Abstract
Nanotechnology is giving us a glimpse into a nascent field of nanopharmacology that deals with pharmacological phenomena at molecular scale. This review presents our perspective on the use of scanning probe microscopy techniques with special emphasis to multidimensional atomic force microscopy (m-AFM) to explore this new field with a particular emphasis to define targets, design therapeutics, and track outcomes of molecular-scale pharmacological interactions. The approach will be to first discuss operating principles of m-AFM and provide representative examples of studies to understand human health and disease at the molecular level and then to address different strategies in defining target macromolecules, screening potential drug candidates, developing and characterizing of drug delivery systems, and monitoring target–drug interactions. Finally, we will discuss some future directions including AFM tip-based parallel sensors integrated with other high-throughput technologies which could be a powerful platform for drug discovery.
Electronic supplementary material
The online version of this article (doi:10.1208/s12248-010-9232-y) contains supplementary material, which is available to authorized users.
Key words: atomic force microscopy, drug discovery, multimodal AFM, nanopharmacology, nanotherapeutics
INTRODUCTION
Jonathan Swift’s Gulliver’s Travels, first published in 1726, concerned, in part, his adventures after being shipwrecked and awakening as a prisoner of a tiny race of people who inhabit the country of Lilliput. The authors of this review (RL and MFA) have been having adventures together since 1991 in their own Lilliputian world of applying nanotechnology to the biomedical science (1) using atomic force microscopy (AFM). This review will focus on the application of multidimensional atomic force microscope (m-AFM) to nanopharmacology in defining the structural targets for drug action, in drug design and delivery systems, in monitoring target–drug interaction and in tracking their outcomes at the molecular level. We will begin this review with a brief introduction to AFM principles, imaging modes, and their unique advantages.
Operating Principle of AFM
Gerd Binnig and Heinrich Rohrer received the Nobel Prize in physics in 1986 for inventing the scanning tunneling microscope 5 years after describing the method. In 1986, Binnig et al. (2) developed the atomic force microscope, a technique that evolved into a series of scanning probe microscopes. The genius of the approach for AFM, like other related scanning probe microscopes, is that it uses the physical interactions between a cantilevered probe (tip) and the specimen for imaging as it scans over the specimen’s features. A very sharp cantilevered tip passively senses the localized interaction forces between the atoms of the scanning tip and the specimen surface as either the tip or the specimen is raster scanned in the x-y plane over its complement (tip or sample). The raster scanning is accomplished either by attaching the sample or the cantilevered tip itself to a scanner. The cantilever is commonly made of silicon or silicon nitride, although other materials can be used, and the radius of curvature of the tip is in the order of nanometers. The scanner consists of piezoelectric ceramic elements with a sub-nanometer hardware resolution. The z (vertical) position of the sample (or tip) is monitored by an electronic feedback circuit that maintains a constant deflection, ensuring a constant force of interaction between the cantilever tip and sample. The deflection of the cantilever is proportional to the interaction force between the sample and the AFM probe. The amount of z variation needed to maintain the interaction force constant is plotted versus the x and y coordinate, thereby producing a topographic image. Most commonly, the cantilever deflection is monitored by bouncing a laser beam on the back of the cantilever to a quadrant photodetector which converts light signal into electrical signal (Fig. 1, top left panel) that is fed to an electronic feedback circuit that generate necessary voltage to the piezocrystal to maintain the tip–specimen distance constant and hence follow the sample surface contour to provide a 3D topographical image.
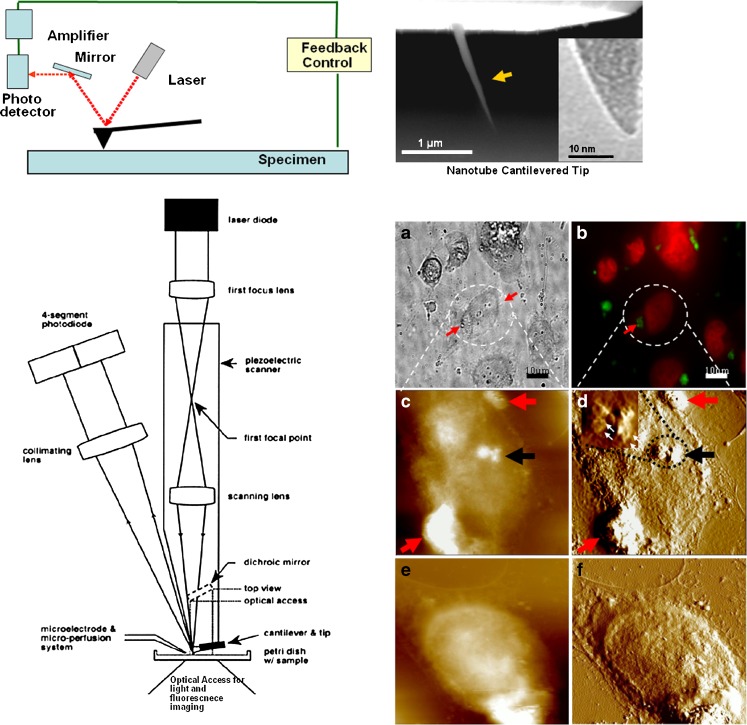
Fig. 1.
Multimodal AFM. Top left Operating principle of AFM. A cantilevered tip is scanned over the sample, maintaining constant force using feedback control to keep cantilever deflection at constant amplitude. Cantilever deflection, in this example, is measured using an optical beam deflection system. Top right Scanning electron microscope image of a microfabricated integrated nanotube cantilevered tip; inset shows the nanometer-sized tip (adapted from Chen et al. (6)). Bottom Left A schematic diagram of the optical components of an AFM combined with a light fluorescence microscope. The optical focusing of the laser light is integrated within the piezotube which provides the lateral movement of the cantilever. The scanning lens is located inside the lower segment, which provides the vertical movement of the cantilever. After the positions of the lenses are adjusted, the scanning focused spot accurately tracks the cantilever, and the zero deflection signal from the four-segment photodiode is independent of position within the scan area. The AFM is placed on the stage of an inverted microscope (adapted from Hansma et al. (15)). Bottom right Internalization of small liposomes: light microscopy (a), fluorescence (b), and tapping mode AFM (c–f) imaging of fixed cells at different time intervals after incubating with cisplatin-encapsulated liposomes. Height mode images (c, e) and error mode images (d, f) of the same cells. Error mode images show better contrast and ultrastructural details. The liposomal position with respect to the cell membrane was determined from the height mode images. For cells incubated with cisplatin-encapsulated liposomes for only 1-h incubation sample, many large liposomes are seen on the cell surface (red arrows in a–d). On the other hand, small cisplatin-encapsulated liposomes appear to be internalized in the cell cytoplasm (black arrows in c, d). The inset in d shows a zoomed-in portion of the encircled area: the internalized liposomes are clusters of small liposomes (white arrows) of ~250-nm diameter. After 16 h of incubation, no defined liposomes were observed (e, f). Scan size of AFM images is 25 μm, adapted from Ramachandran et al. (17)
Tip–Sample Interaction Forces
When the cantilever tip approaches the sample surface, the sum of the attractive and repulsive forces between them determines the deflection of the cantilever in contact mode and the damping of vibration amplitude in the tapping mode (see “Imaging Modes” below). The overlapping of electron orbitals between the atoms of the tip and the sample are the dominant repulsive force sensed by the AFM cantilever. The Van der Waals interaction is the dominant attractive force sensed by the cantilever, which is primarily due to delocalized dipole–dipole interactions (3). The meniscus surface force that exists while imaging in air is another strong attractive force. While imaging in fluids, electrostatic interactions occur between charges of sample and tip, and structural forces such as hydration force, solvation forces, and adhesion forces also come into play (4).
Torsional deflection of the AFM cantilever is used to measure lateral forces (5) and could be used to generate images containing information about the local surface chemistry, like separate lipid and protein clusters in a membrane. In tapping mode, conductive/magnetically coated cantilevers can sense electrostatic/magnetic forces and image, for instance, magnetic domains, surface charge distributions, local surface capacitance, and conductance. In principle, any signal that is spatially resolved (x, y) and that is a function of distance from the sample (z) can be made into a microscope parameter.
Imaging Modes
The two most commonly employed imaging modes for the AFM are the contact and tapping or vibration modes. In the contact mode, an electronic feedback circuit maintains a constant deflection, ensuring a constant force of interaction between the cantilever tip and sample. The amount of z variation required to maintain the interaction force constant is plotted versus the x and y coordinate, thereby producing a topographic image, although images may not be purely topographical (see “Tip–Sample Interaction Forces” above).
In the tapping or vibration mode, the cantilever oscillates up and down near its resonance frequency by a piezoelectric element on the tip holder. The amplitude of cantilever vibration is maintained constant during scanning by a feedback signal. The image is produced by plotting the displacement of the sample relative to the base of the cantilever necessary to keep the amplitude of the vibration constant. The phase lag between the driving circuit and the actual tip vibration is also measured and used for imaging physical properties.
Spatial and Temporal Resolution
Since the interaction between the AFM tip and specimen is not limited by wavelength, molecular, and even atomic resolution is possible. Spatial resolution depends on the sensitivity of the piezoelectric scanner and may be as high as 0.1 nm laterally and 0.01 nm vertically. Resolution also depends on the size and shape of the probing tip as well as on the material properties of the sample under investigation. In general, the sharper the tip, the higher is the resolution. To this end, we have been able to fabricate the first integrated cantilevered nanotip (Fig. 1, top right panel) and were able to obtain both high-resolution structural information as well as the electrical characteristics simultaneously (6). Hard, crystalline samples can be imaged with high resolution, and images with atomic and subatomic features have been achieved (7). Structures <1 nm apart are resolved on non-crystalline arrays (8). Biological samples are soft, so resolution is less. Nevertheless, the resolution can still be remarkable with soft specimens. For example, AFM studies on reconstituted ion channels can resolve the substructure of channels and, at times, the subunits within each channel. We, and others, have distinguished between hexameric and pentameric connexons and have shown that amyloid ion channels range from tetramers to octamers (9–11).
