Coated films (Kodak BioMax MR) were exposed over night or longer at −80°C.
ACKNOWLEDGMENTS
This work was supported by grant SFB-F28 from the Austrian Basic Research Funds (FWF) to R.M., J.-W.K., K.F., A.H., B.K., H.B., V.S., M.M. and E.W.M. and FWF grant WK-001 to F.G. and M.K.
REFERENCES
- 1.Cui Y, Riedlinger G, Miyosh K, Tang W, Li C, Deng CX, Robinson GW, Hennighausen L. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol. 2004;24:8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dolznig H, Grebien F, Deiner EM, Stangl K, Kolbus A, Habermann B, Kerenyi MA, Kieslinger M, Moriggl R, Beug H, Mullner EW. Erythroid progenitor renewal versus differentiation: genetic evidence for cell autonomous, essential functions of EpoR, Stat5 and the GR. Oncogene. 2006;25:2890–2900. doi: 10.1038/sj.onc.1209308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engblom D, Kornfeld JW, Schwake L, Tronche F, Reimann A, Beug H, Hennighausen L, Moriggl R, Schutz G. Direct glucocorticoid receptor-Stat5 interaction in hepatocytes controls body size and maturation-related gene expression. Genes Dev. 2007;21:1157–1162. doi: 10.1101/gad.426007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gatzka M, Piekorz R, Moriggl R, Rawlings J, Ihle JN. A role for STAT5A/B in protection of peripheral T-lymphocytes from postactivation apoptosis: insights from gene expression profiling. Cytokine. 2006;34:143–154. doi: 10.1016/j.cyto.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Hoelbl A, Kovacic B, Kerenyi MA, Simma O, Warsch W, Cui Y, Beug H, Hennighausen L, Moriggl R, Sexl V. Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation. Blood. 2006;107:4898–4906. doi: 10.1182/blood-2005-09-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kieslinger M, Woldman I, Moriggl R, Hofmann J, Marine JC, Ihle JN, Beug H, Decker T. Antiapoptotic activity of Stat5 required during terminal stages of myeloid differentiation. Genes Dev. 2000;14:232–244. [PMC free article] [PubMed] [Google Scholar]
- 7.Li G, Wang Z, Zhang Y, Kang Z, Haviernikova E, Cui Y, Hennighausen L, Moriggl R, Wang D, Tse W, Bunting KD. STAT5 requires the N-domain to maintain hematopoietic stem cell repopulating function and appropriate lymphoid-myeloid lineage output. Exp Hematol. 2007;35:1684–1694. doi: 10.1016/j.exphem.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X, Robinson GW, Hennighausen L. Activation of Stat5a and Stat5b by tyrosine phosphorylation is tightly linked to mammary gland differentiation. Mol Endocrinl. 1996;10:1496–1506. doi: 10.1210/mend.10.12.8961260. [DOI] [PubMed] [Google Scholar]
- 9.Schwaller J, Parganas E, Wang D, Cain D, Aster JC, Williams IR, Lee CK, Gerthner R, Kitamura T, Frantsve J. Stat5 is essential for the myelo- and lymphoproliferative disease induced by TEL/JAK2. Mol Cel. 2000;6:693–704. doi: 10.1016/s1097-2765(00)00067-8. [DOI] [PubMed] [Google Scholar]
- 10.Sims NA, Clement-Lacroix P, Da Ponte F, Bouali Y, Binart N, Moriggl R, Goffin V, Coschigano K, Gaillard-Kelly M, Kopchick J. Bone homeostasis in growth hormone receptor-null mice is restored by IGF-I but independent of Stat5. J Clin Invest. 2000;106:1095–1103. doi: 10.1172/JCI10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Socolovsky M, Fallon AE, Wang S, Brugnara C, Lodish HF. Fetal anemia and apoptosis of red cell progenitors in Stat5a−/−5b−/− mice: a direct role for Stat5 in Bcl-X (L) induction. Cell. 1999;98:181–191. doi: 10.1016/s0092-8674(00)81013-2. [DOI] [PubMed] [Google Scholar]
