Abstract
Most fungal species from marine environments also live on land. It is not clear whether these fungi reach the sea from terrestrial sources as spores or other propagules, or if there are separate ecotypes that live and reproduce in the sea. The emergence of marine diseases has created an urgency to understand the distribution of these fungi. Aspergillus flavus is ubiquitous in both terrestrial and marine environments. This species is an opportunistic pathogen in many hosts, making it a good model to study the relationship between genetic diversity and specificity of marine fungi. In this study, an intraspecific phylogeny of A. flavus isolates based on Amplified Fragment Length Polymorphisms (AFLPs) was used to determine if terrestrial and marine isolates form discrete populations, and to determine if phylogeny predicts substratum specificity. Results suggest lack of population structure in A. flavus. All isolates may compose a single population, with no clade particular to marine environments.
Keywords: AFLP, aspergillosis, Gorgonia, marine fungi, Octocorallia, specificity, sea fans
Introduction
Fungal species in the sea are defined as either obligately marine or facultative: “Obligate marine fungi are those that grow and sporulate exclusively in a marine or estuarine habitat; facultative marine fungi are those from freshwater or terrestrial milieus able to grow (and possibly also to sporulate) in the marine environment” (Kohlmeyer & Volkmann-Kohlmeyer 2003; Shearer et al. 2007). Facultative marine fungi may be carried to the sea by wind, rain or runoff. Given that some are very common in the sea, they may include populations that have evolved adaptations to grow in marine environments, eventually becoming obligately marine. There are certainly precedents for this: colonization of the sea by fungi has happened many times independently (Hibbett & Binder 2001).
The emergence of marine diseases has created an urgency to understand the role and origin of the microbiota associated with marine organisms (Harvell et al. 2007; Rosenberg et al. 2007). Studies of fungi associated with corals have mainly focused on aspergillosis disease of sea fans (Gorgonia ventalina). This disease has been attributed to Aspergillus sydowii, a fungus common in soils (Geiser et al. 1998b), though this finding has recently been questioned (Toledo-Hernández et al. 2007, 2008; Zuluaga-Montero et al. 2010). There is a debate about how inoculum of A. sydowii reaches the Caribbean; the leading theory is that the inoculum is terrestrial, comes from the Sahel, and crosses the Atlantic in dust clouds (Weir-Brush et al. 2004). However, there is evidence for distinct marine and terrestrial populations: marine strains caused aspergillosis when inoculated into sea fans whereas terrestrial strains could not (Geiser et al. 1998b), and carbon utilization profiles showed differences between marine and terrestrial strains (Alker et al. 2001). On the other hand, DNA fingerprints based on microsatellites failed to distinguish between marine and terrestrial strains of A. sydowii (Rypien et al. 2008).
In the present study, we used Aspergillus flavus as a model species to address this marine vs. terrestrial debate. A. flavus is ubiquitous in terrestrial environments and it is increasingly clear that it is ubiquitous in marine environments as well (Koh et al. 2000; Morrison-Gardiner 2002; Zuluaga et al. 2010). Its high salt tolerance and wide range of substrata makes it a logical candidate to adapt to life in the sea. A. flavus has been extensively studied as an opportunistic pathogen in chronic and invasive pulmonary and systemic infections, especially in immune-compromised patients (Hedayati et al. 2007). It can also cause disease in a broad range of organisms other than humans, including birds, insects and plants (Raper et al. 1965; Leger et al. 2000). In addition, A. flavus produces aflatoxins - secondary metabolites that are potent carcinogenic and immunosuppressive toxins in animals when ingested, and pose a significant threat to human health (Pitt 2000; Yu et al. 2005). This fungus frequently invades susceptible crops such as corn, cotton, peanuts and tree nuts before or after harvest, causing aflatoxin contamination (Cotty et al. 1994).
