Abstract
The prolyl peptidases are a family of enzymes characterized by a biochemical preference for cleaving proline-containing peptides. The members of this enzyme family include prolyl endopeptidase, prolyl endopeptidase-like, dipeptidyl peptidase 4 (DPP4), DPP7, DPP8, DPP9, and fibroblast activation protein. DPP4 is the best studied member of the family, due to its role in physiological glucose tolerance, exerted through the regulation of the insulinotropic peptide glucagon-like peptide-1. While other members of the prolyl peptidase family have also been implicated in various (patho)physiological processes, the underlying peptides and pathways regulated by these enzymes are less clear. The identification of endogenous substrates of the prolyl peptidases is an important step in elucidating the molecular mechanisms of these enzymes. Here, we highlight the utility of liquid chromatography–mass spectrometry-based peptidomics to enable the discovery of endogenous prolyl peptidase substrates directly from tissues, and demonstrate the utility of this information in understanding the biochemical and physiological functions of the prolyl peptidases.
Key words: Peptidomics, LC–MS, Prolyl peptidases, DPP4, Prep
INTRODUCTION
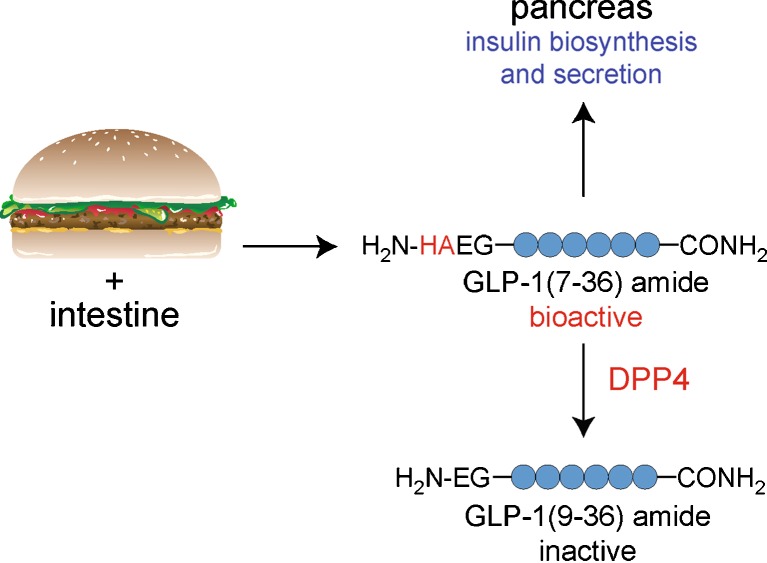
Peptidases play a central role in the production, regulation, and degradation of peptides in vivo (1–4). The elucidation of peptidase substrates is necessary for defining the biochemistry of a peptidase, and this information can also be used to infer the cellular and physiological functions of the enzyme (5,6). For example, the identification of the insulinotropic peptide glucagon-like peptide-1 (GLP-1) as a plasma dipeptidyl peptidase 4 (DPP4) substrate suggested that this enzyme is involved in insulin regulation by controlling levels of bioactive GLP-1 in plasma (6) (Fig. 1). Indeed, mice lacking DPP4 have higher levels of GLP-1 and insulin, and improved glucose tolerance (7). These findings revealed DPP4 to be a potential therapeutic target for the treatment of diabetes, which has subsequently been realized with the approval of DPP4 inhibitors as new antidiabetic drugs (8).
Fig. 1.
DPP4 regulates the levels of bioactive GLP-1. As food enters the intestine, a series of hormones are released that serve to prime the body for the nutrients that are on their way. One peptide, glucagon-like peptide-1 (GLP-1), is an insulinotropic peptide that stimulates insulin biosynthesis and secretion in a glucose-dependent manner. In addition, GLP-1 is rapidly cleaved by dipeptidyl peptidase 4 (DPP4), which removes a dipeptide from the N terminus of GLP-1 to inactivate the peptide. The discovery of this pathway led to the hypothesis that DPP4 might be involved in physiological glucose tolerance, which was later confirmed through the generation of mice lacking DPP4. Furthermore, this pathway revealed the mechanism of GLP-1 degradation, which led to the development of DPP4 inhibitors as a new class of antidiabetic drugs
The importance of DPP4 has spurred the investigation of the biochemical, cellular, and physiological roles of other mammalian dipeptidyl peptidases (DPPs) (3,9). Biochemically, the DPPs are N-terminal dipeptidases with a preference for a proline residue at the penultimate position (i.e., H2N-XaaPro-peptide) of their substrates. DPPs have also been suggested to perform key roles in (patho)physiological processes. For example, fibroblast activation protein (FAP) is upregulated in some cancers (10,11). High levels of proline in the extracellular matrix proteins (collagen) led to the hypothesis that FAP activity might help degrade the extracellular matrix to enable tumor growth and migration. Indeed, FAP has been shown to influence tumor cell growth in vivo through xenograft studies where cells expressing FAP formed larger tumors than cells lacking FAP or cells with a catalytically inactive FAP mutant (12,13). In addition to DPP4 and FAP, there are three additional catalytically active dipeptidyl peptidases, DPP7, DPP8, and DPP9, and the catalytically inactive DPP6 (3).
While DPPs prefer to cleave proline-containing peptides at the N terminus, prolyl endopeptidase (Prep) is able to cleave peptides on the C-terminal side of proline within the peptide sequence (9,14). Together, the DPPs and prolyl endopeptidases are part of the prolyl peptidase family. The members of this enzyme family are serine proteases and share significant sequence homology, and some also have similar structures (3). Like all serine hydrolases, these enzymes use an activated serine nucleophile to attack the scissile amide bond, resulting in peptide cleavage. Prep has been tested against many bioactive peptides that contain proline residues (e.g., substance P and vasopressin), and many of these peptides are excellent in vitro substrates (9). The identification of these substrates led to new hypotheses about Prep function, which were tested in vivo using pharmacological Prep inhibitors. Interestingly, several of these experiments suggested that Prep inhibition would lead to improved cognitive function in mammals, which was later verified (15–18). However, the underlying mechanism for this improvement is still under investigation because some of the Prep substrates predicted to be responsible for this effect were unaffected, or only modestly affected, upon Prep inhibition (19–21).