- 12.Teglund S, McKay C, Schuetz E, van Deursen JM, Stravopodis D, Wang D, Brown M, Bodner S, Grosveld G, Ihle JN. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 13.Udy GB, Towers RP, Snell RG, Wilkins RJ, Park SH, Ram PA, Waxman DJ, Davey HW. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci U S A. 1997;94:7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao Z, Kanno Y, Kerenyi MA, Stephens G, Durant L, Watford WT, Laurence A, Robinson GW, Shevach EM, Moriggl R, Hennighausen L, Wu C, O'Shea JJ. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–4375. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kazansky AV, Spencer DM, Greenberg NM. Activation of signal transducer and activator of transcription 5 is required for progression of autochthonous prostate cancer: evidence from the transgenic adenocarcinoma of the mouse prostate system. Cancer Res. 2003;63:8757–8762. [PubMed] [Google Scholar]
- 16.Liu X, Robinson GW, Gouilleux F, Groner B, Hennighausen L. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc Natl Acad Sci U S A. 1995;92:8831–8835. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moriggl R, Gouilleux-Gruart V, Jahne R, Berchtold S, Gartmann C, Liu X, Hennighausen L, Sotiropoulos A, Groner B, Gouilleux F. Deletion of the carboxyl-terminal transactivation domain of MGF-Stat5 results in sustained DNA binding and a dominant negative phenotype. Mol Cell Biol. 1996;16:5691–5700. doi: 10.1128/mcb.16.10.5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang D, Moriggl R, Stravopodis D, Carpino N, Marine JC, Teglund S, Feng J, Ihle JN. A small amphipathic alpha-helical region is required for transcriptional activities and proteasome-dependent turnover of the tyrosinephosphorylated Stat5. Embo J. 2000;19:392–399. doi: 10.1093/emboj/19.3.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang D, Stravopodis D, Teglund S, Kitazawa J, Ihle JN. Naturally occurring dominant negative variants of Stat5. Mol Cell Biol. 1996;16:6141–6148. doi: 10.1128/mcb.16.11.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramos HL, O'Shea JJ, Watford WT. STAT5 isoforms: controversies and clarifications. Biochem J. 2007;404:e1–2. doi: 10.1042/BJ2007420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark DE, Williams CC, Duplessis TT, Moring KL, Notwick AR, Long W, Lane WS, Beuvink I, Hynes NE, Jones FE. ERBB4/HER4 potentiates STAT5A transcriptional activity by regulating novel STAT5A serine phosphorylation events. J Biol Chem. 2005;280:24175–24180. doi: 10.1074/jbc.M414044200. [DOI] [PubMed] [Google Scholar]
- 22.Weaver AM, Silva CM. S731 in the transactivation domain modulates STAT5b activity. Biochem Biophys Res Commun. 2007;362:1026–1030. doi: 10.1016/j.bbrc.2007.08.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levy DE, Lee CK. What does Stat3 do? J Clin Invest. 2002;109:1143–1148. doi: 10.1172/JCI15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 25.Iavnilovitch E, Groner B, Barash I. Overexpression and forced activation of stat5 in mammary gland of transgenic mice promotes cellular proliferation, enhances differentiation, and delays postlactational apoptosis. Mol Cancer Res. 2002;1:32–47. [PubMed] [Google Scholar]
- 26.Iavnilovitch E, Eilon T, Groner B, Barash I. Expression of a carboxy terminally truncated Stat5 with no transactivation domain in the mammary glands of transgenic mice inhibits cell proliferation during pregnancy, delays onset of milk secretion, and induces apoptosis upon involution. Mol Reprod Dev. 2006;73:841–849. doi: 10.1002/mrd.20479. [DOI] [PubMed] [Google Scholar]