Aspergillus flavus, together with other congenerics, is commonly isolated from marine substrata, including sponges (Holler et al. 2000), sclerectinian corals (Kendrick et al. 1982), and soft corals (Koh et al. 2000). We found A. flavus is common in diseased tissue of the sea fan Gorgonia ventalina in Puerto Rico, suggesting a possible role in sea fan aspergillosis (Toledo-Hernández et al. 2008; Zuluaga-Montero et al. 2010). However, the biology of marine isolates of A. flavus has scarcely been explored.
In the present study, we used Amplified Fragment Length Polymorphisms (AFLPs) to identify intraspecific relationships among A. flavus isolates from terrestrial and marine sources. We tested the hypothesis that marine isolates will be more closely related to other marine isolates than to terrestrial isolates, suggesting that some clades have adaptations for life in the sea. On the other hand, if the source of marine isolates is terrestrial input, marine isolates are not expected to form distinct clades. In addition, we tested the hypothesis that isolates from diseased tissue of sea fans form a clade apart from isolates from healthy tissue, which would suggest that certain genotypes are associated with disease.
Materials and Methods
Fungal isolates and DNA sequencing
Thirty isolates of Aspergillus flavus were obtained from different environmental sources (Table 1). Isolates from seawater and from healthy and diseased sea fan (Gorgonia ventalina) tissue were collected from different reefs around Puerto Rico (Zuluaga-Montero et al. 2010). Other isolates from soil, dried, green coffee beans and air were included for comparison. All isolates were cultured on Glucose Peptone Yeast Agar (GPYA, Difco Labs) incubated at 25 °C and transferred to liquid medium (potato-dextrose broth) for DNA extraction.
Table 1.
Isolates of Aspergillus flavus used in this study with substratum, site of isolation and GenBank accession number of ITS sequence.
| # ID isolate | Substratum | Site of isolation | GenBank accession number |
|---|---|---|---|
| A1 | coffee | PR | HM167490 |
| A2 | algae | PR | EU645653 |
| A3 | air, walnut orchard | Wolfskill, Winters, CA | HM167491 |
| A4 | Soil, walnut orchard | Wolfskill, Winters, CA | HM167492 |
| B1 | Soil | PR | HM167494 |
| B2 | Soil | PR | EU645681 |
| B3 | Soil | Nigeria | HM167488 |
| B4 | Soil | Nigeria | HM167489 |
| B5 | Soil | Nigeria | HM167495 |
| DT1 | Diseased sea fan tissue | PR | HM178946 |
| DT2 | Diseased sea fan tissue | PR | EU554579 |
| DT3 | Diseased sea fan tissue | PR | EU554578 |
| DT4 | Diseased sea fan tissue | PR | |
| DT5 | Diseased sea fan tissue | PR | EU554582 |
| DT6 | Diseased sea fan tissue | PR | EU554577 |
| HT1 | Healthy sea fan tissue | PR | EU554586 |
| HT2 | Healthy sea fan tissue | PR | HM167496 |
| HT3 | Healthy sea fan tissue | PR | HM167497 |
| HT4 | Healthy sea fan tissue | PR | HM178947 |
| HT5 | Healthy sea fan tissue | PR | HM167499 |
| HT6 | Healthy sea fan tissue | PR | EU554573 |
| HT7 | Healthy sea fan tissue | PR | HM167500 |
| HT8 | Healthy sea fan tissue | PR | HM167501 |
| HT9 | Healthy sea fan tissue | PR | HM167502 |
| SW1 | Seawater | PR | EU645713 |
| SW2 | Seawater | PR | EU645706 |
| SW3 | Seawater | PR | EU645692 |
| SW4 | Seawater | PR | EU645703 |
| SW5 | Seawater | PR | EU645702 |
| SW6 | Seawater | PR | HM167493 |
DNA was extracted using a Plant Mini Extraction Kit (Qiagen Sciences). To ensure that all isolates were Aspergillus flavus, the nuclear ribosomal ITS region was amplified using primers ITS 1F and ITS 4 (White et al. 1990; Gardes & Bruns 1993), and sequenced in the University of Puerto Rico Sequencing and Genotyping Facility (UPR SGF). Sequences were assembled and manually examined for errors using Sequencher software (version 3.1), and aligned using CLUSTALX (Version 1.8, Thompson et al. 1997) in BioEdit 7.0 with default settings (full multiple alignment, 1000 bootstrap replicates on NJ tree).