These results highlight the difficulty in predicting endogenous substrates solely based on in vitro experiments, because a number of factors which play a role in enzyme–substrate interactions in vivo are difficult to replicate in vitro. For example, in vitro assays with recombinant enzyme cannot account for protein–protein interactions or spatial localization that can promote or prevent interactions between peptidases and candidate substrates (22). To overcome these challenges, recent studies have begun to look at the direct impact of peptidase activity on the peptides in cells and tissues through the application of mass spectrometry (MS)-based peptidomics (23–28). In these experiments, changes in the peptide profile as a function of peptidase activity can reveal substrates, products, and pathways regulated by a peptidase in vivo.
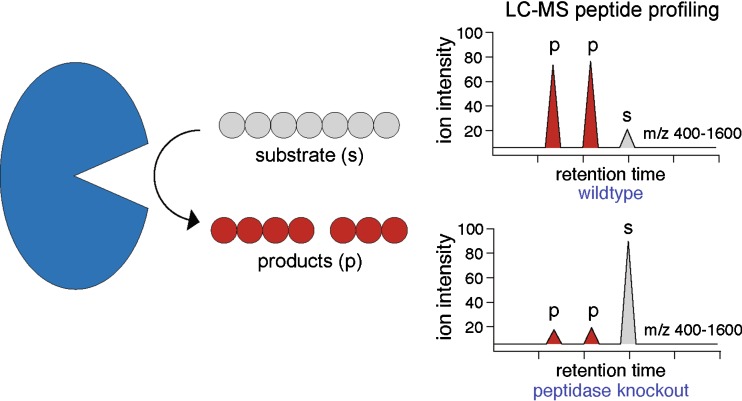
Peptidomics approaches attempt to detect and quantify peptides directly from cells and tissues. Through liquid chromatography (LC)–MS-based peptidomics experiments, the necessary protocols for the isolation and detection of peptides from tissues have been developed (23,24,26,29–32). In such experiments, known endogenous peptides and, importantly, previously unidentified peptides, including posttranslationally modified versions of known peptides, can be measured (29). Recently, peptidomics has been applied to identify peptidase-regulated peptides, including substrates, by quantifying changes in the tissue peptidome as a function of peptidase activity (23–25,27,28,31,32) (Fig. 2). Using this approach, many of the challenges associated with translating the findings from in vitro assays into biological insights that are applicable in vivo can be overcome. Fricker and colleagues, who studied the neuropeptides regulated by carboxypeptidase E (CPE), provided the first examples of the successful use of peptidomics to study peptidases (26,27). In these experiments, comparison of the peptidomes from tissue samples from mice with or without CPE revealed peptides and pathways regulated by CPE (Fig. 2). Since these initial efforts, similar approaches have been used to study other peptidases as well (23–28,31,32).
Fig. 2.
Identification of peptidase substrates through global LC–MS-based peptidomics. As with any enzyme, understanding the substrate profile of a peptidase is an important question that must be answered. Toward this goal, the use of peptidomics to identify changes in tissue peptides as a function of a peptidase activity can overcome many of the challenges associated with in vitro assays. In these experiments, comparison of the peptide profiles from wild-type (peptidase active) and knockout (peptidase inactive) samples can identify peptides regulated by the enzyme as changes in the ion abundance in the LC–MS chromatogram. For example, substrates often accumulate in the absence of a peptidase (bottom panel) resulting in a more abundant peak in the LC–MS chromatogram (gray peak). Through this approach, the substrate selectivity of an enzyme can be determined, and peptidase substrates can be identified, which might provide clues about the role of the enzyme in these tissues
In the analysis of prolyl peptidases, pharmacology (23,28) and genetics (23,24) have both been employed to inactivate the enzymes. The benefit of genetic knockouts is that they are highly specific, effectively remove all enzyme activity, and can be used to target any peptidase without needing any specific reagents. In certain cases of genetic knockouts, however, compensatory activities can emerge that mask the loss of the peptidase of interest, potentially confounding the results obtained. To date, this appears to be more the exception than the rule since peptidase knockouts of CPE and prolyl peptidases do show reasonable changes in the peptidome. Since peptidases can be inhibited with small molecules, pharmacology provides an additional way to regulate peptidase activity. The primary challenge with a pharmacological approach is related to the discovery of a peptidase inhibitor that is selective and able to function within the context of a cell, tissue, and organism. Once such an inhibitor is obtained, it can be used to study the changes in the peptidome that emerge from acute inhibition (typically hours) of a peptidase. Moreover, pharmacologically inhibited samples overcome the challenge of compensation because of the shorter timescale of inhibition. In either strategy, peptides that differ between samples should be tested in vitro to distinguish substrates and products of the enzyme from more indirect changes.
Below, we review the application of peptidomics to the study of the prolyl peptidases Prep and DPP4. These examples highlight the utility of peptidomics in defining peptidase specificity, natural substrates of the enzyme, and unappreciated biochemical crosstalk between peptidase pathways. With Prep, these studies led to an improved understanding of the biochemistry of the enzyme, and illustrated the difficulty in predicting peptidase substrates or cleavage sites based on peptide primary sequence alone (23). In the case of DPP4, peptidomics provided biochemical information about this enzyme and also revealed the existence of interlinked biochemical pathways involved in renal peptide catabolism (25). Lastly, peptide changes regulated by DPP4 were useful as a metric for increasing the coverage of the peptidomics platform through a systematic analysis of the key steps in the workflow (24). Together, these examples highlight the utility of peptidomics in revealing the endogenous functions of the prolyl peptidases.
PEPTIDOMICS OF PREP IN THE CENTRAL NERVOUS SYSTEM
Prep was first identified and partially purified in 1971 as the enzyme responsible for the oxytocin cleaving activity in the human uterus (33). The unique biochemical activity of Prep, namely the ability to cleave peptides at proline amino acids, has prompted a great deal of research into the biochemical, cellular, and physiological functions of this protein (9). In vitro assays have identified Prep substrates that range from the tripeptide, thyrotropin-releasing hormone, to a 31 amino acid peptide, beta endorphin (9,34), which led to new hypotheses about the physiological role of Prep, including a role in cognitive function (15–18).
The identification of endogenous Prep substrates is a major part of the effort to elucidate Prep-regulated pathways related to cognition (9,23,28,35). These studies integrate pharmacological inhibition of Prep with postmortem measurements of peptide levels in the central nervous system. The most common method for measuring peptide levels has been to use antibody-based approaches such as immunoassays to detect and quantify changes in levels of specific peptides as a function of Prep inhibition (19–21). Because immunoassays are necessarily targeted experiments, they are used to test hypotheses, but cannot uncover unknown or unanticipated substrates. For example, elevated levels of substance P in the brain upon Prep inhibition by S17092 were measured by immunohistochemistry (19).
