Abstract
PseudoBase is a database containing structural, functional and sequence data related to RNA pseudoknots. It can be reached at http://wwwbio.LeidenUniv.nl/∼Batenburg/PKB.html. For each pseudoknot, thirteen items are stored, for example the relevant sequence, the stem positions of the pseudoknot, the EMBL accession number of the sequence and the support that can be given regarding the reliability of the pseudoknot. Since the last publication, information on sizes of the stems and the loops in the pseudoknots has been added. Also added are alternative entries that produce surveys of where the pseudoknots are, sorted according to stem size or loop size.
INTRODUCTION
Pseudoknots are widely occurring structural motifs in RNA. First described in the early eighties as part of tRNA-like structures in plant viral RNAs, pseudoknots were recognized as a general principle of RNA folding (1,2). Pseudoknots play an important role in a number of RNA functions (e.g. in ribosomal frameshifting, regulation of translation and splicing) and they are also essential elements of the topology of many structural RNAs such as, for example, ribosomal RNAs or ribozymes. Therefore, a database with a collection of pseudoknot structures has been created (3). This database can be useful for many studies dealing with structure–function relationships in RNA molecules.
RECENT DEVELOPMENTS
The sizes of stems and loops are very important structural characteristics of pseudoknots. Therefore, we have decided to include this information in all database entries; see labels ‘Stem sizes’ and ‘Loop sizes’ in Figure 1.
Figure 1.
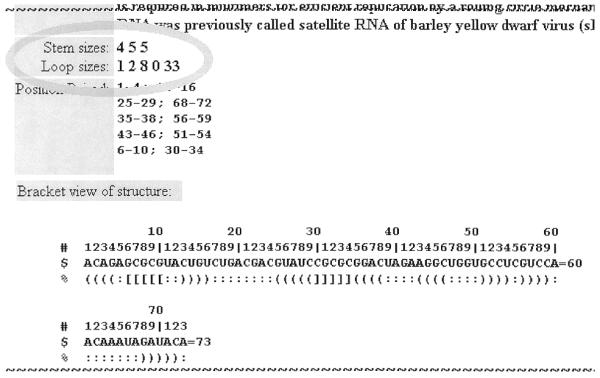
Part of a pseudoknot data-item illustrating the new topics of stem sizes and loop sizes.
Determination of stem- and loop-size in classical or so-called H-(hairpin) pseudoknots is straightforward. However for more complicated structures several decisions had to be made for a consistent nomenclature.
Zero-size regions
Unfortunately, there is not yet a standard nomenclature for pseudoknot structural elements, even for relatively simple structures. In principle, the simplest pseudoknot, the classical or so-called H-pseudoknot, may contain two stems (regions A and C in Fig. 2) and three loops (regions B, D and E in Fig. 2).
Figure 2.
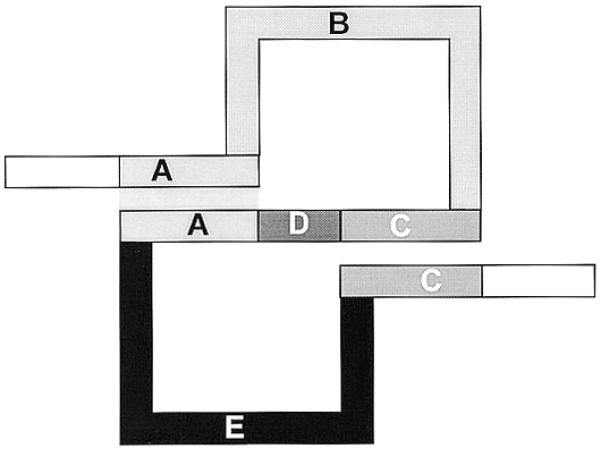
Schematic drawing of the classical pseudoknot. In our definition, sequence region A is called S1, region C is called S2, region B is called L1, region D is called L2 and region E is called L3.
Such stems and loops can be numbered in the 5′→3′ direction as S1, S2 and L1, L2, L3 (for example see 4). However, the most studied type of pseudoknot is with coaxial stacking of stems so that L2 (region D in Fig. 2) is absent. In that case, region E is mostly called L2 instead of L3 (for examples see 2,5–8). Such a shift in nomenclature can even occur within one publication where this loop is called L3 on one page (for pseudoknots with nucleotides present in the D region) and called L2 on another page when there are no nucleotides in region D (for example see 4,9). We have decided always to assign label L2 to region D for reasons of consistent and unambiguous nomenclature in our database. If both stems are adjacent this would be L2 with size = 0; this ensures that structurally similar regions E are always called L3.
A similar consistency is applied to more complex pseudoknots with coaxial stacking where we recognized such areas as Li with size 0. See for example Figure 3 where L1, L2 and L3 are regions with proper nucleotides but L4 has size 0.
Figure 3.
A sequence with two complex pseudoknots demonstrating stem- and loop-labeling.
