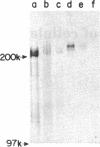
Abstract
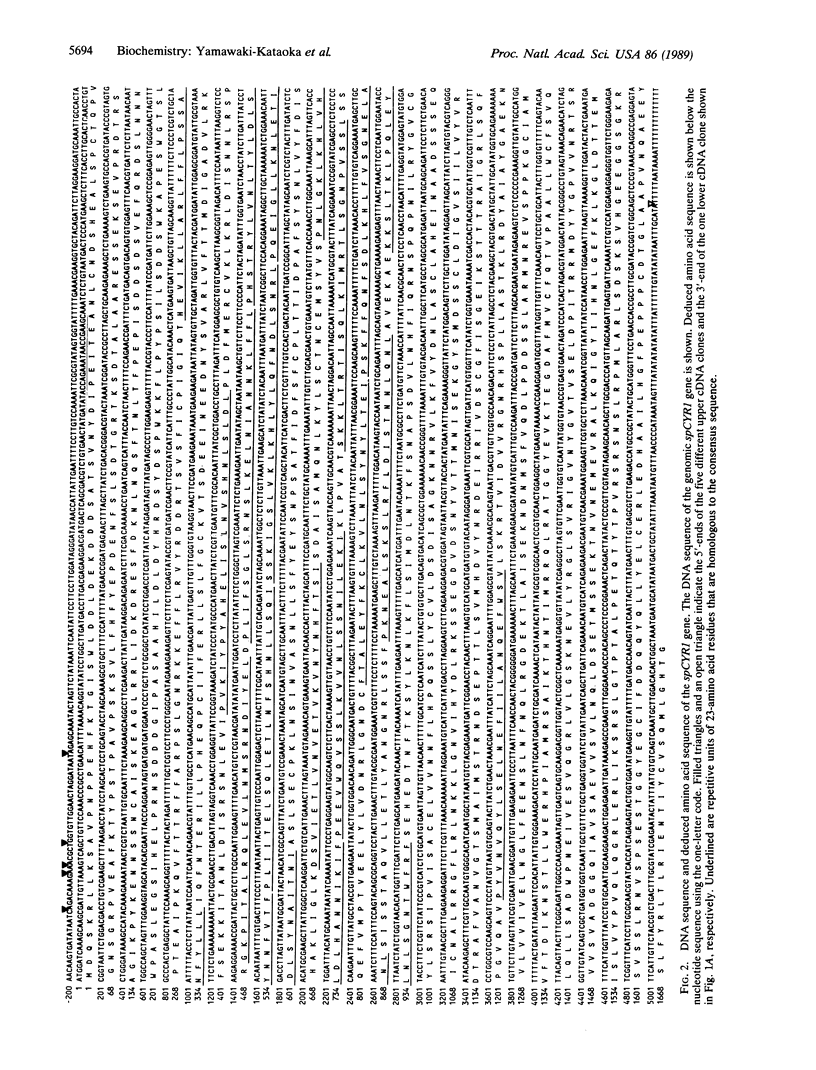
A Schizosaccharomyces pombe gene encoding adenylate cyclase has been cloned by cross-hybridization with the Saccharomyces cerevisiae adenylate cyclase gene. The protein encoded consists of 1692 amino acids and has adenylate cyclase activity that cannot be activated by the Sa. cerevisiae RAS2 protein. Sc. pombe cyclase has a high degree of homology (approximately 60%) with the catalytic domain of Sa. cerevisiae cyclase precisely mapped by a gene-deletion analysis. A 25-40% identity is observed throughout the middle segments of approximately 1000 residues of both cyclases, large parts of which are composed of repetitions of a 23-amino acid motif similar to those found in human glycoproteins, Drosophila chaoptin, and Toll gene product. However, a segment corresponding to the NH2-terminal 620 residues of Sa. cerevisiae cyclase appears lost from Sc. pombe cyclase, and the COOH-terminal 140 residues are not well conserved between the two yeast species. Deletions involving the COOH-terminal residues of Sa. cerevisiae cyclase cause loss of activation by the RAS2 protein. These results suggest that Sc. pombe cyclase may have lost the ability to interact with RAS proteins by the loss of a regulatory site.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beckner S. K., Hattori S., Shih T. Y. The ras oncogene product p21 is not a regulatory component of adenylate cyclase. Nature. 1985 Sep 5;317(6032):71–72. doi: 10.1038/317071a0. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broek D., Samiy N., Fasano O., Fujiyama A., Tamanoi F., Northup J., Wigler M. Differential activation of yeast adenylate cyclase by wild-type and mutant RAS proteins. Cell. 1985 Jul;41(3):763–769. doi: 10.1016/s0092-8674(85)80057-x. [DOI] [PubMed] [Google Scholar]
- Field J., Broek D., Kataoka T., Wigler M. Guanine nucleotide activation of, and competition between, RAS proteins from Saccharomyces cerevisiae. Mol Cell Biol. 1987 Jun;7(6):2128–2133. doi: 10.1128/mcb.7.6.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y., Kozasa T., Kaziro Y., Takeda T., Yamamoto M. Role of a ras homolog in the life cycle of Schizosaccharomyces pombe. Cell. 1986 Jan 31;44(2):329–336. doi: 10.1016/0092-8674(86)90767-1. [DOI] [PubMed] [Google Scholar]
- Gilman A. G. G proteins: transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Hashimoto C., Hudson K. L., Anderson K. V. The Toll gene of Drosophila, required for dorsal-ventral embryonic polarity, appears to encode a transmembrane protein. Cell. 1988 Jan 29;52(2):269–279. doi: 10.1016/0092-8674(88)90516-8. [DOI] [PubMed] [Google Scholar]
- Heideman W., Casperson G. F., Bourne H. R. Adenylyl cyclase in yeast. Hydrodynamic properties and activation by trypsin. J Biol Chem. 1987 May 25;262(15):7087–7091. [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T., Broek D., Wigler M. DNA sequence and characterization of the S. cerevisiae gene encoding adenylate cyclase. Cell. 1985 Dec;43(2 Pt 1):493–505. doi: 10.1016/0092-8674(85)90179-5. [DOI] [PubMed] [Google Scholar]