Two recent studies have applied MS-based peptidomics to study changes in the peptidome upon Prep inhibition in the central nervous system (CNS) of rodents (23,28). Interestingly, while both reports are similar in concept, they differ greatly in the details, including the animal model, inhibitor, time of inhibition, and MS approach. The first study utilized the commercially available inhibitor, Z-ProProlinal, in a rat model (28). After a 4-h treatment with inhibitor, the rats were sacrificed, their brains isolated, and peptides were extracted. To quantify differences between the two samples, an isotopic labeling method termed iTRAQ was employed (36). In this approach, peptides are labeled with a reagent that places distinct isotopic tags on the different sample groups (e.g., inhibitor and vehicle). The two samples are then mixed and analyzed simultaneously by MS, where quantitation is accomplished by calculating the ratio of the different isotopes during the MS analysis. By analyzing the inhibitor- and vehicle-treated groups simultaneously, iTRAQ improves the accuracy of the quantitation by reducing run-to-run variation that might appear using label-free approaches.
Using this approach, Tenorio-Laranga and colleagues were able to identify a number of novel Prep substrates in the brains of inhibitor-treated rats. The largest change corresponded to a fragment of NADH dehydrogenase 1 alpha subunit, which was elevated 1.9-fold in the vehicle-treated animal, suggesting that this is a product of Prep activity whose levels are reduced in the presence of Prep inhibitor. In addition, a fragment of the known Prep substrate thymosin beta4 was similarly elevated in the vehicle-treated sample, demonstrating the ability to identify known substrates (37). A majority of the peptides identified as putative Prep substrates contained a proline, making them likely Prep substrates, although their ability to serve as Prep substrates was not explicitly tested. On the basis of the number and diversity of candidate proline-containing substrates identified, the authors concluded that Prep is involved in the CNS catabolism of peptides, including bioactive peptides.
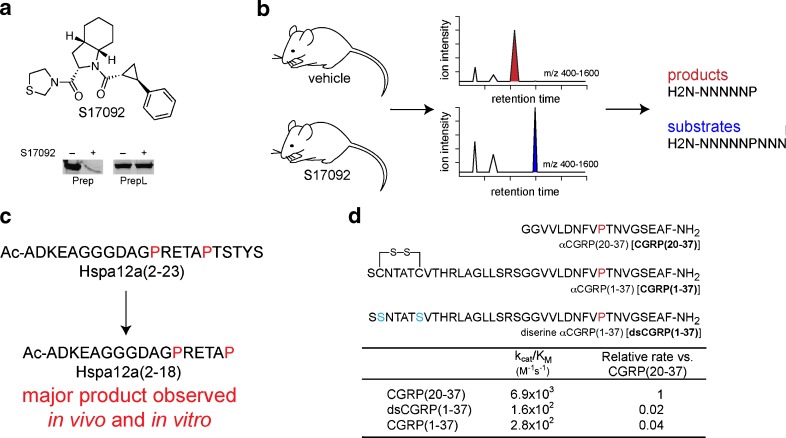
Similarly, we utilized a pharmacological inhibitor of Prep, S17092, in a mouse model to study the impact of Prep inhibition on the CNS peptidome (23) (Fig. 3). In contrast to the approach of Tenorio-Laranga and colleagues, we utilized a label-free strategy to quantify differences between samples. Importantly, a final comparison of the outcomes of the two Prep studies revealed some of the same Prep-regulated peptides, which indicates that the differences between the two approaches did not preclude similar results from being obtained.
Fig. 3.
Peptidomics of Prep. a S17092 is a Prep inhibitor which was shown to be selective for Prep over the closely related peptidase, prolyl endopeptidase-like (PrepL). b Workflow for the identification of Prep substrates in the CNS consisted of examining tissues (brain, spinal cord and hypothalamus) from mice treated with S17092 or vehicle. Differences in ion abundance between the two samples revealed Prep-regulated peptides, including substrates and products of the enzyme. c The identification of natural substrates, such as Hspa12a(12-23), revealed that Prep can show cleavage site selectivity between two proline amino acids, which would not have been predicted based on the known substrate selectivity of Prep. d Prep also showed a length dependence, preferring shorter substrates, which allows it to cleave the degradation fragment of CGRP, CGRP(20-37), without cleaving the bioactive species CGRP(1-37)
In our study, peptides from S17092- and vehicle-treated mice (i.e., control samples with no inhibitor added) were prepared and analyzed by LC–MS (Fig. 3). These data were then processed using XCMS software (38,39), which aligns, quantifies, and statistically ranks changes in ions between two datasets, in this case S17092 and vehicle. XCMS had previously been used to identify differences in metabolomics datasets, and this example demonstrated its utility in peptidome analysis. After XCMS analysis, Sequest (40,41) or Mascot (42,43) can be used to identify the peptide sequences of any changing ions. In the analysis of Prep substrates in the nervous system, we found that it was important to account for posttranslational modifications including acetylation, phosphorylation, and C-terminal amidation because many of the Prep-regulated peptides were modified (23,29).
Analysis of the spinal cord, brain, and hypothalamus revealed 19 Prep-regulated peptides with some overlap between these tissues. Interestingly, some of the changes were specific to one region of the CNS (23). For example, substance P was observed in all tissues, but only changed significantly in the brain. As expected, many of the S17092-elevated peptides, which are potential Prep substrates, contain a proline residue. Moreover, this label-free approach also identified some known Prep substrates, namely substance P and thymosin beta4. Importantly, not every peptide containing a proline residue is a Prep substrate in vivo. For example, we observed a proline-containing cytochrome C fragment that is unchanged between S17092- and vehicle-treated samples.
On the basis of the diversity of substrates discovered, the conclusion that Prep might be involved in the catabolism of proline-containing peptides, first proposed by Tenorio-Laranga and colleagues (28), is very reasonable. The diversity in the peptide sequences of natural substrates, many of which had multiple proline residues, prompted us to use these natural substrates to study the in vitro biochemistry of Prep. Of particular interest was the fact that some of the Prep substrates appeared to be processed in a manner that could have not been predicted from the known substrate selectivity of the peptidase. In particular, some peptides containing multiple proline residues seemed to be cleaved at only a single site, which was evident because only one of several possible product peptides was found at higher levels in the vehicle-treated sample.