Temporal resolution depends on the speed of the feedback electronics as well as the scan size of the specimen. Examination of dynamic behavior requires higher scan speeds. Improving temporal resolution is a rapidly evolving area of AFM design. Current AFMs can image repeatedly and reliably a small line or a point over a sample surface such as a subunit of an ion channel or receptor in a millisecond, a small area such as an ion channel or receptor in less than a second, and an array of 20 × 20 channels in a second or two. The new AFMs under development hold promise to improve the temporal resolution by a factor of two or more in near future.
Advantages of AFM
Important features of AFM include: (1) The physical interaction that underlies AFM imaging is not constrained by wavelength and, therefore, permits the resolution for molecular and even atomic imaging. (2) AFM permits the 3D imaging of surfaces and biomolecules with molecular and submolecular resolution. (3) AFM does not require destructive pretreatment or imaging conditions such as occurs in electron microscopy, nor does it require crystallization as needed for high-resolution X-ray imaging that may affect physiology. (4) AFM can image specimens in a physiologic solution, thereby allowing the investigation of dynamic processes involving molecules, organelles, and other structures in living cells. (5) AFM can be modified to serve as a sensor of intermolecular interaction forces, charge, pH, and other physicochemical properties. (6) AFM cantilever tips can be functionalized chemically or biochemically to allow the identification of specific molecules and the measurement of specific interactions, such as ligand–receptor interactions. (7) Precise physical manipulation of biomolecules, dissection of biological structures and tissues, delivery of drugs, ligands, and nanoparticles to specific locations, and the measurement of interactions at specific sites are possible because of the precise nanometer control of the position and the piconewton control of the force of the cantilever. (8) The open architecture of AFM apparatus allows integrating with other complementary techniques, including conventional light and fluorescence microscopy, total internal reflection fluorescence (TIRF), Förster resonance energy transfer (FRET) microscopies, electrophysiology, opto-electronic manipulators, nano-micro fluidics, etc., to obtain complete structure–function relationship of biomolecules/structures at molecular resolution in their native physiological environment. This flexibility makes it possible to convert the AFM into a powerful multidimensional AFM (m-AFM) platform. (9) Very small amounts of drug/reagents are required, which is an enormous advantage in pharmacology where the molecules are often quite precious.
AFM AND NANOPHARMACOLOGY
Nanopharmacology could be defined as the application of nanotechnologies for drug design and drug delivery to selective molecular targets with improved pharmacodynamic and kinetic profiles for safer and effective drug treatment. This includes understanding the individual nanoscale molecular targets for rational drug design, design, development, and manufacture of nanoscale drug carriers like nanotubes, nanoshells, nanospheres, etc., nanostructured materials for drug storage and release in the body, and the ability to call back the drug after its intended effect is achieved via nanosponge, for example.
The importance of understanding pharmacological targets at their molecular level cannot be overemphasized for the advancement of nanopharmacology. AFM by itself, or along with other complementary techniques (m-AFM), would enable investigators to understand the structure–function of both the targets and effectors (drugs) at their molecular scale. For the sake of simplicity and clarity, applications of AFM techniques in nanopharmacology will be categorized broadly into the following sections for this review:
Defining targets
Development of drugs and carrier systems
Studying target–drug interactions
Monitoring the target–drug interaction outcomes and
Future directions
MULTIMODAL AFM FOR DEFINING TARGETS
Identification of features on biological samples can be difficult since many features look very similar at very high resolutions (<100 nm). One approach has been to use well-defined and highly purified biological materials, such as gap junctions or reconstituted ion channel proteins and receptors as studied in (9,10,12). Another approach has been to use complementary molecules for proper identification. A powerful feature of the AFM is that the cantilever tip can be functionalized to react very specifically with a target molecule. For example, linking of specific antibodies to the tip via a spacer allows the recognition of its antigen on the surface of a specimen (13,14). Such images reflect both topography and the strength of interaction between the antibody on the AFM tip and the antigen on the sample. Multimodal imaging also provides an approach for the direct confirmation of structural identity, such as combining AFM imaging with other techniques such as fluorescence microscopy that provides complementary information specific to each target.
AFM Combined with Optical Microscopy
As mentioned before, multimodal imaging provides an approach for the direct confirmation of targets being imaged by AFM by various optical microscopy techniques through fluorescent labeling. The open architecture of AFM allows such integration, and, as shown in Fig. 1 (bottom left), the AFM has been integrated with optical microscopy. Since its adaptation (15), simultaneous fluorescence and AFM imaging has been used to study complex biological systems, including cells cultured on Petri dishes (16). The approach has been used to link changes in cell mechanical properties, assessed by AFM, to the functionality of hemichannels, determined by fluorescence. Similar approaches could be utilized for studying the effect of drugs on cells/targets. Figure 1 (bottom right panel) shows a compelling evidence of multimodal imaging application for nanopharmacology. Cisplatin-encapsulated liposomes are imaged to show their real-time uptake by cells in culture by fluorescence as well as by AFM. Significantly, cells allowing the uptake of liposomes containing cisplatin did not survive. AFM imaging revealed that smaller liposome’s (<200 nm) were effectively internalized by cells compared to larger preparations (17).
AFM Combined with Tools to Assay Ion Conductance or Permeability
Another powerful application of m-AFM is combining imaging with the assessment of physiologic function, which could be used for target identification. An example of a combined AFM and ion conductance measurement system is the one first developed by Proksch et al. (18) in which a few-nanometer-thick glass electrode serves two purposes. First, it acts like an AFM cantilever tip and, second, acts as a nanoelectrode for recording ion conductance across a bilayer or a cell membrane. Using this system on a synthetic membrane filter (nucleopore membrane), conductance through 200-nm pores was measured when a potential difference was applied. Such an approach allow identifying molecular targets in unstained cells/tissue, which is otherwise impossible with conventional techniques at a single molecular level.
As an extension of such a combined AFM and ion conductance measuring system, AFM has recently been combined with an advanced nanochip-supported, double-chamber permeability and transport assay system (Electronic Supplementary Material (ESM) Fig. S1). This combined system allows studying the activity of ion channels and pores and would facilitate high-throughput screening of channel activity modifiers and thus would be a valuable tool for developing therapeutics for channelopathies. ESM Fig. S1 shows an example of one such design in which conductance is measured for ion channels formed by gramicidin that have been reconstituted in a lipid bilayer supported over an ~70-nm diameter nanopore in a silicon chip. Gramicidin is a complex mixture of linear polypeptide antibiotics produced by Bacillus brevis that induces ion channel-like activity when in contact with the cell membrane and is a good test structure for defining membrane permeability and transport. In the same setup, images of the nanopore and bilayer were also imaged with the AFM. Significantly, this study also shows that the AFM imaging force is soft enough to image delicate and fragile biological membranes that could be used for identifying targets.
AFM tips can be made conductive that would allow studying the conformational changes in ion channels, as would occur in response to ligands and other therapeutics. It can also be used for characterizing many advanced materials with wide biomedical applications. Using this conducting AFM, Ionescu-Zanetti et al. (19) obtained the direct structure–function analysis of conjugated polymer blends. Polymer blends have wide biological applications, including micro-actuators (20), chemical sensors (21), and light-emitting diodes (22). Optically and electrically conjugated polymer blends are currently being tested for their potential application in defining pathological markers and for the targeted delivery of therapeutics.
AFM FOR TARGET IDENTIFICATION: IMAGING ION CHANNELS AND DEFINING CHANNELOPATHIES
Conventional Channelopathies
Traditionally, channelopathies are considered to have an altered structure and/or function of ion channels. A long and rapidly increasing list of diseases fall into this category, such as the long QT syndrome and its associated potentially lethal arrhythmias that involve mutations in sodium and potassium channels, myotonia congenita involving voltage-dependent chloride channels, some forms of retinitis pigmentosa that involve non-specific ligand-gated channels, cystic fibrosis with defective chloride channel, and a long list of other diseases (23). m-AFM would be an ideal and indispensable platform to study and understand the pathogenesis of these disorders (9,10). An example of the potential application of AFM to conventional channelopathies mediated by gap junctions and hemichannels is provided below.
Gap Junctions and Hemichannels
Many years ago, we described the use of what we termed “force dissection,” which is the application of force with the cantilever to expose heretofore inaccessible surfaces. Using this approach, we dissected gap junctions, exposing both the extracellular and cytoplasmic faces of the gap junctions, which could then be imaged (1). In the earlier years of collaboration, we also assessed the electrophysiologic characteristics of gap junctions (24). One of us in the early 1990s called AFM the “dream machine.” We dreamed of measuring conductances while imaging came into being, and now, m-AFM can potentially make the dream come true.
Hemiplaques and hemichannels are non-junctional connexons that are composed of connexin hexamer. In 1995, we observed what we believed were hemiplaques or hemichannels in heart gap junction preparations, but could only speculate on their function (25). There is increasing evidence that hemichannels are involved in cell volume regulation and in programmed cell death (26).
Hemichannels have more recently been imaged in lipid bilayers after reconstitution in liposomes (11). A distinct difference was observed between two populations of channel-like structures that correspond to the extracellular and cytoplasmic side of the hemichannels Furthermore, by changing calcium levels in the imaging buffer, hemichannels could be imaged both in their open (low calcium) and closed (normal calcium) state with sub-nanometer resolution (Fig. 2, bottom panel). Using phase imaging, where the phase lag between the tip oscillation in tapping mode and the driving signal is monitored, (11) and surface energy measurements using AFM, a spike in the phase signal of open channels was observed, most likely due to the exposed hydrophobic domains of the hemichannel in open state.
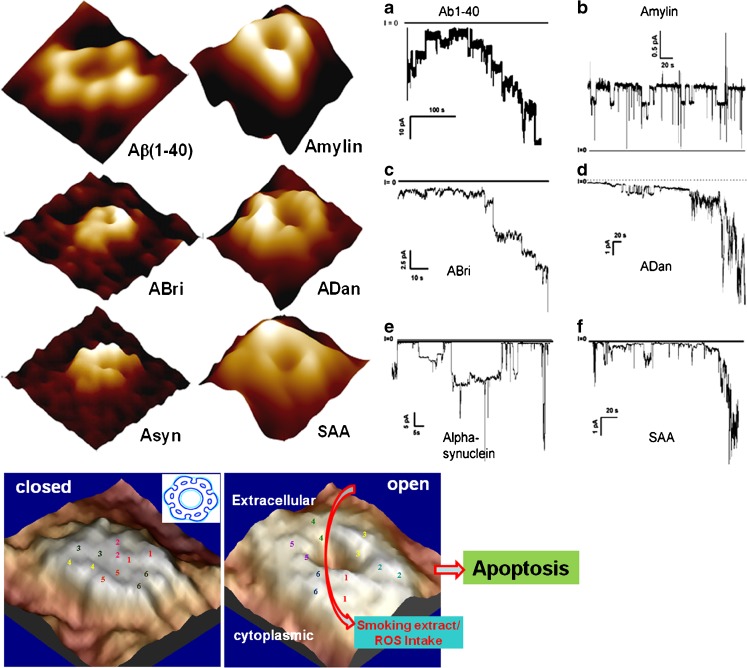
Fig. 2.