AFLPs
Genomic DNA (250 ng) was digested with EcoRI and MseI restriction enzymes, and adapters were ligated to the fragments following instructions of the AFLP Microbial Fingerprinting Kit (Invitrogen). Pre-selective amplification used EcoRI and MseI universal primers (Voss et al. 1995). Selective amplification used 10 primer combinations with two additional nucleotides (Eco-Mse): CC-AC, CC-CA, CC-CC, CC-CG, CC-CT, CG-CA, CG-CG, CG-AC, CG-AG, and CG-GA (Table 2). These combinations were chosen because they showed a high number of polymorphisms when 33 primer combinations were compared in preliminary tests (unpublished data). EcoRI primers were fluorescently labeled with FAM (Applied Biosystems). An aliquot of the selective amplification product was mixed with 1.0 μl of Genescan-500 ROX (6-carboxy-x-rhodamin) length standard and analyzed on an ABI Prism 310 Genetic Analyzer (Applied Biosystems) in the UPR SGF.
Table 2.
Different primer combinations of AFLPs used in this study with the number of polymorphic alleles analyzed. (-) means combinations not used.
| Mse \ Eco | CA | CC | CG |
|---|---|---|---|
| AC | - | 39 | - |
| CA | 35 | 41 | 23 |
| CC | - | 19 | - |
| CG | - | 32 | 31 |
| CT | - | - | - |
| AG | - | 59 | 24 |
| GA | - | - | 20 |
Fragments between 90 and 510 bp were scored as either present or absent using GeneMapper software version 3.5 (Applied Biosystems). For each primer combination, a default AFLP analysis was used to generate a panel that assigned allele markers (bins); peak height threshold was set at 100 and the bin assignment and allele calls were reviewed manually for accuracy of identification of dimorphic alleles (see Karudapuram & Larson 2005). Seven isolates were run twice (including culturing, DNA extraction, AFLP and analysis) for three different AFLP primer combinations, to test reproducibility and consistency of results. The results were reported as a binary matrix of presence-absence of dimorphic alleles. A neighbour-joining distance analysis (NJ) was performed with PAUP (Version 4.0b 10, Swofford 2002), and the resulting tree was visualized as an unrooted phylogram. For additional support, a Jackknife with heuristic search was done with 50% deletion of the total characters. In addition, a maximum parsimony analysis (Swofford 2002) was done to find the most parsimonious tree (MPT) using 100-replicate heuristic search and all characters with equal weight. Bootstrapping for NJ and parsimony was done with 1000 replicates. Since MPT and NJ trees were almost identical, the MPT was chosen to test the monophyly hypotheses: (1) that isolates from diseased gorgonian tissue form a monophyletic group, and (2) that marine isolates form a monophyletic group. For this, a constraint tree with either disease-associated isolates or marine-isolated isolates forced to group as a single clade was drawn using McClade (Madison & Madison 1992). Significant differences between the two topologies for each hypothesis were tested with a Templeton test (Wilcoxon signed-ranks) implemented in PAUP.
Population differentiation was assessed by grouping the samples by source (diseased sea fan tissue, healthy sea fan tissue, seawater, soil, other). Wright’s F statistics (FST) (Whitlock & McCauley 1999) were calculated with Structure 2.2.3 (Falush et al. 2007). The K values (1-4) were estimated on the log likelihood score and posterior probability of K (Pritchard et al. 2007). Structure was set as follows: length of burn in period = 1 million; number of iterations for the Markov Chain Monte Carlo (MCMC) = 300,000; an admixture model was chosen with lambda constant. Correlated allele frequencies among populations were assumed, and information on the population origin was not used.
Results
Phylogenetic relationships estimated by AFLPs and ITS sequence analysis
There was little variation in ITS sequence among isolates. Only two haplotypes with only one base pair difference were found across all samples. The highest BLAST hits in GenBank were A. flavus, with in most cases >98% similarity.