- 27.Sultan AS, Xie J, LeBaron MJ, Ealley EL, Nevalainen MT, Rui H. Stat5 promotes homotypic adhesion and inhibits invasive characteristics of human breast cancer cells. Oncogene. 2005;24:746–760. doi: 10.1038/sj.onc.1208203. [DOI] [PubMed] [Google Scholar]
- 28.Kelly JA, Spolski R, Kovanen PE, Suzuki T, Bollenbacher J, Pise-Masison CA, Radonovich MF, Lee S, Jenkins NA, Copeland NG, Morse HC, 3rd, Leonhard WJ. Stat5 synergizes with T cell receptor/antigen stimulation in the development of lymphoblastic lymphoma. J Exp Med. 2003;198:79–89. doi: 10.1084/jem.20021548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee TK, Man K, Poon RT, Lo CM, Yuen AP, Ng IO, Ng KT, Leonard W, Fan ST. Signal transducers and activators of transcription 5b activation enhances hepatocellular carcinoma aggressiveness through induction of epithelial-mesenchymal transition. Cancer Res. 2006;66:9948–9956. doi: 10.1158/0008-5472.CAN-06-1092. [DOI] [PubMed] [Google Scholar]
- 30.Leong PL, Xi S, Drenning SD, Dyer KF, Wentzel AL, Lerner EC, Smithgall TE, Grandis JR. Differential function of STAT5 isoforms in head and neck cancer growth control. Oncogene. 2002;21:2846–2853. doi: 10.1038/sj.onc.1205385. [DOI] [PubMed] [Google Scholar]
- 31.Sanchez-Ceja SG, Reyes-Maldonado E, Vazquez-Manriquez ME, Lopez-Luna JJ, Belmont A, Gutierrez-Castellanos S. Differential expression of STAT5 and Bcl-xL, and high expression of Neu and STAT3 in non-small-cell lung carcinoma. Lung Cancer. 2006;4:163–168. doi: 10.1016/j.lungcan.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 32.Tan SH, Dagvadorj A, Shen F, Gu L, Liao Z, Abdulghani J, Zhang Y, Gelmann EP, Zellweger T, Culig Z, Visakorpi T, Bubendorf L, Kirken RA, Narras J, Nevalainen MT. Transcription factor Stat5 synergizes with androgen receptor in prostate cancer cells. Cancer Res. 2008;68:236–248. doi: 10.1158/0008-5472.CAN-07-2972. [DOI] [PubMed] [Google Scholar]
- 33.Tsuruyama T, Nakamura T, Jin G, Ozeki M, Yamada Y, Hiai H. Constitutive activation of Stat5a by retrovirus integration in early pre-B lymphomas of SL/Kh strain mice. Proc Natl Acad Sci U S A. 2002;99:8253–8258. doi: 10.1073/pnas.112202899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burchill MA, Goetz CA, Prlic M, O'Neil JJ, Harmon IR, Bensinger SJ, Turka LA, Brennan P, Jameson SC, Farrar MA. Distinct effects of STAT5 activation on CD4+ and CD8+ T cell homeostasis: development of CD4+CD25+ regulatory T cells versus CD8+ memory T cells. J Immunol. 2003;171:5853–5864. doi: 10.4049/jimmunol.171.11.5853. [DOI] [PubMed] [Google Scholar]
- 35.Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol. 2007;178:280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- 36.Moriggl R, Sexl C, Piekorz R, Topham D, Ihle JN. Stat5 activation is uniquely associated with cytokine signaling in peripheral T cells. Immunity. 1999;11:225–230. doi: 10.1016/s1074-7613(00)80097-7. [DOI] [PubMed] [Google Scholar]
- 37.Moriggl R, Topham DJ, Teglund S, Sexl V, McKay C, Wang D, Hoffmeyer A, van Deursen J, Sangster MY, Bunting KD, Grosveld GC, Ihle JN. Stat5 is required for IL-2-induced cell cycle progression of peripheral T cells. Immunity. 1999;10:249–259. doi: 10.1016/s1074-7613(00)80025-4. [DOI] [PubMed] [Google Scholar]