- Kataoka T., Powers S., Cameron S., Fasano O., Goldfarb M., Broach J., Wigler M. Functional homology of mammalian and yeast RAS genes. Cell. 1985 Jan;40(1):19–26. doi: 10.1016/0092-8674(85)90304-6. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Lopez J. A., Chung D. W., Fujikawa K., Hagen F. S., Papayannopoulou T., Roth G. J. Cloning of the alpha chain of human platelet glycoprotein Ib: a transmembrane protein with homology to leucine-rich alpha 2-glycoprotein. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5615–5619. doi: 10.1073/pnas.84.16.5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Uno I., Ishikawa T. Identification of the structural gene and nonsense alleles for adenylate cyclase in Saccharomyces cerevisiae. J Bacteriol. 1984 Jan;157(1):277–282. doi: 10.1128/jb.157.1.277-282.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke R., Krantz D. E., Yen D., Zipursky S. L. Chaoptin, a cell surface glycoprotein required for Drosophila photoreceptor cell morphogenesis, contains a repeat motif found in yeast and human. Cell. 1988 Jan 29;52(2):291–301. doi: 10.1016/0092-8674(88)90518-1. [DOI] [PubMed] [Google Scholar]
- Russell P. R., Hall B. D. Structure of the Schizosaccharomyces pombe cytochrome c gene. Mol Cell Biol. 1982 Feb;2(2):106–116. doi: 10.1128/mcb.2.2.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüther U., Müller-Hill B. Easy identification of cDNA clones. EMBO J. 1983;2(10):1791–1794. doi: 10.1002/j.1460-2075.1983.tb01659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Takahashi Y., Putnam F. W. Periodicity of leucine and tandem repetition of a 24-amino acid segment in the primary structure of leucine-rich alpha 2-glycoprotein of human serum. Proc Natl Acad Sci U S A. 1985 Apr;82(7):1906–1910. doi: 10.1073/pnas.82.7.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titani K., Takio K., Handa M., Ruggeri Z. M. Amino acid sequence of the von Willebrand factor-binding domain of platelet membrane glycoprotein Ib. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5610–5614. doi: 10.1073/pnas.84.16.5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Uno I., Ishikawa T., Powers S., Kataoka T., Broek D., Cameron S., Broach J., Matsumoto K., Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985 Jan;40(1):27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno I., Mitsuzawa H., Tanaka K., Oshima T., Ishikawa T. Identification of the domain of Saccharomyces cerevisiae adenylate cyclase associated with the regulatory function of RAS products. Mol Gen Genet. 1987 Dec;210(2):187–194. doi: 10.1007/BF00325683. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Hol W. G. Predicted nucleotide-binding properties of p21 protein and its cancer-associated variant. Nature. 1983 Apr 28;302(5911):842–844. doi: 10.1038/302842a0. [DOI] [PubMed] [Google Scholar]