Pseudoknots with internal structures
Classical pseudoknots have simple loops where all nucleotides are unpaired. Consequently the word loop is appropriate for this part of the pseudoknot, so there is no argument over defining loop size as the total number of nucleotides in that region. However, in more complicated tertiary structures a loop can contain substructures. Such a region between two pseudoknot stems can have several stems with their own internal loops, hairpin loops and multibranch loops. Here one can even argue whether ‘loop’ is a suitable name or not. Nevertheless, for the sake of simplicity we have decided to consider this whole region a ‘pseudoknot loop’ and abbreviate this in the context of PseudoBase as Li.
Consequently, stems that are not ‘pseudoknotted’ with other stems and their corresponding loops are not included in our PseudoBase enumeration of stems and loops. For example, L5 in Figure 3 contains a hairpin with an internal loop that is not registered in PseudoBase as part of the stem and loop enumeration.
Furthermore, the nucleotides in such complicated loops are all counted as contributions to the size of the particular pseudoknot loop, whether they are unpaired or not. For example, L5 in Figure 3 is equal to 33 nt although only 17 of them are single stranded.
Stems with internal loops
Stems raised another nomenclature problem. Classical pseudoknot nomenclature is based on two simple, regular stems, the first one beginning at the 5′-end is called S1 and the second one ending nearest to the 3′-end is called S2 (for examples see 1,2,5). For more complex structures the question arises: what are the pseudoknot stems, what are their stem numbers and what are their sizes? Should we count a stem with a bulge-loop halfway as one stem or two? And what about a stem ‘interrupted’ by an interior loop, or even by a multibranch loop?
Again we decided to opt for a simple nomenclature and discard bulges, interior loops and multibranch loops. For the size of such stems we did not count the nucleotides as this would raise dilemmas when one stem half contains a bulge, but counted the number of interactions instead, that is to say the number of nucleotide pairs. In other words, if a pseudoknot stem could be considered as ‘interrupted’ with two or more duplexes forming a pseudoknot with some other stem, its stem size is defined as the total number of base pairs in the duplexes. By this decision we do not have to consider whether to include the bulge and loop nucleotides or not, and we think that this number is a more suitable statistic than the total number of nucleotides in ‘interrupted’ stems. For illustration see Figure 3; the first stem S1 is simple and has size 4, but the next stem S2 has size 5 because the unpaired nucleotides 8 and 34 in the internal loop are ignored.
Sorting of pseudoknots
Using the adopted definitions we have developed a program that computes the Si and Li sizes for the submitted pseudoknots and added this information into each pseudoknot data page.
Furthermore, this program also generated html pages with surveys of the available pseudoknot items sorted on stem sizes or loop sizes.
ACCESS
The database is freely accessible at http://wwwbio.leidenuniv.nl/~Batenburg/PKB.html. This presents the introduction page, which leads to the pages for retrieval or submission or an ‘About’ page for more information. Users of PseudoBase are requested to cite this publication and are encouraged to provide corrections and suggestions for improvement to the first author by email (batenburg@rulsfb.leidenuniv.nl) and to submit new pseudoknot data using the automatic submission form that is supplied on the ‘Submission’-page (http://wwwbio.leidenuniv.nl/~Batenburg/PKBPut.html).
Acknowledgments
ACKNOWLEDGEMENT
A.P.G. was supported by the European Commission (project BIO4-98-0189).
References
- 1.Rietveld K., van Poelgeest,R., Pleij,C.W.A., van Boom,J.H. and Bosch,L. (1982) The tRNA-like structure at the 3′ terminus of turnip yellow mosaic virus RNA. Differences and similarities with canonical tRNA. Nucleic Acids Res., 10, 1929–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pleij C.W.A., Rietveld,K. and Bosch,L. (1985) A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res., 13, 1717–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Batenburg F.H.D., Gultyaev,A.P., Pleij,C.W.A., Ng,J. and Oliehoek,J. (2000) PseudoBase: a database with RNA pseudoknots. Nucleic Acids Res., 28, 201–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hilbers C.W., Michiels,P.J.A. and Heus,H.A. (1998) New developments in structure determination of pseudoknots. Biopolymers, 48, 137–153. [DOI] [PubMed] [Google Scholar]
- 5.Abrahams J.P., van den Berg,M., van Batenburg,E. and Pleij,C. (1990) Prediction of RNA secondary structure, including pseudoknotting, by computer simulation. Nucleic Acids Res., 18, 3035–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deiman B.A.L.M. and Pleij,C.W.A. (1997) Pseudoknots: a vital feature in viral RNA. Semin. Virol., 8, 166–175. [Google Scholar]
- 7.Mans R.M.W. and Pleij,C.W.A. (1993) RNA pseudoknots. In Eckstein,F. and Lilley,D.M.J. (eds) Nucleic Acids and Molecular Biology. Springer Verlag, Berlin, Vol.7, pp. 250–270.
- 8.Pleij C.W.A. (1994) RNA pseudoknots. Curr. Opin. Struct. Biol., 4, 337–344. [Google Scholar]
- 9.ten Dam E., Pleij,K and Draper,D. (1992) Structural and functional aspects of RNA pseudoknots. Biochemistry, 31, 11665–11676. [DOI] [PubMed] [Google Scholar]