- 38.Staerk J, Lacout C, Sato T, Smith SO, Vainchenker W, Constantinescu SN. An amphipathic motif at the transmembrane-cytoplasmic junction prevents autonomous activation of the thrombopoietin receptor. Blood. 2006;107:1864–1871. doi: 10.1182/blood-2005-06-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor DK, Walsh PT, LaRosa DF, Zhang J, Burchill MA, Farrar MA, Turka LA. Constitutive activation of STAT5 supersedes the requirement for cytokine and TCR engagement of CD4+ T cells in steady-state homeostasis. J Immunol. 2006;177:2216–2223. doi: 10.4049/jimmunol.177.4.2216. [DOI] [PubMed] [Google Scholar]
- 40.Harir N, Pecquet C, Kerenyi MA, Sonneck K, Kovacic B, Nyga R, Brevet M, Dhennin T, Gouilleux-Gruart V, Beug H, Valent P, Lassoued K, Moriggl R, Gouilleux F. Constitutive activation of Stat5 promotes its cytoplasmic localization and association with PI3-kinase in myeloid leukemias. Blood. 2007;109:1678–1686. doi: 10.1182/blood-2006-01-029918. H. [DOI] [PubMed] [Google Scholar]
- 41.Nyga R, Pecquet C, Harir N, Gu H, Dhennin-Duthille I, Regnier A, Gouilleux-Gruart V, Lassoued K, Gouilleux F. Activated STAT5 proteins induce activation of the PI 3-kinase/Akt and Ras/MAPK pathways via the Gab2 scaffolding adapter. Biochem J. 2005;390:359–366. doi: 10.1042/BJ20041523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Diaz-Flores E, Li G, Wang Z, Kang Z, Haviernikova E, Rowe S, Qu CK, Tse W, Shannon KM, Bunting KD. Abnormal hematopoiesis in Gab2 mutant mice. Blood. 2007;110:116–124. doi: 10.1182/blood-2006-11-060707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bunting KD. STAT5 signaling in normal and pathologic hematopoiesis. Front Biosci. 2007;12:2807–2820. doi: 10.2741/2274. [DOI] [PubMed] [Google Scholar]
- 44.Bunting KD, Bradley HL, Hawley TS, Moriggl R, Sorrentino BP, Ihle JN. Reduced lymphomyeloid repopulating activity from adult bone marrow and fetal liver of mice lacking expression of STAT5. Blood. 2002;99:479–487. doi: 10.1182/blood.v99.2.479. [DOI] [PubMed] [Google Scholar]
- 45.Lockyer HM, Tran E, Nelson BH. STAT5 is essential for Akt/p70S6 kinase activity during IL-2-induced lymphocyte proliferation. J Immunol. 2007;179:5301–5308. doi: 10.4049/jimmunol.179.8.5301. [DOI] [PubMed] [Google Scholar]
- 46.Wofford JA, Wieman HL, Jacobs SR, Zhao Y, Rathmell JC. IL-7 promotes Glut1 trafficking and glucose uptake via STAT5-mediated activation of Akt to support T cell survival. Blood. 2007 doi: 10.1182/blood-2007-06-096297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herrmann A, Vogt M, Monnigmann M, Clahsen T, Sommer U, Haan S, Poli V, Heinrich PC, Muller-Newen G. Nucleocytoplasmic shuttling of persistently activated STAT3. J Cell Sci. 2007;120:3249–3261. doi: 10.1242/jcs.03482. [DOI] [PubMed] [Google Scholar]
- 48.Iyer J, Reich NC. Constitutive nuclear import of latent and activated STAT5a by its coiled coil domain. Faseb J. 2007 doi: 10.1096/fj.07-8965com. [DOI] [PubMed] [Google Scholar]
- 49.Bunting KD, Xie XY, Warshawsky I, His ED. Cytoplasmic localization of phosphorylated STAT5 in human acute myeloid leukemia is inversely correlated with Flt3-ITD. Blood. 2007;110:2775–2776. doi: 10.1182/blood-2007-05-090969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sallmyr A, Fan J, Datta K, Kim KT, Grosu D, Shapiro P, Small D, Rassool F. Internal tandem duplication of FLT3 (FLT3/ITD) induces increased ROS production, DNA damage and misrepair: implications for poor prognosis in AML. Blood. 2008 doi: 10.1182/blood-2007-05-092510. [DOI] [PubMed] [Google Scholar]