Recombinant expression of Prep and chemical synthesis of some of these Prep substrates enabled the investigation of Prep selectivity in vitro. The cleavage of the neurosecretory protein VGF (490-507) (Vgf(490-507)), Vgf(24-39), preproenkephalin 1 (114-133) (Penk(114-133)), and acetylated heat shock 70-kDa protein 12a(2-23) (Ac-Hspa12a(2-23)) were studied with the recombinant enzyme. Two of these peptides, Vgf(24-39) and Penk(114-133), were cleaved at multiple proline sites in the peptide, which was expected for Prep. In contrast, the other two peptides, Vgf(490-507) and Ac-Hspa12a(2-23), showed cleavage patterns that could not be predicted or explained using known models for Prep selectivity. For example, Prep cleaves the peptide Ac-Hspa12a(2-23) predominantly at one specific proline, even though there are two potential Prep cleavage sites in this peptide, one after each proline (Fig. 3).
Overall, there was good agreement between in vitro and in vivo results, suggesting that the cleavage patterns observed in vivo were a direct result of Prep activity, and not other confounding factors that might have been present in tissues. Additional evidence that Prep does not unconditionally cut at any proline in a peptide can be found in the in vivo data. Here, many of the vehicle-elevated peptides, which are potential Prep cleavage products, still contain proline, indicating that Prep does not cut at every possible proline.
We also compared the cleavage of CGRP(20-37), an endogenous Prep substrate, and full-length CGRP(1-37) to investigate the role of peptide length and cleavage site context in Prep biochemistry. Pioneering work by Polgár and coworkers had previously revealed a length dependence in Prep substrate selectivity, namely that Prep clearly preferred shorter peptides. Similarly, our experiments demonstrate that Prep favors the shorter peptide, CGRP(20-37), as a substrate, with a specificity constant (kcat/KM) that is 20-fold higher than that for CGRP(1-37) (Fig. 3). In vivo, this selectivity would allow Prep to cleave peptides of a certain length, e.g. CGRP(20-37), while having no impact on longer peptides, such as the full-length bioactive CGRP(1-37), whose levels do not change between S17092- and vehicle-treated samples. More generally, this result suggests that peptidases in the proteome might be stratified by a length dependence in addition to a sequence preference. This provides an added layer of selectivity which might be difficult to assess using in vitro assays with unnatural substrates alone.
In aggregate, our results reveal the value of integrating peptidomics analysis with biochemical studies to define features of enzymes, such as site selectivity, that are not evident using model substrates. Thus, these studies provide insight into the biochemical substrates of Prep in the nervous system and, like any discovery-based approach, reveal gaps in our current knowledge that will stimulate future studies with Prep.
PEPTIDOMICS OF DPP4 IN THE KIDNEY
The best studied member of the prolyl peptidase family is DPP4. To better understand the full range of physiological roles that DPP4 might be involved in, recent efforts have attempted to identify new DPP4 substrates. Initial efforts in this area relied on in vitro assays using purified enzymes and peptide substrate libraries (44). However, the difficulty in using in vitro substrate profiles to predict endogenous substrates is demonstrated by the lack of a preferred cleavage site (H2N-Xaa-Pro) in certain DPP4 substrates such as GLP-1 and peptide YY (PYY) (45). Additionally, the realization that the in vivo peptide pool is composed of many unknown peptides makes the challenge of substrate prediction even more daunting (30). In an effort to circumvent the problem of using nonnatural substrates, Yates and colleagues performed in vitro MS-based peptidomics, by adding recombinant DPP4 to a pool of plasma peptides and measuring changes in the peptidome, an approach referred to as differential mass spectrometry (46). These studies were able to identify a number of DPP4 substrates in the plasma, including the bioactive peptide bradykinin (47).
Similarly, Jost and colleagues expanded on this approach by looking at changes in plasma peptides upon pharmacological inhibition of DPP4 in rats (48). In this work, they used a method termed differential peptide display, which relies on quantitative MALDI–MS to identify physiological DPP4-regulated peptides. These studies were able to find a number of peptide substrates of DPP4, including collagen fragments and the BRI peptide, an amyloid-causing peptide. As these examples demonstrate, the role of DPP4 in plasma peptide regulation has been investigated using MS-based peptidomics, and we wondered whether analysis of DPP4 in other tissues could provide insights into other roles of the enzyme.
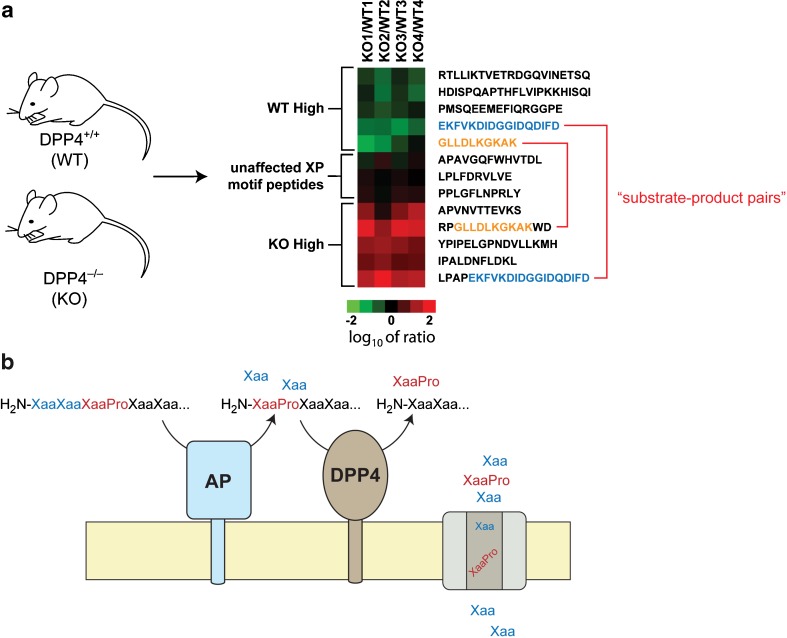
In these experiment, tissue samples from wild-type (DPP4+/+) and DPP4 null (DPP4−/−) (7) mice were compared by peptidomics to identify DPP4 substrates in the kidney (25). The peptides found to be regulated by DPP4 in the kidney were not classical hormones but fragments from different precursor proteins. Interestingly, few of the peptides regulated by DPP4 in the kidney had previously been reported in the literature, which is a testament to the complexity of the peptidome and also serves to highlight the value of unbiased mass spectrometry methods.