A total of 323 variable AFLP markers were detected among 30 isolates using ten primer combinations, which generated between 19 and 59 polymorphic alleles each (Table 2). When different cultures of the same isolate were compared as a control, results were identical. The NJ tree and maximum parsimony tree had similar topologies (Figs.1, 2). The bootstrap and jackknife analyses showed >90% support for many nodes. Differences among individuals was between 3%-50% (minimum=9, maximum=167). Three pairs of closely related isolates (B1-B2, A3-A4, and H7-H8) were isolated from the same place and the same substratum, but no two isolates were identical. The population genetic analysis showed low levels of differentiation among isolates from different substrata (Fst = 0.002).
Figure 1.
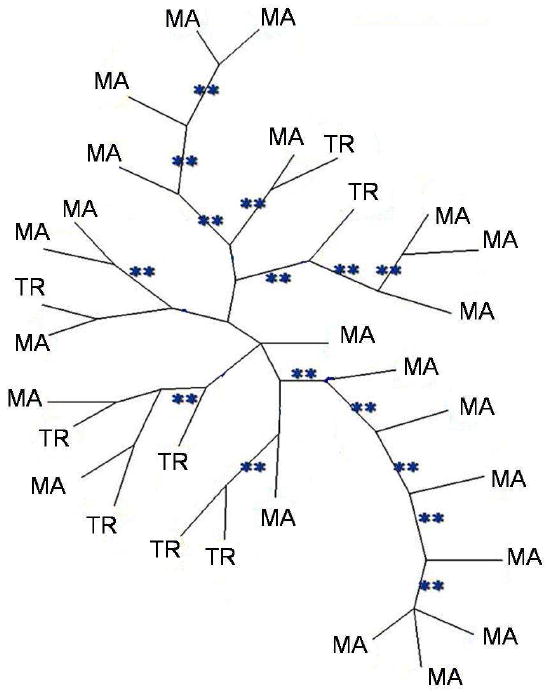
Unrooted neighbour-joining trees of Aspergillus flavus showing distribution of of marine isolates (MA) vs. terrestrial (TR). The tree is based on presence/absence of 323 AFLPs. Bootstrap and Jackknife values ≥ 90% are represented by (**).
Figure 2.
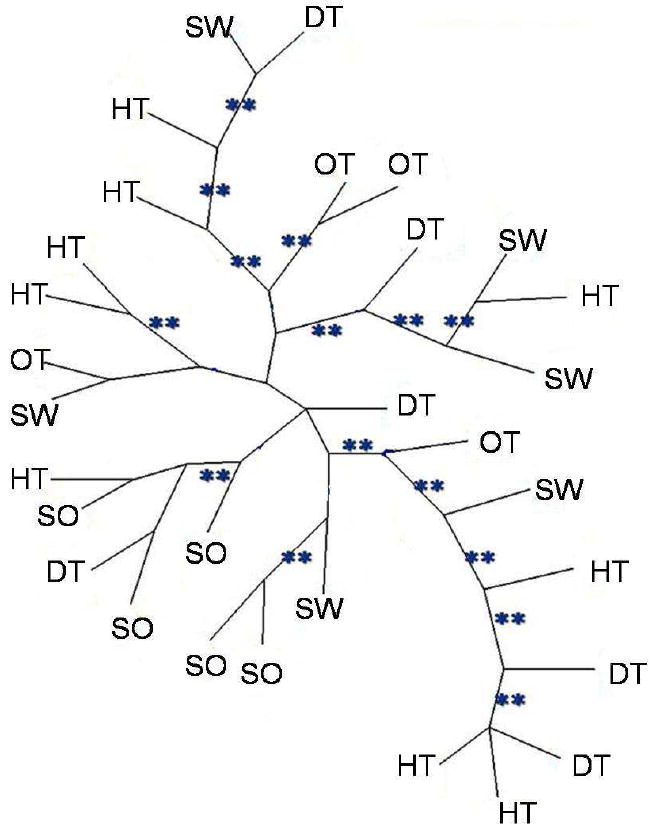
Unrooted neighbour-joining trees of Aspergillus flavus based on AFLPs. Letters represent the substrata from which isolates were collected: (SW) seawater, (DT) diseased sea fan tissue, (HT) healthy sea fan tissue, (SO) soil, (OT) other sources.