Top left panels Imaging single amyloid channels and hemichannels. Amyloid β(1-40), Amylin, ABri, ADan, σ-synuclein, and serum amyloid A all show structures resembling ion channels after reconstitution in lipid bilayer and top right panels (a–f) show single ion channel activity as indicated by parallel electrical recording. For details, see Quist et al. (10). Bottom panels Imaging open–closed conformation of hemichannels. Bottom left panel shows 3D molecular topography of a single Cx43 hemichannel with six subunits and a closed central pore. Bottom right panel shows the open conformation of the same hemichannel when the calcium concentration was reduced in the imaging media. A clear open pore is visible. For details, see Thimm et al. (11). These open hemichannels allow the direct uptake of oxidative stress-inducing molecules and cell apoptosis (Ramachandran et al. (27)
Significantly, these open hemichannels are known to regulate isosmotic cell volume and cell mechanics for normal tissue growth (16) and are known to facilitate external oxidative stress signal to induce apoptosis, as would occur in response to environmental stress and lifestyle (e.g., smoking) choices (27,28). Thus, m-AFM has been used to define molecular targets in health and disease (Fig. 2, bottom panel). Similar approach could be used to reveal novel targets in our pursuit of understanding human health and disease.
Channelopathies Mediated by Ion Channels Formed from Misfolded Proteins
We now turn to what we believe is a true paradigm shift. The ability of the AFM to image with high resolution has led to a paradigm shift in amyloidogenic diseases and holds promise to do the same for a number of other diseases. Misfolded proteins can form channels in the cellular membranes and alter ionic homeostasis, which can cause stress and injury to electrically sensitive cells in the body, including for example cardiomyocytes, neurons, and muscle cells. This mechanism has opened an exciting new approach to the understanding of these diseases.
A series of AFM studies of many amyloids, including amyloid beta protein, amylin, alpha-synuclein, ADan, ABri, and serum amyloid A, revealed ion channel-like structures that show single ion channel activity when reconstituted in membranes (Fig. 2, top left panels) (10,29). The resolution of AFM images has been sufficient to resolve the channel structure and distinguish different subunit arrangements. Electrical recording from these preparations show that these amyloid channels are active and conduct ionic currents (Fig. 2, top right panels). Cell calcium loading and cell degeneration studies showed that these amyloids induce a pathological response via their ion channel formation, a finding that provides a paradigm shifting perspective of amyloid-dependent pathologies. The pathogenetic mechanism of amyloidosis, at least in part, involves the creation of new channels that allow the toxic flow of ions. These diseases that form channels are not really covered by the classic concept of channelopathies. Yet, with the apparent similarity, we consider them to be “channelopathies of misfolded proteins.”
In defining a specific structural substrate (ion channels vs. undefined features such as plaques and non-fibrillar intermediates), AFM provides a structural basis for understanding the mechanistic details of neurodegenerative (e.g., Alzheimer’s disease, Parkinson’s disease) and systemic (e.g., diabetes, cancer, heart disease) diseases and for designing drugs and therapeutic evaluations—m-AFM is a versatile tool to study the high-resolution structures of these toxic ion channels and could provide a technology platform for screening potential drug candidates for treating these diseases by monitoring their action under real time with structural evidence.
m-AFM FOR DRUG DISCOVERY AND DELIVERY SYSTEMS
AFM Cantilever-Based Parallel Array of Sensors for Drug Screening
Emerging technologies are generating AFM cantilever-based array sensors being utilized for many purposes ranging from high-throughput screening of genetic materials to the identification of individual protein/peptides that rely on specific interactions using antibodies and peptides labeled with fluorescence markers. For assaying drugs for “channelopathies” and for identifying biomarkers of diseases, two powerful avenues involve patch clamp(s) on chips and microfluidics. Significantly, AFM cantilever-based array sensors can be combined with both techniques easily and thus provide powerful screening/diagnostic platform.
Patch Clamp on a Chip
Patch clamp recording is one of the main techniques employed in electrophysiological studies and commonly used in the study of channelopathies (30,31,32). The technique, however, is slow with low throughput, making it undesirable for use in drug discovery; an automated patch clamp that uses disposable devices would be a most useful outcome. An example of such an on-chip microchannel planar patch clamp is shown in ESM Fig. S2. The middle panels show a HeLa cell in the micro-fluidic system and being trapped at one of the channels. Cell deformation can be observed, and current traces show a seal resistance of 144 MΩ (30). Such devices using a cell reservoir linked to patch channels allows for the simultaneous optical and electrical recording which will facilitate studying the role of ion channels with respect to cellular functions. Future devices can be designed with an improved patch pore geometry and surface treatment to obtain a better seal necessary for single-channel conductance studies. Since the cell reservoir is linked to many patch clamp pores, a parallel readout is possible, analyzing multiple cells simultaneously. Also, there are many new variations of this technique commonly being developed, the most important being an array of patch clamping on nanochips with integrated microfluidics.
Microfluidic Viscosity, Velocity, and Molecular Affinity Sensors
Parallel readout techniques based on AFM cantilever arrays allow for the analysis of several analytes with exquisite intermolecular interaction sensitivity. When combined with micro- (and nano-) fluidic chambers, these array nanosensors become high-throughput screening devices for biomarkers and therapeutic agents. They can probe for instance multiple live cells for their elastic properties or the presence of targets in the cell membrane. Similarly, coating a parallel array of cantilever with a different material or reagent on each lever results in a “chemical nose” that can sense a variety of chemicals or toxins in very small volumes.
Figure 3 (Top left and right panels) shows the schematics of cantilevered microfluidic sensing tools in which a piezoelectric cantilever senses the viscosity and velocity of fluids with different compositions at extremely small volumes. Such a device can be easily adapted for screening biomarkers in blood and body fluids (Fig. 3, bottom panel). When combined with local fluorescence sensors (e.g., SPR, photo-sensitive nanoparticles), electrical sensors (e.g., piezoelectric circuitry), and mechanical sensors (e.g., cantilever deflection), an array of cantilevers with specific complements can provide a powerful and highly sensitive tool for high-throughput screening of pathologies and therapeutics. This scheme would allow the identification of an array of biomarkers and potential drug candidate molecules from a very small (micro- to milliliter) amount of biofluid.
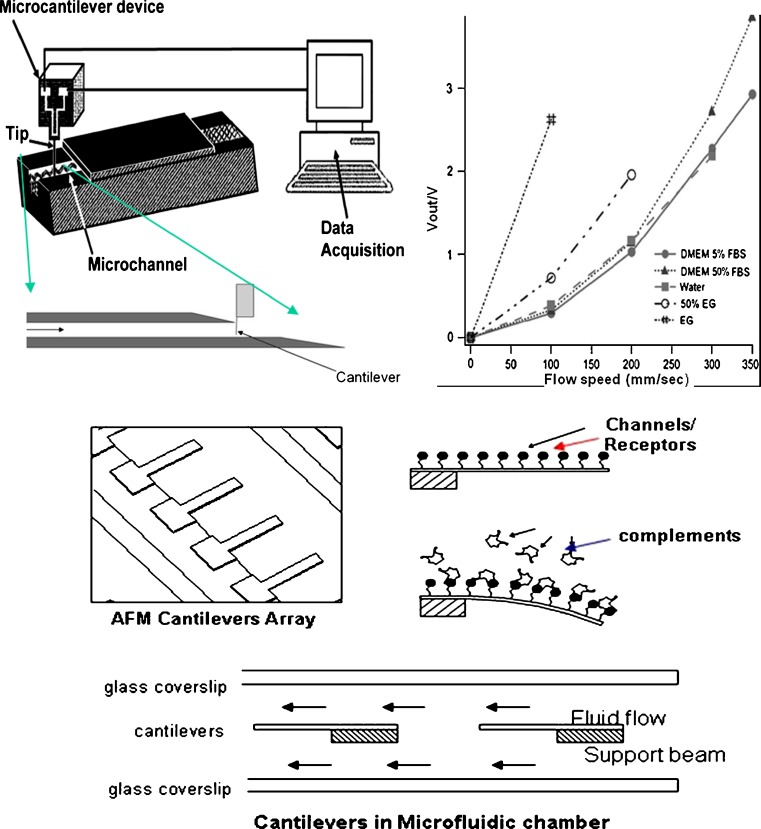
Fig. 3.
Nanosensors for biomarkers and therapeutic agents. Top left panel Schematics of cantilevered microfluidic sensor. Setup of the stainless steel needle. The cantilever is inserted and aligned using a micromanipulator and held with the base nearly touching the upper edge of the channel opening. Middle left panel FIB-milled cantilevers serve as flow and viscosity sensors for biological fluids. Top right panel Comparison of voltage readout at different flow speeds. High-viscosity fluids such as ethylene glycol saturate the amplifier at relatively low flow speeds. At higher flow speeds, the sensor distinguished between DMEM media with 5% and 50% fetal bovine serum. The protein content of blood serum is a major contributor to the viscosity of biological fluids. For details, see Quist et al. (62). Bottom panel Cantilever-based array detectors for complements and biomarkers assay in a microfluidic chamber
Cell Mechanics
Regulated biochemical processes as well as cytoskeletal structural integrity play important roles in cell growth, migration, and development (33). The local mechanical properties of a cell are closely associated with biochemical gradients across the membrane, but most techniques that have been used to study single cells averaged over the entire cell. AFM allows the measurement of mechanical properties of cells and portions of cells with high spatial resolution. For instance, rat atrial myocytes were imaged showing clearly the cytoskeletal network beneath the cell membrane and myofibrillar structure (34). Using a constant cantilever deflection maintained by feedback, contractile activity, and the change in contractile activity using perturbations in the buffer environment (Fig. 4), cell stiffness was examined. Different cantilever deflections were observed under different buffer conditions for the same imaging force. This demonstrates the possibility to quantify the coupling between extracellular substrates to cellular functions such as contraction, migration, growth, and differentiation.