- 51.Ripperger JA, Fritz S, Richter K, Hocke GM, Lottspeich F, Fey GH. Transcription factors Stat3 and Stat5b are present in rat liver nuclei late in an acute phase response and bind interleukin-6 response elements. J Biol Chem. 1995;270:29998–30006. doi: 10.1074/jbc.270.50.29998. [DOI] [PubMed] [Google Scholar]
- 52.Haspel RL, Salditt-Georgieff M, Darnell JE., Jr The rapid inactivation of nuclear tyrosine phosphorylated Stat1 depends upon a protein tyrosine phosphatase. Embo J. 1996;15:6262–6268. [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Y, Dai X, Haas AL, Wen R, Wang D. Proteasome-dependent down-regulation of activated Stat5A in the nucleus. Blood. 2006;108:566–574. doi: 10.1182/blood-2005-12-4777. D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Q, Wang HY, Liu X, Wasik MA. STAT5A is epigenetically silenced by the tyrosine kinase NPM1-ALK and acts as a tumor suppressor by reciprocally inhibiting NPM1-ALK expression. Nat Med. 2007;13:1341–1348. doi: 10.1038/nm1659. [DOI] [PubMed] [Google Scholar]
- 55.Chatterjee-Kishore M, Wright KL, Ting JP, Stark GR. How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. Embo J. 2000;19:4111–4122. doi: 10.1093/emboj/19.15.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang J, Chatterjee-Kishore M, Staugaitis SM, Nguyen H, Schlessinger K, Levy DE, Stark GR. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res. 2005;65:939–947. [PubMed] [Google Scholar]
- 57.Yang J, Liao X, Agarwal ML, Barnes L, Auron PE, Stark GR. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev. 2007;21:1396–1408. doi: 10.1101/gad.1553707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.LeBaron MJ, Xie J, Rui H. Evaluation of genome-wide chromatin library of Stat5 binding sites in human breast cancer. Mol Cancer. 2005;4:6. doi: 10.1186/1476-4598-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao Z, Cui Y, Watford WT, Bream JH, Yamaoka K, Hissong BD, Li D, Durum SK, Jiang W, Bhandoola A, Hennighausen L, O'Shea JJ. Stat5a/b are essential for normal lymphoid development and differentiation. Proc Natl Acad Sci U S A. 2006;103:1000–1005. doi: 10.1073/pnas.0507350103. A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stoecklin E, Wissler M, Gouilleux F, Groner B. Functional interactions between Stat5 and the glucocorticoid receptor. Nature. 1996;383:726–728. doi: 10.1038/383726a0. [DOI] [PubMed] [Google Scholar]
- 61.Moriggl R, Sexl V, Kenner L, Duntsch C, Stangl K, Gingras S, Hoffmeyer A, Bauer A, Piekorz R, Wang D, Bunting KD, Wagner EF, Sonneck K, Valent P, Ihle JN, Beug H. Stat5 tetramer formation is associated with leukemogenesis. Cancer Cell. 2005;7:87–99. doi: 10.1016/j.ccr.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 62.Liu X, Robinson GW, Wagner KU, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 63.Murray PJ. The JAK-STAT signaling pathway: input and output integration. J Immunol. 2007;178:2623–2629. doi: 10.4049/jimmunol.178.5.2623. [DOI] [PubMed] [Google Scholar]
- 64.Dai X, Chen Y, Di L, Podd A, Li G, Bunting KD, Hennighausen L, Wen R, Wang D. Stat5 is essential for early B cell development but not for B cell maturation and function. J Immunol. 2007;179:1068–1079. doi: 10.4049/jimmunol.179.2.1068. L. [DOI] [PubMed] [Google Scholar]
- 65.Cui Y, Hosui A, Sun R, Shen K, Gavrilova O, Chen W, Cam MC, Gao B, Robinson GW, Hennighausen L. Loss of signal transducer and activator of transcription 5 leads to hepatosteatosis and impaired liver regeneration. Hepatology. 2007 doi: 10.1002/hep.21713. [DOI] [PubMed] [Google Scholar]