Many highly supported nodes included both marine and terrestrial isolates (Fig. 1). The hypothesis of a monophyletic origin of marine isolates was rejected by the Templeton (Wilcoxon signed-ranks) test (T=83.5, P=0.0001, the constraint topology being 970 steps longer than MPT.
The second hypothesis of monophyly, isolates from diseased tissue forming a single clade, was also rejected. The constraint topology was 169 steps longer than MPT (1108 steps) (Wilcoxon signed-ranks test, T=1048.5, P<0.0001).
Discussion
AFLPs provided sufficient resolution to differentiate all Aspergillus flavus isolates tested. Some evidence of clonality was expected in isolates from the same place and substratum, as reported for A. flavus isolates from a single cotton field in Arizona (Bayman & Cotty 1993), but this was not observed. Isolates of A. flavus with identical multilocus haplotypes were isolated from soil populations in Arizona and Texas (Grubisha & Cotty 2010). The fact that identical haplotypes were isolated in that study but not the present study may reflect the much more intensive sampling done by Grubisha & Cotty (2010), the greater resolving power of AFLPs compared to microsatellites, or both. One advantage of AFLPs is the high number of polymorphic characters that can be generated - 323 in this study.
All samples, including marine and terrestrial isolates, comprised one population, with no specific clade significantly more common in sea fans (Figs. 1, 2). This suggests that the high prevalence of A. flavus in marine substrata is not due to particular adaptations in any clade of A. flavus, but rather to its weediness and ability to colonize a wide range of substrata. Similarly, no clade was particularly associated with diseased sea fan tissue vs. healthy tissue. This neither precludes nor supports the idea that A. flavus may be an opportunistic pathogen of sea fans (Toledo-Hernández et al. 2008) and it does not support the hypothesis that the strains found in diseased tissue have particular adaptations for pathogenesis compared to strains found in healthy tissue. As pathogens, A. flavus strains appear to be generalists: when strains isolated as pathogens from humans, insects and plants were inoculated into insect larvae and corn kernels, there was no evidence of adaptation to certain hosts (Leger et al. 2000).
In general, there is a lack of information regarding fungi in marine ecosystems. For instance, it is unknown whether Aspergillus sporulates in the water (Smith et al. 1996; Shinn et al. 2000) or how long it can survive pelagically. However, Aspergillus spores and mycelial fragments are capable of growth under simulated deep-sea conditions (Damare et al. 2008). It is assumed that Aspergillus colonies in the sea are the accidental result of the arrival of inoculum from land, and that the most common species are basically terrestrial (Geiser et al. 1998b), a theory which is supported by the lack of differences between marine and terrestrial isolates found here. In terrestrial environments, A. flavus produces numerous airborne conidia, which are readily dispersed by wind, water and insects (Abarca 2000). However, since the ocean lacks barriers for microbial dispersion, the rate of dispersal in the sea can be faster than in terrestrial systems, due to strongly directional ocean currents that run along the coastlines (McCallum et al. 2003).
The lack of population structure of A. flavus in this study suggests there is sufficient gene flow to prevent population differentiation among different places and substrata. This supports the classical theory of microbial biogeography most succinctly stated by Bass-Becking in 1934: “Everything is everywhere; the environment selects” (Whitfield 2005, Martiny et al. 2006). This theory holds that species of microorganisms, because of their small propagule size and tremendous capacity for dispersal, may grow wherever environmental conditions favor them--in contrast to larger organisms, whose populations show more biogeographic structure.