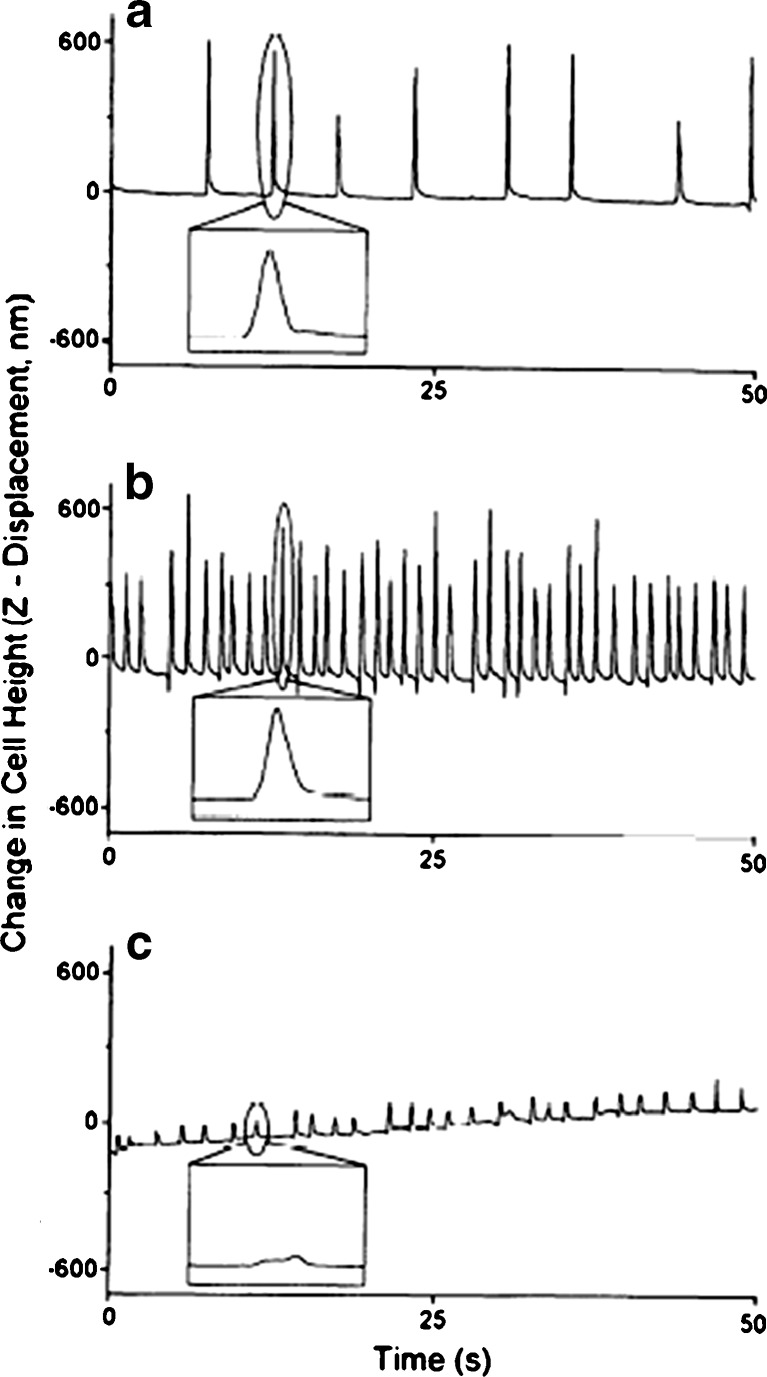
Fig. 4.
Local contractile activity of a beating atrial cell recorded with AFM tip under 1.8 mM (a) and 5 mM (b) calcium and 4 mM butanedione monoxime with high calcium (c). For details, see Shroff et al. (34)
Such monitoring of cell mechanics can be used effectively to monitor the efficacy of therapeutics that is expected to correct abnormal cell mechanics. For example, the efficacy of novel inotropes, chronotropes on cardiac myocytes, endothelial barrier function (34,35), etc., could be assessed at single cell with high spatial resolution, thus serving a good drug screening tool as well as assessing the efficacy of a candidate drug.
Tissue Nanoelasticity and Nanopatterning
Nanomechanical properties of tissues depend on underlying ultrastructure of their constituents. A detailed understanding of tissue elasticity can be used for the early diagnosis and for monitoring the disease progression and their treatment outcomes, especially diseases of bones and joints. Indeed, there are some new diagnostic tools being tested to diagnose bone diseases (36,37) that resulted from our understanding of the tissue mechanics at the molecular level (38). The correlation between the mechanical properties and the heterogeneous subunits on a nanoscale has been limited. Using AFM, such correlation studies are possible (39), as shown in ESM Fig. S3.
AFM has been able to show the features of a cross-sectioned wool fiber with similar amount of detail as TEM (ESM Fig. S3, left panels). To study the mechanical properties of the different constituents of the fibers, AFM force mapping has been used. The force curves give information about the stiffness and elastic modulus of the sample regions. For instance, it was shown that the exo-cuticle part of wool fibers has the highest elastic modulus (19.8 GPa), while the endo-cuticle and cortical regions of the fiber cross-section have significantly lower modulus (3.8 and 4.0 GPa, respectively). The indentations made by a diamond tipped cantilever, all made with the same indentation force, look indeed distinctly different for the different regions (ESM Fig. S3, right panel). Furthermore, the AFM can give conclusive evidence of the role of disulfide bonds in the fiber stiffness. Reduction of such bonds, abundant in the exo-cuticle, using dithiothreitol, resulted in a reduction of modulus to 0.3 GPa (39). This has shed new light on the role of the exo-cuticle (small in mass proportion) to the rigidity of wool fibers.
Of what use are these studies on wool to nanopharmacology? Using the unique feature of defined nanoindentation, the compactness of drug molecules in a pellet, which is critical for their release kinetics in body fluids, could be used as a quality control measure in drug preparations. Significantly, such nanopatterned features would be helpful for creating security watermarks to fight the counterfeit drugs.
AFM for Drug Delivery
Targeted delivery of therapeutics has long been an ideal, and a number of different approaches have been tried in the past. Ideally, therapy could be efficient if selective delivery of drugs to the diseased tissue is achieved. Current bulk delivery of drugs by oral or parenteral routes is inefficient since this requires a larger dosage and often has deleterious side effects. Advanced molecular dynamic simulations and nanostructured materials are providing new avenues for drug discovery and delivery systems. Several nanoscale carrier systems, including nanotubes, nanoshells, nanoliposomes, and dendrimers, are being evaluated for effective and targeted delivery. These carriers are designed to contain target guidance molecules that are specific to cell/tissue surface.
AFM allows the design, characterization, and quality control of these carriers. Integrated multimodal AFM also allows imaging real-time transport of these carriers with specific drugs and their effect on cells (17). One such successful example of AFM application for characterizing nanocarriers is shown in (Fig. 1). Small cisplatin-encapsulated liposomes (<200 nm) are effective in killing cells due to their efficient internalization by cell endocytosis. An AFM-fluorescence microscopy study was able to demonstrate that the most effective liposomes that were able to deliver optimal dosage for cell toxicity are those with a diameter of 100–150 nm. Much smaller liposomes were unable to contain sufficient amount of cisplatin to induce toxicity, and larger liposomes were unstable and not internalized by the cells efficiently.
Recently, cell-penetrating peptides, including TAT peptide, have been used to deliver various nano-particular pharmaceutical carriers including liposomes, micelles, and nanoparticles (40,41). Nanocarrier systems incorporated with stimuli-sensitive properties are being developed. These stimuli may include pH, temperature, and redox potential. Even carbon nanotubes filled with a chemotherapeutic agent are being developed for the treatment of tumors (42). These and other nanomedicinals will likely use m-AFM in design, imaging, assessing pharmacologic design, targeting, measuring interactions, and the like.
ASSESSING TARGET–DRUG INTERACTIONS: AFM NANOSENSING
Multimodal AFM allows us to define targets, such as channels, receptors, etc., and their functional activity. The ability to modify the cantilevered tip biochemically adds another dimension to the versatility of the m-AFM techniques. By suitable modification of the tip, one could get a wealth of information about intermolecular interactions at the single molecular level (nanosensing). Attaching a drug molecule directly or indirectly through spacers to the tip will allow studying selective and specific interaction between the target under investigation and the drug molecule. This will be an indispensable tool in drug design and development. Here, we highlight some of the examples where this approach has been adopted to understand intermolecular interactions in our lab.
Structure, Density, Distribution, and Functional Viability of Membrane Receptors and Channels
Rational drug design should consider the potential biological targets of drugs. As mentioned throughout this review, channels, receptors, and other nanoscale structures are smaller than the current resolution of light microscopic imaging. Electron microscopy and other types of high-resolution imaging cannot be used for live imaging, and hence, their utility is limited. m-AFM, however, withdraws the resolution veil and allows us to see new structures and new dynamic processes in the Lilliputian nanopharmacological world.
In one of the earliest example of AFM mapping of cell membrane receptors, the AFM was used to identify individual nicotinic acetylcholine receptor (nAchR) that was expressed in Xenopus oocytes (12). The structure was determined without fixation or other preparations that might denature the sample, as is often required for EM and other comparable imaging techniques. The density, distribution, clustering, and functional viability of these AChR receptors were confirmed by other relevant techniques, including channel conductance measurements and ligand binding assays (ESM Fig. S4). Using ion conductance microscopy (43) and intermolecular force mapping (44–47) (see below) capability, m-AFM can provide density, distribution, clustering, and functional viability of most of the cell membrane channels/pores.