- 66.Lee JY, Gavrilova O, Davani B, Na R, Robinson GW, Hennighausen L. The transcription factors Stat5a/b are not required for islet development but modulate pancreatic beta-cell physiology upon aging. Biochim Biophys Acta. 2007 doi: 10.1016/j.bbamcr.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klover P, Hennighausen L. Postnatal body growth is dependent on the transcription factors signal transducers and activators of transcription 5a/b in muscle: a role for autocrine/paracrine insulin-like growth factor I. Endocrinology. 2007;148:1489–1497. doi: 10.1210/en.2006-1431. [DOI] [PubMed] [Google Scholar]
- 68.Imada K, Bloom ET, Nakajima H, Horvath-Arcidiacono JA, Udy GB, Davey HW, Leonard WJ. Stat5b is essential for natural killer cell-mediated proliferation and cytolytic activity. J Exp Med. 1998;188:2067–2074. doi: 10.1084/jem.188.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grebien F, Kerenyi MA, Kovacic B, Kolbe T, Becker V, Dolznig H, Pfeffer K, Klingmuller U, Muller H, Beug H, Mullner EW, Moriggl R. Stat5 activation enables erythropoiesis in the absence of EpoR and Jak2. Blood. 2008 doi: 10.1182/blood-2007-07-102848. Epub ahead. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fievez L, Desmet C, Henry E, Pajak B, Hegenbarth S, Garze V, Bex F, Jaspar F, Boutet P, Gillet L, Vanderplasschen A, Knolle PA, Leo O, Moser M, Lekeux P, Bureau F. STAT5 is an ambivalent regulator of neutrophil homeostasis. PLoS ONE. 2007;2:e727. doi: 10.1371/journal.pone.0000727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barnstein BO, Li G, Wang Z, Kennedy S, Chalfant C, Nakajima H, Bunting KD, Ryan JJ. Stat5 expression is required for IgE-mediated mast cell function. J Immunol. 2006;177:3421–3426. doi: 10.4049/jimmunol.177.5.3421. G. [DOI] [PubMed] [Google Scholar]
- 72.Zhu Y, Chen L, Huang Z, Alkan S, Bunting KD, Wen R, Wang D, Huang H. Cutting edge: IL-5 primes Th2 cytokine-producing capacity in eosinophils through a STAT5-dependent mechanism. J Immunol. 2004;173:2918–2922. doi: 10.4049/jimmunol.173.5.2918. [DOI] [PubMed] [Google Scholar]
- 73.Berchtold S, Moriggl R, Gouilleux F, Silvennoinen O, Beisenherz C, Pfitzner E, Wissler M, Stocklin E, Groner B. Cytokine receptor-independent, constitutively active variants of STAT5. J Biol Chem. 1997;272:30237–30243. doi: 10.1074/jbc.272.48.30237. [DOI] [PubMed] [Google Scholar]
- 74.Onishi M, Nosaka T, Misawa K, Mui AL, Gorman D, McMahon M, Miyajima A, Kitamura T. Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Mol Cell Biol. 1998;18:3871–3879. doi: 10.1128/mcb.18.7.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Woelfle J, Billiard J, Rotwein P. Acute control of insulin-like growth factor-I gene transcription by growth hormone through Stat5b. J Biol Chem. 2003;278:22696–22702. doi: 10.1074/jbc.M301362200. [DOI] [PubMed] [Google Scholar]
- 76.Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21:43–48. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mallette FA, Gaumont-Leclerc MF, Huot G, Ferbeyre G. Myc down-regulation as a mechanism to activate the Rb pathway in STAT5A-induced senescence. J Biol Chem. 2007;282:34938–34944. doi: 10.1074/jbc.M707074200. [DOI] [PubMed] [Google Scholar]
- 78.Paukku K, Yang J, Silvennoinen O. Tudor and nuclease-like domains containing protein p100 function as coactivators for signal transducer and activator of transcription 5. Mol Endocrinol. 2003;17:1805–1814. doi: 10.1210/me.2002-0256. [DOI] [PubMed] [Google Scholar]