The DPP4-regulated peptides elevated in the DPP4−/− samples contained the canonical DPP4 cleavage site (i.e., H2N-XaaPro-peptide), suggesting that these peptides were being regulated directly by DPP4, which was confirmed through in vitro cleavage assays (Fig. 4). Importantly, a number of endogenous peptides that have a penultimate proline at the N terminus were not regulated by DPP4 in vivo; however, these peptides were excellent in vitro substrates, demonstrating the difficulty in using in vitro assays alone to identify physiological substrates.
Fig. 4.
Peptidomics of DPP4 in the kidney. a Comparison of kidney tissue from DPP4+/+ and DPP4−/− mice revealed a number of peptides regulated by the enzyme in vivo. All the peptides elevated in the DPP4−/− kidney contained the canonical DPP4 cleavage site (H2N-XaaPro-peptide), and these peptides were confirmed as substrates for the enzyme. Importantly, not all H2N-XaaPro-peptides were regulated by DPP4, demonstrating the value of in vivo measurements in identifying physiologically relevant substrates. b Further studies with brush border membranes revealed that aminopeptidase activity is required to generate DPP4 substrates, providing a mechanism and a pathway for peptide catabolism in the kidney
Prior work indicated a role for DPP4 in renal protein and peptide catabolism (4). In this model, DPP4 is part of the proteolytic system that converts proteins and peptides into amino acids and dipeptides, which are then transported into cells through amino acid and dipeptide transporters, respectively (4). Amino acid and peptide measurements of urine from rats lacking a functional copy of DPP4 revealed lower levels of proline and higher levels of peptide-bound proline in the absence of DPP4 activity, demonstrating a role for DPP4 in liberating proline from peptides in vivo (4).
Additional support for this model comes from the LC–MS peptidomics data. For instance, in the peptidomics data, there are multiple fragments of the diazepam binding inhibitor (DBI) protein, which suggests that the protein is being catabolized in the kidney. Of the three DBI peptides detected, only the proline-containing peptide is found at a higher level in DPP4−/− kidneys, which indicates a specific role for DPP4 in proline peptide catabolism in the kidney. In addition, the DPP4 substrates are derived from precursor proteins with differing cellular localizations (e.g., intracellular, extracellular, nuclear, etc.), which is consistent with the notion that extracellular DPP4 has a broad role in renal protein/peptide catabolism, which occurs extracellularly.
While the peptidomics data supported a physiological role for DPP4 in renal peptide catabolism, it was still unclear how the DPP4 substrates themselves were generated. The mechanism of DPP4 substrate generation was investigated through in vitro experiments with synthetic peptides and brush border membrane preparations from kidneys of DPP4+/+ and DPP4−/− mice. The brush border is the region within the proximal tubule of the kidney which contains the proteolytic and peptidolytic activity for protein catabolism. It is the site where DPP4 and several other peptidases are located (4,49).
Synthetic versions of the endogenous DPP4 substrates were used to study, in greater detail, the mechanism of renal peptide catabolism. In these experiments, a peptide is added to the brush border membrane lysate and subsequently analyzed by LC–MS to identify any degradation products. The products identified provide insight into the underlying enzymatic activities that generated the peptides. For example, addition of the DPP4 substrate derived from the meprin β protein, Mepβ(21-41), to brush border membranes from DPP4+/+ and DPP4−/− mice resulted in different peptide profiles. In the DPP4+/+ sample, the peptide was cleanly processed to give the dipeptide-truncated products, Mepβ(23-41) and Mepβ(25-41), while the levels of these peptides were significantly lower in the DPP4−/− sample. As expected, this experiment demonstrates an irreplaceable role for DPP4 in the processing of proline-containing peptides in the kidney.
In addition, Mepβ(25-41), the DPP4 cleavage product which lacks proline, was also assayed in this experiment. Unlike the proline-containing peptide, there was no genotype-dependent processing of Mepβ(25-41). Instead, the N-terminal processing of this peptide was mediated by aminopeptidase activity, which produced a ladder of singly truncated N-terminal products. Thus, the brush border experiments showed that aminopeptidase (AP) and DPP4 activities are both important pathways for N-terminal peptide degradation and that DPP4 products are AP substrates. Interestingly, aminopeptidase activity adjacent to a proline residue (i.e., XaaPro-peptide) was not seen even in the absence of DPP4 activity, indicating that kidney aminopeptidases do not accept substrates with a penultimate proline. This insight, provided by the lack of a particular product, in turn suggested a potential mechanism for DPP4 substrate production.
In this model for DPP4 substrate production, any proline-containing peptide is first processed by aminopeptidase activity until a penultimate proline is reached and the peptide is no longer an aminopeptidase substrate. At this point, the peptide is cleaved by DPP4 to liberate a proline-containing dipeptide. In this model, AP and DPP4 activities result in interlinked pathways that together enable N-terminal degradation of proline-containing peptides. A mechanism is thus also provided for the generation of DPP4 substrates in the kidney (Fig. 4). The model requires that kidney proteolytic activity is able to generate a DPP4 substrate (i.e., a penultimate proline-containing peptide) from peptides with an internal proline. This was tested using two peptides with proline at position 3 and position 5 with respect to the N terminus. These peptides were incubated with DPP4+/+ and DPP4−/− brush border membranes, which led to the accumulation of H2N-XaaPro-containing peptides in the DPP4−/− brush border membranes, demonstrating that kidney proteolytic activity is able to generate DPP4 substrates from peptides with internal proline residues.
Next, brush border membranes from mice lacking both aminopeptidase A and N (25,50,51), the two predominant kidney aminopeptidases (52), were used to test whether DPP4 substrate production is aminopeptidase-dependent. The same two peptides with proline at position 3 and position 5 with respect to the N terminus were added to brush border membranes from wild-type (APN/A+/+) and aminopeptidase A and N null (APN/A−/−) mice. The production of penultimate proline-containing peptides was measured by LC–MS, revealing a clear and significant decrease in the amount of H2N-XaaPro-containing peptide in the absence of aminopeptidase activity. Together, these brush border experiments support the hypothesis that kidney aminopeptidase activity is responsible for the production of DPP4 substrates and lead to a model where the interlinked activities of aminopeptidase and DPP4 combine to degrade proline-containing peptides and produce amino acids and dipeptides during renal catabolism.