However, recent studies have challenged “everything is everywhere” based on extensive sampling of natural populations and improved taxonomic resolution. While some well-known species of free-living ciliates do indeed appear to be everywhere, many consist of similar but distinct, geographically restricted groups (Foissner et al. 2008). However, the issue is still in dispute: according to ciliatologist Bland Finlay, “There is no biogeography for anything smaller than 1 millimeter” (Whitfield 2005). In fungi, in contrast, multiple gene genealogies and a phylogenetic species concept have shown biogeographic structure and endemism in many fungi previously thought to be universally distributed (Jacobson et al. 2004; Aa et al. 2006; Taylor et al. 2006). A notable exception cited by Taylor et al. (2006) is Aspergillus fumigatus, which is truly cosmopolitan (Pringle et al. 2005; Rydholm et al. 2006). Lack of geographic population structure has also been reported for a closely related species, A. sydowii (but sample size for this study was more restricted) (Rypien et al. 2008). These Aspergillus species share features such as high production of conidia, rapid growth and weediness (ruderal characteristics) that promote ubiquity and cosmopolitanism. A. flavus shares these features and apparently belongs to this group of species whose distributions support ‘everything is everywhere.’ Although A. flavus may contain cryptic species (Geiser et al. 1998a), a single clade defined by high resolution markers such as AFLPs can include isolates from substrata as diverse as soil and sea fan tissue (Figs. 1, 2). Facultative marine fungi provide an interesting test for the ‘everything is everywhere’ theory.
Acknowledgments
This study was supported by grants from UPR Sea Grant (NOAA award NA16RG2278, project R- R-92-2-08), NOAA-CRES, NIH-SCORE (S06GM08102), and NSF-CREST (HRD0734826). A. Zuluaga-Montero thanks the International Society for Reef Studies (ISRS) for an ISRS/TOC Coral Reef Conservation Award (2006) and the Graduate Studies and Research Dean’s Office of UPR Río Piedras for a PBDT fellowship (2007). Sequencing at UPR SGF was supported in part by NCRR-AABRE Grant #P20 RR16470. We are grateful to Jenny Acevedo, Karla Maldonado, Ligia Lebrón and Manuel Ramírez for assistance in the AFLP analysis.
Footnotes
Genetic variability in A. flavus
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aa E, Townsend JP, Adams RI, Nielsen KM, Taylor JW. Population structure and gene evolution in Saccharomyces cerevisiae. FEMS Yeast Research. 2006;6:702–715. doi: 10.1111/j.1567-1364.2006.00059.x. [DOI] [PubMed] [Google Scholar]
- Abarca ML. Taxonomía e identificación de especies implicadas en las aspergilosis nosocomial. Revista Iberoamericana de Micología. 2000;17:S79–S84. [PubMed] [Google Scholar]
- Alker AP, Smith GW, Kim K. Characterization of Aspergillus sydowii (Thom et Church), a fungal pathogen on Caribbean sea fan corals. Hydrobiologia. 2001;460:105–111. [Google Scholar]
- Bayman P, Cotty PJ. Genetic diversity in Aspergillus flavus: association with aflatoxins production and morphology. Canadian Journal of Botany. 1993;71:23–31. [Google Scholar]
- Cotty PJ, Bayman P, Egel D, Elias K. Agriculture, Aspergillus, and aflatoxins. In: Powell KA, Renwick A, Peberdy JF, editors. The Genus Aspergillus. Plenum Press; NY: 1994. pp. 1–272. [Google Scholar]
- Damare SR, Nagarajan M, Raghukumar C. Spore germination of fungi belonging to Aspergillus species under deep-sea conditions. Deep-Sea Research. 2008;55:670–678. [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissner W, Chao A, Katz LA. Diversity and geographic distribution of ciliates (Protista: Ciliophora) Biodiversity and Conservation. 2008;17:345–363. [Google Scholar]