The possibility of linking molecules to the probing tip provides a range of options to probe molecular interactions between the probe on the AFM cantilever and the sample surface (48,49,50). In this way, membrane proteins can be probed on living cells. Using such modified and functionalized cantilevers, adhesion forces have been measured and mapped between ligands and receptors on the surface of living cells (ESM Fig. S5). Using force volume imaging (13), regional distribution as well as ligand- or antibody-induced clustering of VEGF receptors have been reported (ESM Fig. S5) (13). Unbinding forces of 60–70 pN were observed, which could be reduced by competitive inhibition using antibodies.
Force mapping can also be useful for volume measurement on cells too spherical or weakly attached to the substrate surface. Using m-AFM with fluorescence microscopy, Quist et al. (16) have shown that volume regulation of several cell lines occurs through hemichannels. The added benefit of using force mapping is that not only are data obtained with respect to unbinding forces, but, simultaneously with imaging, one can also map the stiffness of the cell membrane. The stiffness is obtained by looking at the approach force distance curves in each pixel, which results in an elasticity map. Almqvist et al. (13) showed clustering of receptors using this technique, and (16) showed the cytoskeletal stiffness change induced by volume change in the cell.
Intermolecular interaction force maps can be used to define specific domains of an effector (e.g., channels and receptors). In one such example, Liu et al. (50) used AFM tip conjugated with antibodies specific to different regions of a hemichannel peptide, connexin43 (Cx43), to map the specific Cx43 epitopes that open and close the hemichannel in response to changing calcium concentration in the medium (ESM Fig. S6). The amount of force required to unwind the peptide necessary for opening the channel correlated with the mobility and folding of specific portions of Cx43.
Similar approaches could be utilized to investigate a candidate drug molecule against its target in terms of its efficacy, which would be a boon in drug screening, drug design, as well as in monitoring their pharmacologic action.
MEASURING THERAPEUTIC EFFICACY-TRACKING OUTCOMES
Most drugs have a tight therapeutic range; toxicities arise due to variations in metabolism, clearance rate, preexisting conditions, interactions with other drugs (cross-reactivity) and idiosyncrasies, etc. Currently, the therapeutic efficacy of medications is determined by bulk assays that have no molecular resolution. Molecular understanding of the drug–target interactions could provide important insights into some of these issues. As discussed, single molecules can be imaged routinely with AFM, and it is quite feasible to use AFM or m-AFM to monitor real-time molecular interactions. Here, we highlight some of the key studies which utilized this approach, which could be of great potential in tracking the outcomes following therapeutic intervention.
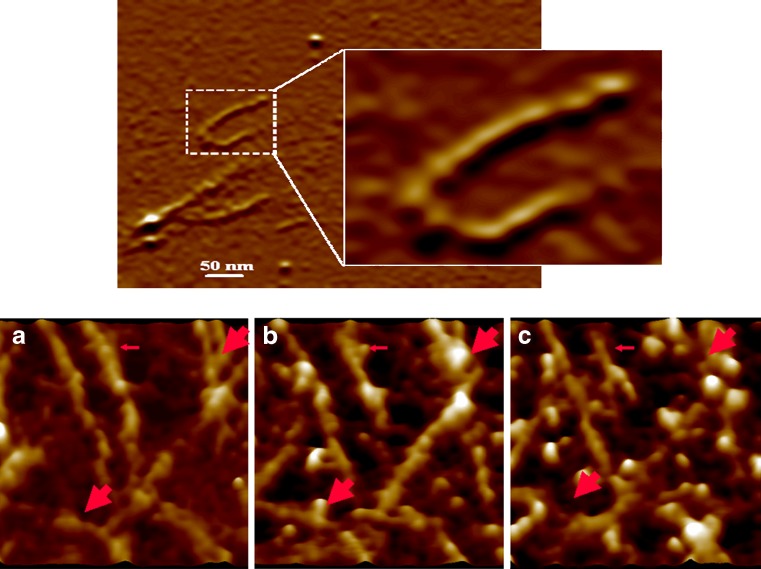
An interesting example is the proteolysis of collagen that has been observed using AFM (Fig. 5) (51). Single collagenase binding to collagen could be imaged and its digestion was followed in real time. Significantly, as shown in panels b and c, very often, collagenase molecules would bind to collagen and dissociate without cutting the collagen molecule. At present, such processes are typically monitored using SDS-PAGE, which gives little information about the real-time activity and the 3D structural information of the binding of the enzyme to its substrate. The dissociation constant derived from single-molecule proteolysis is similar to what is observed from the bulk assay, but clearly not all molecules are cleaved. This is an important issue, for example in a drug cocktail, as used in the treatment of HIV/AIDs where the efficacy of individual molecules cannot be determined from bulk assay.
Fig. 5.
Imaging of triple helical collagen I with AFM. The inset magnifies one collagen molecule. Bottom panels Collagenase binding to collagen molecules. Images are taken before collagenase addition (a), immediately after collagenase addition (b), and 4 min later (c). Globular particles bound to collagen are indicated by arrows; thick arrows show sites where collagen was broken after collagenase binding. For details, see Lin et al. (51)
With the possibility of monitoring single molecules comes the advantage of being able to manipulate the single molecules as well. As described above (ESM Fig. S6), by tethering antibodies to the AFM tip, and after bringing the tip in contact with the sample of interest, the antigen in the sample cannot only be detected but also unfolded, as reported above. Using this approach, mechanistic details of calcium-mediated opening/closing of Cx43 hemichannels were identified using antibodies specific for different regions of the Cx43 cytoplasmic tail. The precise estimate of the energy needed to unfold (fold) and amount of stretch (contraction) in the channel unfolding measured by AFM shows the promises of using AFM for better measure of the therapeutic efficacy of drugs and pharmacological agents with single-molecule resolution. For example, a similar approach could be used to assess the efficacies of channel blockers by measuring their unbinding force.
A number of years ago, we used AFM to detect microcrystals in synovial fluid from patients with crystal-induced arthritis (52). The study was undertaken to develop a system of “fingerprinting” microcrystals with AFM, and, to a large extent, we succeeded. Some specimens contained more than one type of crystal, while other specimens contained microcrystals that were undetected by polarized light microscopy and by transmission electron microscopy. Crystallization is also important in nephrolithiasis and renal disease. AFM can also be used to study the development of crystals which, in turn, could be a system for assessing the effect of pharmacological intervention (53–56).
Another example would be the use of AFM in studying diseases of the bone. Paul Hansma, one of the pioneers in AFM, and his colleagues, recently wrote a very interesting paper on imaging of bone ultrastructure using atomic force microscopy (57). They point out that bone is a “remarkable natural nano-composite material.” They reviewed much of the literature and indicate how little has been studied at the nanoscale level while emphasizing the potential of the approach of imaging of bone in a functional manner using AFM.
Damage to and degeneration of cellular and subcellular elements is a common pathophysiological feature in a variety of diseases. For example, the loss of synapses and dendrites with eventual neurite degeneration characterize several neurodegenerative diseases. AFM can be used to image and track changes in cellular and subcellular morphology. This, especially if combined with a fluorescence or confocal microscope, gives the opportunity to track changes in the cellular morphology and function induced by various perturbations. For instance, amyloid beta protein-induced changes in endothelial cells were tracked (9) (ESM Fig. S7). Since AFM can operate under biological liquid environments, reagents can be added and the physiologic solutions changed during imaging. In this way, the therapeutic capability of a potential therapeutic candidate can be evaluated directly with structural evidence at nanometer resolution. For example, zinc ions and extracellular calcium concentrations have been changed in real time to prevent pathological amyloid beta-induced changes in cellular morphology (ESM Fig. S7, right panels).
A multimodal AFM combined with fluorescence microscopy can give valuable additional information and conformation of processes observed with the AFM. For instance, Calcein dye can be used to track neuronal death and neurite arborization optically during AFM imaging (ESM Fig. S7, left panel). Such optical techniques have better temporal resolution since the AFM imaging of a whole cell is time-consuming. Further developments in fast-scanning AFM systems will be able to study such events with sub-second time resolution, a large improvement over the conventional AFM scans that take several minutes to collect.
AFM to Assess Micro-rheological Properties
Fluid dynamics at tissue microcirculation, including fluid velocity, viscosity, shear stress, and shear rate, are important in maintaining normal tissue function and in the pathogenesis of vascular lesions like atherosclerosis, thrombosis, etc. Vascular lesions affect the interaction of therapeutics with their targets due to local alterations in the fluid flow, such as reduced flow or turbulence around a stenotic artery, occluded vein, or tumor growth. Yet we know very little about them. It is difficult to measure fluid viscosity with high precision in small volumes for non-Newtonian fluids like blood. Traditionally, ultrasonic devices are used to measure viscosity (58). They operate at megahertz frequency at which the viscosity of non-Newtonian fluids can be different as compared with low-frequency values, which are of greater biomedical interest (59). Flexural-mode resonance devices, such as microfabricated AFM cantilevers, may be more reliable since they allow measurement at lower frequencies. Using an optical detection in standard AFM equipment, viscous drag was measured using a piezoelectric actuator to vibrate an AFM silicon cantilever (60). Another approach to the AFM measurement of liquid viscosity has been the assessment of torsion in an AFM cantilever while scanning a whisker tip inside the liquid (61). These techniques are indispensable for assessing the effect of blood-thinning drugs like aspirin, coumarin, heparin, dipyridamole, clopidogrel, etc. and in the development of novel therapeutic agents with potential effect on blood viscosity.
OUTLOOK
The salient features of the AFM have allowed us to see beyond the limits of resolution thought possible only a few years ago. We now have molecular and submolecular resolution in physiologic environments; the ability to measure incredibly small interactions, perhaps as small as the hydrogen bond; and a device that can be modified to allow the identification of specific structures and specific interactions, such as ligand–receptor interactions; precise nanometer control of the position and piconewton control of the cantilever that allows the physical manipulation of molecules, the dissection of biological structure, the precise delivery of ligands, drugs and materials, and even nanosynthesis.
The simple design and invariance to the operating environment allows AFM to be integrated with other techniques for simultaneous structure–function correlation studies, such as the integration of the AFM with a fluorescence microscope including confocal, TIRF, FRET modalities, near-field differential scanning optical microscope, electro-optical manipulators, electrophysiologic recordings, etc. These multidimensional and multimode AFM hold great promise for the investigation of nanoscale structure and function in complex biological systems. The AFM goes beyond correlative studies. The functionalized AFM tip, for example, allows the direct observation of ligand–receptor, protein–protein, and other such interactions in physiological solutions. Moreover, the flexibility and high-resolution capability of these integrated tools point toward a unique window through which new and exciting information could be harnessed about and from multiscale biological systems.