OPTIMIZING THE PEPTIDOMICS PLATFORM USING PEPTIDASE SUBSTRATES
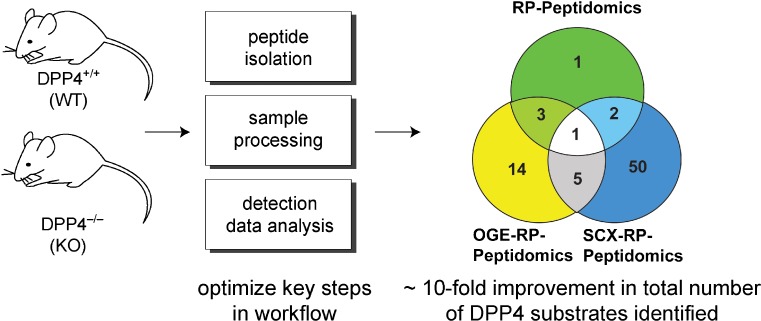
The identification of peptidase substrates and peptidase-mediated pathways required an LC–MS peptide-profiling platform capable of quantitative analysis across the peptidome. In the case of DPP4, kidney peptidomics studies supported a role for DPP4 in the catabolism of peptides based on the identification of seven DPP4 substrates that were elevated in the DPP4−/− samples. However, we anticipated finding more substrates, since loss of DPP4 activity has been shown to impact global levels of proline in rat urine, suggesting that there are numerous proline-containing peptides that are DPP4 substrates (4). We suspected that our peptide-profiling platform, while functional, needed to be improved to provide greater coverage of the peptidome. Systematic investigation of key steps in the workflow, including tissue preparation, peptide isolation, LC–MS, and data analysis, was performed in order to optimize the peptide-profiling platform (24).
Improvement in the peptide-profiling platform was measured by the number of new DPP4 substrates identified during the comparison of DPP4+/+ and DPP4−/− kidney samples. Many of the changes in the workflow had modest impacts on the number of DPP4 substrates detected. Trypsinization of the samples prior to LC–MS, for example, doubled the number of DPP4-regulated peptides identified. However, by performing an additional orthogonal fractionation approach, a strategy known to reduce sample complexity and improve peptide detection, a big improvement in peptidome coverage was indeed observed. Fractionation of the peptidome using strong cation exchange (SCX) and Offgel electrophoresis (OGE) resulted in a marked improvement in the number of DPP4 substrates identified (24) (Fig. 5). In total, the optimized workflow resulted in a 10-fold improvement in the number of DPP4-regulated peptides (75 vs. 7 previously) identified, and demonstrated the value of the DPP4 model in improving the platform.
Fig. 5.
Optimized peptidomics. By using the number of DPP4 substrates identified, the peptidomics platform was improved. In total, 75 DPP4 substrates were identified, which represents a ∼10-fold improvement over the initial conditions. Separation methods used include reverse phase (RP), strong cation exchange (SCX) and Offgel electrophoresis (OGE)
Importantly, the ability to detect a larger number of DPP4 substrates provided additional data to support an interlinked aminopeptidase–DPP4 pathway for N-terminal processing of proline-containing peptides. Specifically, while the number of DPP4-regulated peptides increased to a total of 75, there were only four peptides that did not contain the canonical DPP4 cut site (H2N-XaaPro) and only one that terminated in a proline, providing strong evidence that aminopeptidases do not process penultimate proline-containing peptides in vivo either. Finally, this improved platform was then applied to analyze DPP4-regulated peptides in the intestine, which revealed a handful of additional DPP4 substrates and demonstrated the generality of this optimized peptidomics platform.
CONCLUSIONS AND OUTLOOK
The application of LC–MS-based peptidomics has had a strong impact on the understanding of the biochemistry and proteolytic pathways regulated by the prolyl peptidases. The application of these approaches to other peptidases, including less well-characterized prolyl peptidases, promises to enrich the understanding of this important class of peptidases. These types of studies will be especially useful in elucidating the roles of peptidases that might emerge from genetic studies linking enzymes to human disease. As a result, the findings from peptide-profiling experiments are important in basic science and could impact medicine as well. Finally, peptidomics can be extended to other types of research, such as biomarker studies or the investigation of the function of other peptide-modifying enzymes.
References
- 1.Beinfeld MC, Funkelstein L, Foulon T, Cadel S, Kitagawa K, Toneff T, et al. Cathepsin L plays a major role in cholecystokinin production in mouse brain cortex and in pituitary AtT-20 cells: protease gene knockout and inhibitor studies. Peptides. 2009;30(10):1882–91. doi: 10.1016/j.peptides.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paul M, Poyan Mehr A, Kreutz R. Physiology of local renin–angiotensin systems. Physiol Rev. 2006;86(3):747–803. doi: 10.1152/physrev.00036.2005. [DOI] [PubMed] [Google Scholar]
- 3.Rosenblum JS, Kozarich JW. Prolyl peptidases: a serine protease subfamily with high potential for drug discovery. Curr Opin Chem Biol. 2003;7(4):496–504. doi: 10.1016/S1367-5931(03)00084-X. [DOI] [PubMed] [Google Scholar]
- 4.Tiruppathi C, Miyamoto Y, Ganapathy V, Roesel RA, Whitford GM, Leibach FH. Hydrolysis and transport of proline-containing peptides in renal brush-border membrane vesicles from dipeptidyl peptidase IV-positive and dipeptidyl peptidase IV-negative rat strains. J Biol Chem. 1990;265(3):1476–83. [PubMed] [Google Scholar]
- 5.Cavasin MA, Liao TD, Yang XP, Yang JJ, Carretero OA. Decreased endogenous levels of Ac-SDKP promote organ fibrosis. Hypertension. 2007;50(1):130–6. doi: 10.1161/HYPERTENSIONAHA.106.084103. [DOI] [PubMed] [Google Scholar]
- 6.Weber AE. Dipeptidyl peptidase IV inhibitors for the treatment of diabetes. J Med Chem. 2004;47(17):4135–41. doi: 10.1021/jm030628v. [DOI] [PubMed] [Google Scholar]