- 79.Pfitzner E, Jahne R, Wissler M, Stoecklin E, Groner B. p300/CREB-binding protein enhances the prolactin-mediated transcriptional induction through direct interaction with the transactivation domain of Stat5, but does not participate in the Stat5-mediated suppression of the glucocorticoid response. Mol Endocrinol. 1998;12:1582–1593. doi: 10.1210/mend.12.10.0180. [DOI] [PubMed] [Google Scholar]
- 80.Gewinner C, Hart G, Zachara N, Cole R, Beisenherz-Huss C, Groner B. The coactivator of transcription CREB-binding protein interacts preferentially with the glycosylated form of Stat5. J Biol Chem. 2004;279:3563–3572. doi: 10.1074/jbc.M306449200. [DOI] [PubMed] [Google Scholar]
- 81.Magne S, Caron D, Charon M, Rouyez MC, Dusanter-Fourt I. STAT5 and Oct-1 form a stable complex that modulates cyclin D1 expression. Mol Cell Biol. 2003;23:8934–8945. doi: 10.1128/MCB.23.24.8934-8945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nakajima H, Brindle PK, Handa M, Ihle JN. Functional interaction of STAT5 and nuclear receptor corepressor SMRT: implications in negative regulation of STAT5-dependent transcription. Embo J. 2001;20:6836–6844. doi: 10.1093/emboj/20.23.6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu M, John S, Berg M, Leonard WJ. Functional association of Nmi with Stat5 and Stat1 in IL-2- and IFNgamma-mediated signaling. Cell. 1999;96:121–130. doi: 10.1016/s0092-8674(00)80965-4. [DOI] [PubMed] [Google Scholar]
- 84.Moriggl R, Kristofic C, Kinzel B, Volarevic S, Groner B, Brinkmann V. Activation of STAT proteins and cytokine genes in human Th1 and Th2 cells generated in the absence of IL-12 and IL-4. J Immunol. 1998;160:3385–3392. [PubMed] [Google Scholar]
- 85.Tronche F, Opherk C, Moriggl R, Kellendonk C, Reimann A, Schwake L, Reichardt HM, Stangl K, Gau D, Hoeflich A, Beug H, Schmid W, Schutz G. Glucocorticoid receptor function in hepatocytes is essential to promote postnatal body growth. Genes Dev. 2004;18:492–497. doi: 10.1101/gad.284704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Neculai D, Neculai AM, Verrier S, Straub K, Klumpp K, Pfitzner E, Becker S. Structure of the unphosphorylated STAT5a dimer. J Biol Chem. 2005;280:40782–40787. doi: 10.1074/jbc.M507682200. [DOI] [PubMed] [Google Scholar]
- 87.Mao X, Ren Z, Parker GN, Sondermann H, Pastorello MA, Wang W, McMurray JS, Demeler B, Darnell JE, Jr, Chen X. Structural bases of unphosphorylated STAT1 association and receptor binding. Mol Cell. 2005;17:761–771. doi: 10.1016/j.molcel.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 88.Ota N, Brett TJ, Murphy TL, Fremont DH, Murphy KM. N-domain-dependent nonphosphorylated STAT4 dimers required for cytokine-driven activation. Nat Immunol. 2004;5:208–215. doi: 10.1038/ni1032. [DOI] [PubMed] [Google Scholar]
- 89.Zhong M, Henriksen MA, Takeuchi K, Schaefer O, Liu B, ten Hoeve J, Ren Z, Mao X, Chen X, Shuai K, Darnell JE., Jr Implications of an antiparallel dimeric structure of nonphosphorylated STAT1 for the activation-inactivation cycle. Proc Natl Acad Sci U S A. 2005;102:3966–3971. doi: 10.1073/pnas.0501063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stocklin E, Wissler M, Gouilleux F, Groner B. Functional interactions between Stat5 and the glucocorticoid receptor. Nature. 1996;383:726–728. doi: 10.1038/383726a0. [DOI] [PubMed] [Google Scholar]