One exciting view through this new window has been the discovery of a new type of toxic channel formed in protein-misfolding diseases such as Alzheimer’s and other neurodegenerative diseases, diabetes, tuberculosis, etc. The next generation of AFMs will be faster and have softer imaging features that would allow us not only to understand real-time conformational changes in nanoscale biological systems but would also allow designing and fabricating such systems, the biomimetics.
In summary, AFM per se or m-AFM and AFM cantilever-based techniques offer a unique opportunity to probe complex biological systems at single molecular level in their native physiological conditions, which makes it a versatile and indispensable tool in the advancement of nanopharmacology. These current and future developments in AFM and its growing application to biomedical and health research will be at the forefront of nanomedicine, including nanodiagnostics, nanotherapeutics, and the development of nanomedical devices.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
AFM integrated with nanochip-separated, double-chamber electrical recording system. Top rows: a AFM image of arrays of nanopores. The pores are produced in a silicon nitride chip using electron beam lithography and range in size from 50 nm (top rows) to 100 nm (rows on the right). b After deposition of lipid bilayers, AFM image in PBS shows a bilayer covering the pores. The bilayer is not fully contiguous and has several holes (red arrows); the cross-section on a hole in the bilayer (inset) indicates the thickness of a lipid bilayer ~5.3 nm. Blue arrows indicate 500-nm-wide corner alignment marks; red arrows indicate defects in the bilayer. c Schematic of the liquid cell AFM setup for imaging the silicon nitride chip and bilayer. Bottom rows: Left AFM images (left) and conductance maps (right) over a pore in a microfabricated silicon chip for different applied bias potentials (−2.0, −1.0, −0.5 V, respectively, for the top, middle, and bottom panels). Far right: I–V curves (applied potential –−0.5–0.5 V) obtained for 10 s (top), 2 min (middle), and 4 min (bottom) upon addition of gramicidin (0.2 mg/ml) to the bilayer, the overall current across pores increases and conductance increases from 25 pS (bilayer only) to 1 nS (at 4 min), suggesting the formation of hundreds of gramicidin ion channels. For details, see Quist et al., 2007 (63) (TIFF 4,121 kb)
Schematic drawing of conventional patch clamp and an on-chip patch clamp (left). Cells can be pulled into one of the on-chip patch pores (middle top panel), resulting in membrane deformation and patch formation (middle bottom panel). Resulting current measured before (top right top panel) and after cell attachment (right bottom panel). For details, see Seo et al. (29) (TIFF 3,897 kb)
Left panels: Schematic diagram of wool fiber (a). TEM image (b) (higher magnification in d) and AFM image (c) (higher magnification in e) of the same wool fiber region. AFM and TEM images correlate well with respect to identified subcellular structures. Center panels Force curves on cortex (top), exo-cuticle (middle), and embedding resin (bottom). Solid line is collected on hard glass surface. Larger shaded area indicates more elastic surface. Right panels: Top right image shows the corresponding AFM image. Bottom right image shows surface after indentation with diamond tip to create specific patterns on wool fiber. For details, see Parbhu et al. (37) (TIFF 5,356 kb)
AFM imaging or nAChR expressed in Xenopus oocytes. AFM imaging of expressed AChR. After recording ACh-sensitive electrical current in an oocyte, its vitelline membrane was removed. The cell was cut into two halves and one half was imaged. Top left AFM image of the plasma membrane. A crack in the membrane is shown. Middle left High-resolution low-pass-filtered image showing the irregular spacing of receptor cluster. Bottom left High-resolution image of a control oocyte not injected with any mRNA. Top right Line graph of an image of a single AChR channel (~10.5-nm diameter) showing the contour map of subunits and pore. The five subunits (~1- to 1.5-nm diameter) with a central pore are shown. Middle right ACh-sensitive current measured in an oocyte after the AChR expression. The dose-dependent ACh-sensitive current was reversible. Bottom right α-Bungarotoxin binding assay revealed a high level of AChR expression. The channel density calculated from AFM, electrophysiological, and pharmacological studies were comparable; for details, see Lal and Yu (12) (TIFF 6,167 kb)
Top half (left) Adhesion forces between VEGF receptor and antibody (a), with distribution of unbinding forces (b). c Blocking peptide prevents the interaction. d Tapping mode image of VEGF receptors on mica. e, f Adhesion forces between VEGF receptor on endothelial cell and antibody on tip without (e) and with (f) the presence of blocking peptide. Bottom half (right) AFM images of endothelial cells before (a) and after (b) the addition of VEGF showing cytoskeletal reorganization. Fluorescence images showing the presence of Flk-1 receptors (c, d) and control (e) with nonspecific antibody. Bottom half Simultaneous acquisition of topography and elasticity images before and 10 and 45 min after adding VEGF antibody. Clustering of receptors can be observed both in topography and elasticity maps (spots labeled 1–4). Top histograms show the distribution of adhesion forces. For details, see Almqvist et al. (13) (TIFF 5,640 kb)
AFM antibody-conjugated tip pulling experiment: Sensing intermolecular interaction for mapping conformational epitopes. Left half Model of a gap junction plaque with apposing cell membranes with hexagonally packed hemichannels (left). Schematic of force spectroscopy measurement showing the binding of antibody connected to the AFM tip with flexible PEG spacers to Cx43 hemichannels reconstituted in lipid bilayer (middle) and the schematic of Cx43 membrane topology with two extracellular loops, one cytoplasmic loop, and the cytoplasmic carboxyl-terminal domain and locations of antigenic binding sites for anti-CT252–270 and anti-CT360–382 (right) are shown. Right half Probability histograms of the rupture forces of the measured anti-CT252–270-Cx43 (a), anti-CT360–382-Cx43 (b), and GAP26-Cx43 interactions (c). d Representative force–extension curve showing specific avidin–biotin interaction in the PEG spacer extension test system. e Average extension of PEG spacer stretching (~28 nm). f Histograms of measured tether extensions in anti-CT252–270-Cx43, anti-CT360–382-Cx43, and GAP26-Cx43, respectively. For details, see Liu et al. (50) (TIFF 4,683 kb)
Amyloid beta channel-induced cell death. Left half Loss of synaptic arborization and neuronal death imaged with calcein dye fluorescence before and after addition of Aβ peptide to the buffer solution (compare a, c, e, g (control) with panels b, d, f, g (after the addition of nano-to-micromolar abeta peptide). Right half Simultaneously, AFM can be used to image the cytoskeletal structure of cells to track the effects of Aβ peptide on the cells and to test various blocking agents for their performance in blocking toxicity effects. Yellow arrows compare the changes in cytoskeleton upon the addition of amyloid beta peptides or other reagents before and after their addition. For details, see Lin et al. (9) (TIFF 5,570 kb)
Acknowledgment
This work was supported by the research grants (R01DA025296 and RO1DA024871) from National Institute on Drug Abuse (NIDA).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
This work is dedicated to the memory of Prof. Morton F. Arnsdorf, who passed away on 06/09/10.
References
- 1.Hoh JH, Lal R, John SA, Revel JP, Arnsdorf MF. Atomic force microscopy and dissection of gap-junctions. Science. 1991;253(5026):1405–1408. doi: 10.1126/science.1910206. [DOI] [PubMed] [Google Scholar]
- 2.Binnig G, Quate CF, Gerber C. Atomic force microscope. Phys Rev Lett. 1986;56(9):930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 3.Goodman FO, Garcia N. Roles of the attractive and repulsive forces in atomic-force microscopy. Phys Rev B. 1991;43(6):4728–4731. doi: 10.1103/PhysRevB.43.4728. [DOI] [PubMed] [Google Scholar]
- 4.Mechler A, Kokavecz J, Heszler P, Lal R. Surface energy maps of nanostructures: atomic force microscopy and numerical simulation study. Appl Phys Lett. 2003;82(21):3740–3742. doi: 10.1063/1.1577392. [DOI] [Google Scholar]
- 5.Meyer G, Amer NM. Simultaneous measurement of lateral and normal forces with an optical-beam-deflection atomic force microscope. Appl Phys Lett. 1990;57(20):2089–2091. doi: 10.1063/1.103950. [DOI] [Google Scholar]
- 6.Chen IC, Chen LH, Ye XR, Daraio C, Jin S, Orme CA, et al. Extremely sharp carbon nanocone probes for atomic force microscopy imaging. Appl Phys Lett. 2006;88(15):153102. doi: 10.1063/1.2193435. [DOI] [Google Scholar]
- 7.Giessibl FJ, Hembacher S, Bielefeldt H, Mannhart J. Subatomic features on the silicon (111)–(7 × 7) surface observed by atomic force microscopy. Science. 2000;289(5478):422–425. doi: 10.1126/science.289.5478.422. [DOI] [PubMed] [Google Scholar]
- 8.Ohnesorge F, Binnig G. True atomic-resolution by atomic force microscopy through repulsive and attractive forces. Science. 1993;260(5113):1451–1456. doi: 10.1126/science.260.5113.1451. [DOI] [PubMed] [Google Scholar]
- 9.Lin H, Bhatia R, Lal R. Amyloid beta protein forms ion channels: implications for Alzheimer’s disease pathophysiology. FASEB J. 2001;15(13):2433–2444. doi: 10.1096/fj.01-0377com. [DOI] [PubMed] [Google Scholar]
- 10.Quist A, Doudevski L, Lin H, Azimova R, Ng D, Frangione B, et al. Amyloid ion channels: a common structural link for protein-misfolding disease. Proc Natl Acad Sci USA. 2005;102(30):10427–10432. doi: 10.1073/pnas.0502066102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thimm J, Mechler A, Lin H, Rhee S, Lal R. Calcium-dependent open/closed conformations and interfacial energy maps of reconstituted hemichannels. J Biol Chem. 2005;280(11):10646–10654. doi: 10.1074/jbc.M412749200. [DOI] [PubMed] [Google Scholar]
- 12.Lal R, Yu L. Atomic-force microscopy of cloned nicotinic acetylcholine-receptor expressed in xenopus-oocytes. Proc Natl Acad Sci USA. 1993;90(15):7280–7284. doi: 10.1073/pnas.90.15.7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Almqvist N, Bhatia R, Primbs G, Desai N, Banerjee S, Lal R. Elasticity and adhesion force mapping reveals real-time clustering of growth factor receptors and associated changes in local cellular rheological properties. Biophys J. 2004;86(3):1753–1762. doi: 10.1016/S0006-3495(04)74243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kienberger F, Kada G, Mueller H, Hinterdorfer P. Single molecule studies of antibody-antigen interaction strength versus intra-molecular antigen stability. J Mol Biol. 2005;347(3):597–606. doi: 10.1016/j.jmb.2005.01.042. [DOI] [PubMed] [Google Scholar]