- 7.Marguet D, Baggio L, Kobayashi T, Bernard AM, Pierres M, Nielsen PF, et al. Enhanced insulin secretion and improved glucose tolerance in mice lacking CD26. Proc Natl Acad Sci USA. 2000;97(12):6874–9. doi: 10.1073/pnas.120069197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thornberry NA, Weber AE. Discovery of JANUVIA (Sitagliptin), a selective dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes. Curr Top Med Chem. 2007;7(6):557–68. doi: 10.2174/156802607780091028. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Horsman JA, Mannisto PT, Venalainen JI. On the role of prolyl oligopeptidase in health and disease. Neuropeptides. 2007;41(1):1–24. doi: 10.1016/j.npep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Kelly T. Fibroblast activation protein-alpha and dipeptidyl peptidase IV (CD26): cell-surface proteases that activate cell signaling and are potential targets for cancer therapy. Drug Resist Updat. 2005;8(1-2):51–8. doi: 10.1016/j.drup.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Scanlan MJ, Raj BK, Calvo B, Garin-Chesa P, Sanz-Moncasi MP, Healey JH, et al. Molecular cloning of fibroblast activation protein alpha, a member of the serine protease family selectively expressed in stromal fibroblasts of epithelial cancers. Proc Natl Acad Sci USA. 1994;91(12):5657–61. doi: 10.1073/pnas.91.12.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng JD, Valianou M, Canutescu AA, Jaffe EK, Lee HO, Wang H, et al. Abrogation of fibroblast activation protein enzymatic activity attenuates tumor growth. Mol Cancer Ther. 2005;4(3):351–60. doi: 10.1158/1535-7163.MCT-04-0269. [DOI] [PubMed] [Google Scholar]
- 13.Cheng JD, Dunbrack RL, Jr, Valianou M, Rogatko A, Alpaugh RK, Weiner LM. Promotion of tumor growth by murine fibroblast activation protein, a serine protease, in an animal model. Cancer Res. 2002;62(16):4767–72. [PubMed] [Google Scholar]
- 14.Polgar L. pH-dependent mechanism in the catalysis of prolyl endopeptidase from pig muscle. Eur J Biochem. 1991;197(2):441–7. doi: 10.1111/j.1432-1033.1991.tb15930.x. [DOI] [PubMed] [Google Scholar]
- 15.Morain P, Lestage P, De Nanteuil G, Jochemsen R, Robin JL, Guez D, et al. S 17092: a prolyl endopeptidase inhibitor as a potential therapeutic drug for memory impairment. Preclinical and clinical studies. CNS Drug Rev. 2002;8(1):31–52. doi: 10.1111/j.1527-3458.2002.tb00214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider JS, Giardiniere M, Morain P. Effects of the prolyl endopeptidase inhibitor S 17092 on cognitive deficits in chronic low dose MPTP-treated monkeys. Neuropsychopharmacology. 2002;26(2):176–82. doi: 10.1016/S0893-133X(01)00307-4. [DOI] [PubMed] [Google Scholar]
- 17.Marighetto A, Touzani K, Etchamendy N, Torrea CC, De Nanteuil G, Guez D, et al. Further evidence for a dissociation between different forms of mnemonic expressions in a mouse model of age-related cognitive decline: effects of tacrine and S 17092, a novel prolyl endopeptidase inhibitor. Learn Mem. 2000;7(3):159–69. doi: 10.1101/lm.7.3.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toide K, Iwamoto Y, Fujiwara T, Abe H. JTP-4819: a novel prolyl endopeptidase inhibitor with potential as a cognitive enhancer. J Pharmacol Exp Ther. 1995;274(3):1370–8. [PubMed] [Google Scholar]
- 19.Bellemere G, Morain P, Vaudry H, Jegou S. Effect of S 17092, a novel prolyl endopeptidase inhibitor, on substance P and alpha-melanocyte-stimulating hormone breakdown in the rat brain. J Neurochem. 2003;84(5):919–29. doi: 10.1046/j.1471-4159.2003.01536.x. [DOI] [PubMed] [Google Scholar]
- 20.Bellemere G, Vaudry H, Morain P, Jegou S. Effect of prolyl endopeptidase inhibition on arginine-vasopressin and thyrotrophin-releasing hormone catabolism in the rat brain. J Neuroendocrinol. 2005;17(5):306–13. doi: 10.1111/j.1365-2826.2005.01308.x. [DOI] [PubMed] [Google Scholar]
- 21.Toide K, Okamiya K, Iwamoto Y, Kato T. Effect of a novel prolyl endopeptidase inhibitor, JTP-4819, on prolyl endopeptidase activity and substance P- and arginine-vasopressin-like immunoreactivity in the brains of aged rats. J Neurochem. 1995;65(1):234–40. doi: 10.1046/j.1471-4159.1995.65010234.x. [DOI] [PubMed] [Google Scholar]
- 22.Saghatelian A, Trauger SA, Want EJ, Hawkins EG, Siuzdak G, Cravatt BF. Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry. 2004;43(45):14332–9. doi: 10.1021/bi0480335. [DOI] [PubMed] [Google Scholar]
- 23.Nolte WM, Tagore DM, Lane WS, Saghatelian A. Peptidomics of prolyl endopeptidase in the central nervous system. Biochemistry. 2009;48(50):11971–81. doi: 10.1021/bi901637c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tinoco AD, Tagore DM, Saghatelian A. Expanding the dipeptidyl peptidase 4-regulated peptidome via an optimized peptidomics platform. J Am Chem Soc. 2010;132(11):3819–30. doi: 10.1021/ja909524e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tagore DM, Nolte WM, Neveu JM, Rangel R, Guzman-Rojas L, Pasqualini R, et al. Peptidase substrates via global peptide profiling. Nat Chem Biol. 2009;5(1):23–5. doi: 10.1038/nchembio.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Che FY, Lim J, Pan H, Biswas R, Fricker LD. Quantitative neuropeptidomics of microwave-irradiated mouse brain and pituitary. Mol Cell Proteomics. 2005;4(9):1391–405. doi: 10.1074/mcp.T500010-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Che FY, Yuan Q, Kalinina E, Fricker LD. Peptidomics of Cpe fat/fat mouse hypothalamus: effect of food deprivation and exercise on peptide levels. J Biol Chem. 2005;280(6):4451–61. doi: 10.1074/jbc.M411178200. [DOI] [PubMed] [Google Scholar]