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Molecular Ecology. 1993;2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- Geiser DM, Pitt JI, Taylor JW. Cryptic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus. PNAS. 1998a;95:388–393. doi: 10.1073/pnas.95.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser DM, Taylor JW, Ritchie KB, Smith GW. Cause of sea fan death in the West Indies. Nature. 1998b;394:137–138. [Google Scholar]
- Grubisha LC, Cotty PJ. Genetic isolation among sympatric vegetative compatibility groups of the aflatoxin-producing fungus Aspergillus flavus. Molecular Ecology. 2010;19:269–280. doi: 10.1111/j.1365-294X.2009.04467.x. [DOI] [PubMed] [Google Scholar]
- Harvell C, Jordán-Dahlgren E, Merkel S, Rosenberg E, Raimundo L, Smith G, Weil E, Willis B. Coral disease, environmental drivers, and the balance between coral and microbial associates. Oceanography. 2007;20:172–195. [Google Scholar]
- Hedayati MT, Pasqualotto AC, Warn PA, Bowyer P, Denning DW. Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiology. 2007;153:1677–1692. doi: 10.1099/mic.0.2007/007641-0. [DOI] [PubMed] [Google Scholar]
- Hibbett DS, Binder M. Evolution of marine mushrooms. The Biological Bulletin. 2001;201:319–322. doi: 10.2307/1543610. [DOI] [PubMed] [Google Scholar]
- Holler U, Wright AD, Matthée GF, Konig GM, Draeger S, Aust H, Schulz B. Fungi from marine sponges: diversity, biological activity and secondary metabolites. Mycological Research. 2000;2:1354–1365. [Google Scholar]
- Jacobson DJ, Powell AJ, Dettman JR, Saenz GS, Barton MM, Hiltz MD, Dvorachek WH, Glass NL, Taylor JW, Natvig DO. Neurospora in temperate forests of western North America. Mycologia. 2004;96:66–74. [PubMed] [Google Scholar]
- Karudapuram S, Larson S. Identification of Hedysarum Varieties Using Amplified Fragment Length Polymorphism on a Capillary Electrophoresis System. Journal of Biomolecular Techniques. 2005;16:316–324. [PMC free article] [PubMed] [Google Scholar]
- Kendrick B, Risk MJ, Michaelides J, Bergman K. Amphibious microborers: bioeroding fungi isolated from live corals. Bulletin of Marine Science. 1982;32:862–867. [Google Scholar]
- Koh L, Tan TK, Chou LM, Goh NK. Fungi associated with gorgonians in Singapore. Proceedings 9th international coral reef symposium. 2000;1:521–526. [Google Scholar]
- Kohlmeyer J, Volkmann-Kohlmeyer B. Fungi from coral reefs: A commentary. Mycological Research. 2003;107:386–387. doi: 10.1017/s0953756203227775. [DOI] [PubMed] [Google Scholar]
- St Leger J, Screen SE, Shams-Pirzadeh B. Lack of host specialization in Aspergillus flavus. Applied and Environmental Microbiology. 2000;1:320–324. doi: 10.1128/aem.66.1.320-324.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny JBH, Bohannan B, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Adams J, Kuske PJ, Morin CR, Naeem S, Øvreås L, Reysenbach A, Smith VH, Staley JT. Microbial biogeography: putting microorganisms on the Map. Nature Reviews Microbiology. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- McCallum H, Harvell D, Dobson A. Rates of spread of marine pathogens. Ecology Letters. 2003;6:1062–1067. [Google Scholar]
- Madison WP, Madison DR. MacClade: analysis of phylogeny and character evolution v3. Sinauer Associates Inc.; Sunderland, MA: 1992. [Google Scholar]
- Morrison-Gardiner S. Dominant fungi from Australian coral reefs. Fungal Diversity. 2002;9:105–121. [Google Scholar]
- Pitt JI. Toxigenic fungi: which are important? Medical Mycology. 2000;38S:17–22. [PubMed] [Google Scholar]
- Pringle A, Baker DM, Platt JL, Wares JP, Latge JP, Taylor JW. Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution. 2005;59:1886–1899. [PubMed] [Google Scholar]
- Pritchard JK, Wen X, Falush D. [18.02.10];Documentation for Structure Software, Version 2.2. 2007 http://pritch.bsd.uchicago.edu/software/structure22/readme.pdf.