- 15.Hansma HG, Hoh JH. Biomolecular imaging with the atomic-force microscope. Annu Rev Biophys Biomol Struct. 1994;23:115–139. doi: 10.1146/annurev.bb.23.060194.000555. [DOI] [PubMed] [Google Scholar]
- 16.Quist AP, Rhee SK, Lin H, Lal R. Physiological role of gap-junctional hemichannels: extracellular calcium-dependent isosmotic volume regulation. J Cell Biol. 2000;148(5):1063–1074. doi: 10.1083/jcb.148.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramachandran S, Quist AP, Kumar S, Lal R. Cisplatin nanoliposomes for cancer therapy: AFM and fluorescence imaging of cisplatin encapsulation, stability, cellular uptake, and toxicity. Langmuir. 2006;22(19):8156–8162. doi: 10.1021/la0607499. [DOI] [PubMed] [Google Scholar]
- 18.Proksch R, Lal R, Hansma PK, Morse D, Stucky G. Imaging the internal and external pore structure of membranes in fluid: tappingmode scanning ion conductance microscopy. Biophys J. 1996;71(4):2155–2157. doi: 10.1016/S0006-3495(96)79416-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ionescu-Zanetti C, Mechler A, Carter SA, Lal R. Semiconductive polymer blends: correlating structure with transport properties at the nanoscale. Advanced Materials. 2004;16(5):385–389. doi: 10.1002/adma.200305747. [DOI] [Google Scholar]
- 20.Jager EWH, Smela E, Inganas O. Microfabricating conjugated polymer actuators. Science. 2000;290(5496):1540–1545. doi: 10.1126/science.290.5496.1540. [DOI] [PubMed] [Google Scholar]
- 21.Hagleitner C, Hierlemann A, Lange D, Kummer A, Kerness N, Brand O, et al. Smart single-chip gas sensor microsystem. Nature. 2001;414(6861):293–296. doi: 10.1038/35104535. [DOI] [PubMed] [Google Scholar]
- 22.Carter SA, Angelopoulos M, Karg S, Brock PJ, Scott JC. Polymeric anodes for improved polymer light-emitting diode performance. Appl Phys Lett. 1997;70(16):2067–2069. doi: 10.1063/1.118953. [DOI] [Google Scholar]
- 23.Kass RS. The channelopathies: novel insights into molecular and genet mechanisms of human disease. J Clin Investig. 2005;115(8):1986–1989. doi: 10.1172/JCI26011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lal R, Arnsdorf MF. Voltage-dependent gating and single-channel conductance of adult mammalian atrial gap-junctions. Circ Res. 1992;71(3):737–743. doi: 10.1161/01.res.71.3.737. [DOI] [PubMed] [Google Scholar]
- 25.Lal R, John SA, Laird DW, Arnsdorf MF. Heart gap junction preparations reveal hemiplaques by atomic-force microscopy. Am J Physiol Cell Physiol. 1995;37(4):C968–C77. doi: 10.1152/ajpcell.1995.268.4.C968. [DOI] [PubMed] [Google Scholar]
- 26.Goodenough DA, Paul DL. Beyond the gap: functions of unpaired connexon channels. Nat Rev Mol Cell Biol. 2003;4(4):285–294. doi: 10.1038/nrm1072. [DOI] [PubMed] [Google Scholar]
- 27.Ramachandran S, Xie LH, John SA, Subramaniam S, Lal R. A novel role for connexin hemichannel in oxidative stress and smoking-induced cell injury. Plos One. 2007;2(8), Aug 8. [DOI] [PMC free article] [PubMed]
- 28.Decrock E, Vinken M, De Vuyst E, Krysko DV, D’Herde K, Vanhaecke T, et al. Connexin-related signaling in cell death: to live or let die[quest]. Cell Death Differ. 2009;16(4):524–36. [DOI] [PubMed]
- 29.Jang H, Arce FT, Ramachandran S, Capone R, Azimova R, Kagan BL, et al. Truncated β-amyloid peptide channels provide an alternative mechanism for Alzheimer’s Disease and Down syndrome. Proc Natl Acad Sci. 2010;107(14):6538–43. [DOI] [PMC free article] [PubMed]
- 30.Seo J, Ionescu-Zanetti C, Diamond J, Lal R, Lee LP. Integrated multiple patch-clamp array chip via lateral cell trapping junctions. Appl Phys Lett. 2004;84(11):1973–1975. doi: 10.1063/1.1650035. [DOI] [Google Scholar]
- 31.Xu J, Wang XB, Ensign B, Li M, Wu L, Guia A, et al. Ion-channel assay technologies: quo vadis? Drug Discov Today. 2001;6(24):1278–1287. doi: 10.1016/S1359-6446(01)02095-5. [DOI] [PubMed] [Google Scholar]
- 32.Sakmann B, Neher E. Single channel recording. New York: Plenum; 1983. [Google Scholar]
- 33.Brady AJ. Mechanical-properties of isolated cardiac myocytes. Physiol Rev. 1991;71(2):413–428. doi: 10.1152/physrev.1991.71.2.413. [DOI] [PubMed] [Google Scholar]
- 34.Shroff SG, Saner DR, Lal R. Dynamic micromechanical properties of cultured rat atrial myocytes measured by atomic-force microscopy. Am J Physiol Cell Physiol. 1995;38(1):C286–C92. doi: 10.1152/ajpcell.1995.269.1.C286. [DOI] [PubMed] [Google Scholar]
- 35.Arce FT, Whitlock JL, Birukova AA, Birukov KG, Arnsdorf MF, Lal R, et al. Regulation of the micromechanical properties of pulmonary endothelium by S1P and thrombin: role of cortactin. Biophys J. 2008;95(2):886–894. doi: 10.1529/biophysj.107.127167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fantner GE, Birkedal H, Kindt JH, Hassenkam T, Weaver JC, Cutroni JA, et al. Influence of the degradation of the organic matrix on the microscopic fracture behavior of trabecular bone. Bone. 2004;35(5):1013–1022. doi: 10.1016/j.bone.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 37.Fantner GE, Hassenkam T, Kindt JH, Weaver JC, Birkedal H, Pechenik L, et al. Sacrificial bonds and hidden length dissipate energy as mineralized fibrils separate during bone fracture. Nat Mater. 2005;4(8):612–616. doi: 10.1038/nmat1428. [DOI] [PubMed] [Google Scholar]
- 38.Thompson JB, Kindt JH, Drake B, Hansma HG, Morse DE, Hansma PK. Bone indentation recovery time correlates with bond reforming time. Nature. 2001;414(6865):773–776. doi: 10.1038/414773a. [DOI] [PubMed] [Google Scholar]
- 39.Parbhu AN, Bryson WG, Lal R. Disulfide bonds in the outer layer of keratin fibers confer higher mechanical rigidity: correlative nano-indentation and elasticity measurement with an AFM. Biochemistry. 1999;38(36):11755–11761. doi: 10.1021/bi990746d. [DOI] [PubMed] [Google Scholar]
- 40.Torchilin VP. Tat peptide-mediated intracellular delivery of pharmaceutical nanocarriers. Adv Drug Deliv Rev. 2008;60(4–5):548–558. doi: 10.1016/j.addr.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 41.Ganta S, Devalapally H, Shahiwala A, Amiji M. A review of stimuli-responsive nanocarriers for drug and gene delivery. J Control Release. 2008;126(3):187–204. doi: 10.1016/j.jconrel.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 42.Hampel S, Kunze D, Haase D, Kramer K, Rauschenbach M, Ritscbel M, et al. Carbon nanotubes filled with a chemotherapeutic agent: a nanocarrier mediates inhibition of tumor cell growth. Nanomedicine. 2008;3(2):175–182. doi: 10.2217/17435889.3.2.175. [DOI] [PubMed] [Google Scholar]
- 43.Shevchuk AI, Hobson P, Lab MJ, Klenerman D, Krauzewicz N, Korchev YE. Endocytic pathways: combined scanning ion conductance and surface confocal microscopy study. Pflügers Arch Eur J Physiol. 2008;456(1):227–235. doi: 10.1007/s00424-007-0410-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baumgartner W, Hinterdorfer P, Ness W, Raab A, Vestweber D, Schindler H, et al. Cadherin interaction probed by atomic force microscopy. Proc Natl Acad Sci USA. 2000;97(8):4005–4010. doi: 10.1073/pnas.070052697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hinterdorfer P, Baumgartner W, Gruber HJ, Schilcher K, Schindler H. Detection and localization of individual antibody–antigen recognition events by atomic force microscopy. Proc Natl Acad Sci USA. 1996;93(8):3477–3481. doi: 10.1073/pnas.93.8.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan CB, Chen A, Kolb P, Moy VT. Energy landscape of streptavidin–biotin complexes measured by atomic force microscopy. Biochemistry. 2000;39(33):10219–10223. doi: 10.1021/bi992715o. [DOI] [PubMed] [Google Scholar]
- 47.Zhang XH, Wojcikiewicz E, Moy VT. Force spectroscopy of the leukocyte function-associated antigen-1/intercellular adhesion molecule-1 interaction. Biophys J. 2002;83(4):2270–2279. doi: 10.1016/S0006-3495(02)73987-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen A, Moy VT. Cross-linking of cell surface receptors enhances cooperativity of molecular adhesion. Biophys J. 2000;78(6):2814–2820. doi: 10.1016/S0006-3495(00)76824-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horton M, Charras G, Lehenkari P. Analysis of ligand–receptor interactions in cells by atomic force microscopy. J Recept Signal Transduct Res. 2002;22(1–4):169–190. doi: 10.1081/rrs-120014594. [DOI] [PubMed] [Google Scholar]
- 50.Liu F, Arce FT, Ramachandran S, Lal R. Nanomechanics of hemichannel conformations—connexin flexibility underlying channel opening and closing. J Biol Chem. 2006;281(32):23207–23217. doi: 10.1074/jbc.M605048200. [DOI] [PubMed] [Google Scholar]
- 51.Lin H, Clegg DO, Lal R. Imaging real-time proteolysis of single collagen I molecules with an atomic force microscope. Biochemistry. 1999;38(31):9956–9963. doi: 10.1021/bi990800q. [DOI] [PubMed] [Google Scholar]
- 52.Blair JM, Sorensen LB, Arnsdorf MF, Lal R. The application of atomic-force microscopy for the detection of microcrystals in synovial-fluid from patients with recurrent synovitis. Semin Arthritis Rheum. 1995;24(5):359–369. doi: 10.1016/S0049-0172(95)80005-0. [DOI] [PubMed] [Google Scholar]
- 53.Ward MD. Bulk crystals to surfaces: combining X-ray diffraction and atomic force microscopy to probe the structure and formation of crystal interfaces. Chem Rev. 2001;101(6):1697–1725. doi: 10.1021/cr000020j. [DOI] [PubMed] [Google Scholar]
- 54.Guo SW, Ward MD, Wesson JA. Direct visualization of calcium oxalate monohydrate crystallization and dissolution with atomic force microscopy and the role of polymeric additives. Langmuir. 2002;18(11):4284–42891. doi: 10.1021/la011754+. [DOI] [Google Scholar]
- 55.Jung T, Sheng XX, Choi CK, Kim WS, Wesson JA, Ward MD. Probing crystallization of calcium oxalate monohydrate and the role of macromolecule additives with in situ atomic force microscopy. Langmuir. 2004;20(20):8587–8596. doi: 10.1021/la0488755. [DOI] [PubMed] [Google Scholar]
- 56.Wesson JA, Ward MD. Pathological biomineralization of kidney stones. Elements. 2007;3(6):415–4121. doi: 10.2113/GSELEMENTS.3.6.415. [DOI] [Google Scholar]
- 57.Thurner PJOE, Jungmann R, Kreutz C, Kindt JH, Schitter G, Okouneva TO, Lauer ME, FAntner GE, Hansma HG, Hansma PK. Imaging of bone ultrastructure using atomic force microscopy, 7th edn. In: Mendez-Vilas A DJ, editors. Badajoz, Spain: Formatex Research Center; 2007.