- 28.Tenorio-Laranga J, Valero ML, Mannisto PT, Sanchez del Pino M, Garcia-Horsman JA. Combination of snap freezing, differential pH two-dimensional reverse-phase high-performance liquid chromatography, and iTRAQ technology for the peptidomic analysis of the effect of prolyl oligopeptidase inhibition in the rat brain. Anal Biochem. 2009;393(1):80–7. doi: 10.1016/j.ab.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 29.Svensson M, Skold K, Svenningsson P, Andren PE. Peptidomics-based discovery of novel neuropeptides. J Proteome Res. 2003;2(2):213–9. doi: 10.1021/pr020010u. [DOI] [PubMed] [Google Scholar]
- 30.Zougman A, Pilch B, Podtelejnikov A, Kiehntopf M, Schnabel C, Kumar C, et al. Integrated analysis of the cerebrospinal fluid peptidome and proteome. J Proteome Res. 2008;7(1):386–99. doi: 10.1021/pr070501k. [DOI] [PubMed] [Google Scholar]
- 31.Minokadeh A, Funkelstein L, Toneff T, Hwang SR, Beinfeld M, Reinheckel T, et al. Cathepsin L participates in dynorphin production in brain cortex, illustrated by protease gene knockout and expression. Mol Cell Neurosci. 2010;43(1):98–107. doi: 10.1016/j.mcn.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Yasothornsrikul S, Greenbaum D, Medzihradszky KF, Toneff T, Bundey R, Miller R, et al. Cathepsin L in secretory vesicles functions as a prohormone-processing enzyme for production of the enkephalin peptide neurotransmitter. Proc Natl Acad Sci USA. 2003;100(16):9590–5. doi: 10.1073/pnas.1531542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walter R, Shlank H, Glass JD, Schwartz IL, Kerenyi TD. Leucylglycinamide released from oxytocin by human uterine enzyme. Science. 1971;173(999):827–9. doi: 10.1126/science.173.3999.827. [DOI] [PubMed] [Google Scholar]
- 34.Moriyama A, Nakanishi M, Sasaki M. Porcine muscle prolyl endopeptidase and its endogenous substrates. J Biochem. 1988;104(1):112–7. doi: 10.1093/oxfordjournals.jbchem.a122404. [DOI] [PubMed] [Google Scholar]
- 35.Brandt I, De Vriendt K, Devreese B, Van Beeumen J, Van Dongen W, Augustyns K, et al. Search for substrates for prolyl oligopeptidase in porcine brain. Peptides. 2005;26(12):2536–46. doi: 10.1016/j.peptides.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 36.Wiese S, Reidegeld KA, Meyer HE, Warscheid B. Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics. 2007;7(3):340–50. doi: 10.1002/pmic.200600422. [DOI] [PubMed] [Google Scholar]
- 37.Cavasin MA, Rhaleb NE, Yang XP, Carretero OA. Prolyl oligopeptidase is involved in release of the antifibrotic peptide Ac-SDKP. Hypertension. 2004;43(5):1140–5. doi: 10.1161/01.HYP.0000126172.01673.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nordstrom A, O'Maille G, Qin C, Siuzdak G. Nonlinear data alignment for UPLC-MS and HPLC-MS based metabolomics: quantitative analysis of endogenous and exogenous metabolites in human serum. Anal Chem. 2006;78(10):3289–95. doi: 10.1021/ac060245f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78(3):779–87. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 40.Eng JK, McCormack AL, Yates Iii JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5(11):976–89. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 41.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 42.Creasy DM, Cottrell JS. Error tolerant searching of uninterpreted tandem mass spectrometry data. Proteomics. 2002;2(10):1426–34. doi: 10.1002/1615-9861(200210)2:10<1426::AID-PROT1426>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 43.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol. 2005;1:0017. doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leiting B, Pryor KD, Wu JK, Marsilio F, Patel RA, Craik CS, et al. Catalytic properties and inhibition of proline-specific dipeptidyl peptidases II, IV and VII. Biochem J. 2003;371(Pt 2):525–32. doi: 10.1042/BJ20021643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mentlein R, Dahms P, Grandt D, Kruger R. Proteolytic processing of neuropeptide Y and peptide YY by dipeptidyl peptidase IV. Regul Pept. 1993;49(2):133–44. doi: 10.1016/0167-0115(93)90435-B. [DOI] [PubMed] [Google Scholar]
- 46.Yates NA, Deyanova EG, Geissler W, Wiener MC, Sachs JR, Wong KK, et al. Identification of peptidase substrates in human plasma by FTMS based differential mass spectrometry. Int J Mass Spectrom. 2007;259(1–3):174–83. [Google Scholar]
- 47.Regoli D, Barabe J. Pharmacology of bradykinin and related kinins. Pharmacol Rev. 1980;32(1):1–46. [PubMed] [Google Scholar]
- 48.Jost MM, Lamerz J, Tammen H, Menzel C, De Meester I, Lambeir AM, et al. In vivo profiling of DPP4 inhibitors reveals alterations in collagen metabolism and accumulation of an amyloid peptide in rat plasma. Biochem Pharmacol. 2009;77(2):228–37. doi: 10.1016/j.bcp.2008.09.032. [DOI] [PubMed] [Google Scholar]
- 49.Biber J, Stieger B, Stange G, Murer H. Isolation of renal proximal tubular brush-border membranes. Nat Protoc. 2007;2(6):1356–9. doi: 10.1038/nprot.2007.156. [DOI] [PubMed] [Google Scholar]
- 50.Lin Q, Taniuchi I, Kitamura D, Wang J, Kearney JF, Watanabe T, et al. T and B cell development in BP-1/6C3/aminopeptidase A-deficient mice. J Immunol. 1998;160(10):4681–7. [PubMed] [Google Scholar]
- 51.Rangel R, Sun Y, Guzman-Rojas L, Ozawa MG, Sun J, Giordano RJ, et al. Impaired angiogenesis in aminopeptidase N-null mice. Proc Natl Acad Sci USA. 2007;104(11):4588–93. doi: 10.1073/pnas.0611653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hooper NM, Turner AJ. Ectoenzymes of the kidney microvillar membrane. Differential solubilization by detergents can predict a glycosyl-phosphatidylinositol membrane anchor. Biochem J. 1988;250(3):865–9. doi: 10.1042/bj2500865. [DOI] [PMC free article] [PubMed] [Google Scholar]