- Raper KB, Fennell DI, Austwick PKC. The Genus Aspergillus. Academic Press; New York: 1965. [Google Scholar]
- Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg L. The role of microorganisms in coral health, disease and evolution. Nature Review Microbiology. 2007;5:355–362. doi: 10.1038/nrmicro1635. [DOI] [PubMed] [Google Scholar]
- Rydholm C, Szakacs G, Lutzoni F. Low genetic variation and no detectable population structure in Aspergillus fumigatus compared to closely related Neosartorya species. Eukaryotic Cell. 2006;5:650–657. doi: 10.1128/EC.5.4.650-657.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rypien KL, Andras JP, Harvell CD. Globally panmictic population structure in the opportunistic fungal pathogen Aspergillus sydowii. Molecular Ecology. 2008;17:4068–4078. doi: 10.1111/j.1365-294X.2008.03894.x. [DOI] [PubMed] [Google Scholar]
- Shearer CA, Descals E, Kohlmeyer B, Kohlmeyer J, Marvanova L, Padgett D, Porter D, Raja HA, Schmit JP, Thorton HA, Voglymayr H. Fungal biodiversity in aquatic habitats. Biodiversity and Conservation. 2007;16:49–67. [Google Scholar]
- Shinn E, Smith GM, Prospero JM, Betzer P, Hayes ML, Garrison V, Barber RT. African dust and the demise of Caribbean coral reefs. Geophysical Research Letter. 2000;27:3129–3032. [Google Scholar]
- Smith GW, Ives LD, Nagelkerken IA, Ritchie KB. Caribbean sea-fan mortalities. Nature. 1996;383:487. [Google Scholar]
- Swofford DL. Phylogenetic analysis using parsimony (* and other methods) Sinauer; Sunderland, Massachusetts: 2002. [Google Scholar]
- Taylor JW, Turner E, Townsend JP, Dettman JR, Jacobson D. Eukaryotic microbes, species recognition and the geographic limits of species: examples from the kingdom Fungi. Philosophical Transactions of the Royal Society B: Biological Sciences. 2006;361:1947–1963. doi: 10.1098/rstb.2006.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo -Hernández C, Bones-González A, Ortiz-Vázquez OE, Sabat AM, Bayman P. Fungi in the sea fan Gorgonia ventalina: diversity and sampling strategies. Coral reefs. 2007;26:725–730. [Google Scholar]
- Toledo -Hernández C, Zuluaga-Montero A, Bones-González A, Sabat AM, Bayman P. Fungi in healthy and diseased sea fans (Gorgonia ventalina): is Aspergillus sydowii always the pathogen? Coral Reef. 2008;27:707–714. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgin DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir -Brush J, Garrison VH, Smith GW, Shinn EZ. The relationship between gorgonian coral (Cnidaria: Gorgonacea) diseases and African dust storms. Aerobiologia. 2004;20:119–126. [Google Scholar]
- White TJ, Bruns TD, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: A guide to methods and applications. Academic Press; New York: 1990. pp. 315–322. [Google Scholar]
- Whitfield J. Biogeography: Is everything everywhere? Science. 2005;5750:960–961. doi: 10.1126/science.310.5750.960. [DOI] [PubMed] [Google Scholar]
- Voss P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock MC, McCauley DE. Indirect measures of gene flow and migration: FST ≠ 1/(4Nm+1) Heredity. 1999;82:117–125. doi: 10.1038/sj.hdy.6884960. [DOI] [PubMed] [Google Scholar]
- Yu J, Cleveland TE, Nierman WC, Bennett JW. Aspergillus flavus genomics: gateway to human and animal health, food safety, and crop resistance to diseases. Revista Iberoamericana de Micología. 2005;22:194–202. doi: 10.1016/s1130-1406(05)70043-7. [DOI] [PubMed] [Google Scholar]
- Zuluaga-Montero A, Toledo-Hernández C, Rodríguez JA, Sabat M, Bayman P. Spatial variation in fungal communities isolated from healthy and diseased sea fans Gorgonia ventalina and seawater. Aquatic Biology. 2010;8:151–160. [Google Scholar]