- 58.Jensenius H, Thaysen J, Rasmussen AA, Veje LH, Hansen O, Boisen A. A microcantilever-based alcohol vapor sensor-application and response model. Appl Phys Lett. 2000;76(18):2615–2617. doi: 10.1063/1.126426. [DOI] [Google Scholar]
- 59.Wee KW, Kang GY, Park J, Kang JY, Yoon DS, Park JH, et al. Novel electrical detection of label-free disease marker proteins using piezoresistive self-sensing micro-cantilevers. Biosens Bioelectron. 2005;20(10):1932–1938. doi: 10.1016/j.bios.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 60.Hauptmann P, Lucklum R, Puttmer A, Henning B. Ultrasonic sensors for process monitoring and chemical analysis: state-of-the-art and trends. Sens Actuators A-Phys. 1998;67(1–3):32–48. doi: 10.1016/S0924-4247(97)01725-1. [DOI] [Google Scholar]
- 61.Mechler A, Piorek B, Lal R, Banerjee S. Nanoscale velocity–drag force relationship in thin liquid layers measured by atomic force microscopy. Appl Phys Lett. 2004;85(17):3881–3883. doi: 10.1063/1.1808504. [DOI] [Google Scholar]
- 62.Quist A, Chand A, Ramachandran S, Cohen D, Lal R. Piezoresistive cantilever based nanoflow and viscosity sensor for microchannels. Lab on a Chip. 2006;6(11):1450–4. [DOI] [PubMed]
- 63.Quist AP, Chand A, Ramachandran S, Daraio C, Jin S, Lal R. Atomic force microscopy imaging and electrical recording of lipid bilayers supported over microfabricated silicon chip nanopores: Lab-on-a-chip system for lipid membranes and ion channels. Langmuir. 2007;23(3):1375–80. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
AFM integrated with nanochip-separated, double-chamber electrical recording system. Top rows: a AFM image of arrays of nanopores. The pores are produced in a silicon nitride chip using electron beam lithography and range in size from 50 nm (top rows) to 100 nm (rows on the right). b After deposition of lipid bilayers, AFM image in PBS shows a bilayer covering the pores. The bilayer is not fully contiguous and has several holes (red arrows); the cross-section on a hole in the bilayer (inset) indicates the thickness of a lipid bilayer ~5.3 nm. Blue arrows indicate 500-nm-wide corner alignment marks; red arrows indicate defects in the bilayer. c Schematic of the liquid cell AFM setup for imaging the silicon nitride chip and bilayer. Bottom rows: Left AFM images (left) and conductance maps (right) over a pore in a microfabricated silicon chip for different applied bias potentials (−2.0, −1.0, −0.5 V, respectively, for the top, middle, and bottom panels). Far right: I–V curves (applied potential –−0.5–0.5 V) obtained for 10 s (top), 2 min (middle), and 4 min (bottom) upon addition of gramicidin (0.2 mg/ml) to the bilayer, the overall current across pores increases and conductance increases from 25 pS (bilayer only) to 1 nS (at 4 min), suggesting the formation of hundreds of gramicidin ion channels. For details, see Quist et al., 2007 (63) (TIFF 4,121 kb)
Schematic drawing of conventional patch clamp and an on-chip patch clamp (left). Cells can be pulled into one of the on-chip patch pores (middle top panel), resulting in membrane deformation and patch formation (middle bottom panel). Resulting current measured before (top right top panel) and after cell attachment (right bottom panel). For details, see Seo et al. (29) (TIFF 3,897 kb)
Left panels: Schematic diagram of wool fiber (a). TEM image (b) (higher magnification in d) and AFM image (c) (higher magnification in e) of the same wool fiber region. AFM and TEM images correlate well with respect to identified subcellular structures. Center panels Force curves on cortex (top), exo-cuticle (middle), and embedding resin (bottom). Solid line is collected on hard glass surface. Larger shaded area indicates more elastic surface. Right panels: Top right image shows the corresponding AFM image. Bottom right image shows surface after indentation with diamond tip to create specific patterns on wool fiber. For details, see Parbhu et al. (37) (TIFF 5,356 kb)
AFM imaging or nAChR expressed in Xenopus oocytes. AFM imaging of expressed AChR. After recording ACh-sensitive electrical current in an oocyte, its vitelline membrane was removed. The cell was cut into two halves and one half was imaged. Top left AFM image of the plasma membrane. A crack in the membrane is shown. Middle left High-resolution low-pass-filtered image showing the irregular spacing of receptor cluster. Bottom left High-resolution image of a control oocyte not injected with any mRNA. Top right Line graph of an image of a single AChR channel (~10.5-nm diameter) showing the contour map of subunits and pore. The five subunits (~1- to 1.5-nm diameter) with a central pore are shown. Middle right ACh-sensitive current measured in an oocyte after the AChR expression. The dose-dependent ACh-sensitive current was reversible. Bottom right α-Bungarotoxin binding assay revealed a high level of AChR expression. The channel density calculated from AFM, electrophysiological, and pharmacological studies were comparable; for details, see Lal and Yu (12) (TIFF 6,167 kb)
Top half (left) Adhesion forces between VEGF receptor and antibody (a), with distribution of unbinding forces (b). c Blocking peptide prevents the interaction. d Tapping mode image of VEGF receptors on mica. e, f Adhesion forces between VEGF receptor on endothelial cell and antibody on tip without (e) and with (f) the presence of blocking peptide. Bottom half (right) AFM images of endothelial cells before (a) and after (b) the addition of VEGF showing cytoskeletal reorganization. Fluorescence images showing the presence of Flk-1 receptors (c, d) and control (e) with nonspecific antibody. Bottom half Simultaneous acquisition of topography and elasticity images before and 10 and 45 min after adding VEGF antibody. Clustering of receptors can be observed both in topography and elasticity maps (spots labeled 1–4). Top histograms show the distribution of adhesion forces. For details, see Almqvist et al. (13) (TIFF 5,640 kb)
AFM antibody-conjugated tip pulling experiment: Sensing intermolecular interaction for mapping conformational epitopes. Left half Model of a gap junction plaque with apposing cell membranes with hexagonally packed hemichannels (left). Schematic of force spectroscopy measurement showing the binding of antibody connected to the AFM tip with flexible PEG spacers to Cx43 hemichannels reconstituted in lipid bilayer (middle) and the schematic of Cx43 membrane topology with two extracellular loops, one cytoplasmic loop, and the cytoplasmic carboxyl-terminal domain and locations of antigenic binding sites for anti-CT252–270 and anti-CT360–382 (right) are shown. Right half Probability histograms of the rupture forces of the measured anti-CT252–270-Cx43 (a), anti-CT360–382-Cx43 (b), and GAP26-Cx43 interactions (c). d Representative force–extension curve showing specific avidin–biotin interaction in the PEG spacer extension test system. e Average extension of PEG spacer stretching (~28 nm). f Histograms of measured tether extensions in anti-CT252–270-Cx43, anti-CT360–382-Cx43, and GAP26-Cx43, respectively. For details, see Liu et al. (50) (TIFF 4,683 kb)
Amyloid beta channel-induced cell death. Left half Loss of synaptic arborization and neuronal death imaged with calcein dye fluorescence before and after addition of Aβ peptide to the buffer solution (compare a, c, e, g (control) with panels b, d, f, g (after the addition of nano-to-micromolar abeta peptide). Right half Simultaneously, AFM can be used to image the cytoskeletal structure of cells to track the effects of Aβ peptide on the cells and to test various blocking agents for their performance in blocking toxicity effects. Yellow arrows compare the changes in cytoskeleton upon the addition of amyloid beta peptides or other reagents before and after their addition. For details, see Lin et al. (9) (TIFF 5,570 kb)