- 91.Stoecklin E, Wissler M, Moriggl R, Groner B. Specific DNA binding of Stat5, but not of glucocorticoid receptor, is required for their functional cooperation in the regulation of gene transcription. Mol Cell Biol. 1997;17:6708–6716. doi: 10.1128/mcb.17.11.6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vinkemeier U, Cohen SL, Moarefi I, Chait BT, Kuriyan J, Darnell JE., Jr DNA binding of in vitro activated Stat1 alpha, Stat1 beta and truncated Stat1: interaction between NH2-terminal domains stabilizes binding of two dimers to tandem DNA sites. Embo J. 1996;15:5616–5626. [PMC free article] [PubMed] [Google Scholar]
- 93.Becker S, Corthals GL, Aebersold R, Groner B, Muller CW. Expression of a tyrosine phosphorylated, DNA binding Stat3beta dimer in bacteria. FEBS Lett. 1998;441:141–147. doi: 10.1016/s0014-5793(98)01543-9. [DOI] [PubMed] [Google Scholar]
- 94.Chen X, Bhandari R, Vinkemeier U, Van Den Akker F, Darnell JE, Jr., Kuriyan J. A reinterpretation of the dimerization interface of the N-terminal domains of STATs. Protein Sci. 2003;12:361–365. doi: 10.1110/ps.0218903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vinkemeier U, Moarefi I, Darnell JE, Jr., Kuriyan J. Structure of the amino-terminal protein interaction domain of STAT-4. Science. 1998;279:1048–1052. doi: 10.1126/science.279.5353.1048. [DOI] [PubMed] [Google Scholar]
- 96.Schmitt-Ney M, Happ B, Ball RK, Groner B. Developmental and environmental regulation of a mammary gland-specific nuclear factor essential for transcription of the gene encoding beta-casein. Proc Natl Acad Sci U S A. 1992;89:3130–3134. doi: 10.1073/pnas.89.7.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ehret GB, Reichenbach P, Schindler U, Horvath CM, Fritz S, Nabholz M, Bucher P. DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J Biol Chem. 2001;276:6675–6688. doi: 10.1074/jbc.M001748200. P. [DOI] [PubMed] [Google Scholar]
- 98.Xu X, Sun YL, Hoey T. Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science. 1996;273:794–797. doi: 10.1126/science.273.5276.794. [DOI] [PubMed] [Google Scholar]
- 99.John S, Vinkemeier U, Soldaini E, Darnell JE, Jr., Leonard WJ. The significance of tetramerization in promoter recruitment by Stat5. Mol Cell Biol. 1999;19:1910–1918. doi: 10.1128/mcb.19.3.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim HP, Kelly J, Leonard WJ. The basis for IL-2-induced IL-2 receptor alpha chain gene regulation: importance of two widely separated IL-2 response elements. Immunity. 2001;15:159–172. doi: 10.1016/s1074-7613(01)00167-4. [DOI] [PubMed] [Google Scholar]
- 101.Soldaini E, John S, Moro S, Bollenbacher J, Schindler U, Leonard WJ. DNA binding site selection of dimeric and tetrameric Stat5 proteins reveals a large repertoire of divergent tetrameric Stat5a binding sites. Mol Cell Biol. 2000;20:389–401. doi: 10.1128/mcb.20.1.389-401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Verdier F, Rabionet R, Gouilleux F, Beisenherz-Huss C, Varlet P, Muller O, Mayeux P, Lacombe C, Gisselbrecht S, Chretien S. A sequence of the CIS gene promoter interacts preferentially with two associated STAT5A dimers: a distinct biochemical difference between STAT5A and STAT5B. Mol Cell Biol. 1998;18:5852–5860. doi: 10.1128/mcb.18.10.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Joliot V, Cornier F, Medyouf H, Alcalde H, Ghysdael J. Constitutive STAT5 activation specifically cooperates with the loss of p53 function in B-cell lymphomagenesis. Oncogene. 2006;25:4573–4584. doi: 10.1038/sj.onc.1209480. [DOI] [PubMed] [Google Scholar]