Abstract
In title compound, [Ni(C9H5NO4S)(H2O)2]n, the NiII atom is coordinated by one N atom and two bridging O atoms from two 8-oxidoquinoline-5-sulfonate ligands, one sulfonate O atom from a third ligand, and two water molecules in a distorted octahedral geometry. The two NiII atoms are linked to each other through the bridging O atoms, forming a dimer. Adjacent dimers are connected through the coordination of the sulfonate O atom into a two-dimensional coordination network parallel to (010). Hydrogen bonds between the coordinated water molecules and the uncoordinated O atoms of the sulfonate groups result in the construction of a three-dimensional supramolecular structure.
Related literature
For related structures, see: Ammor et al. (1992 ▶); Petit et al. (1993a
▶,b
▶); Rao et al. (2003 ▶); Wu et al. (2008 ▶); Xie et al. (1992 ▶).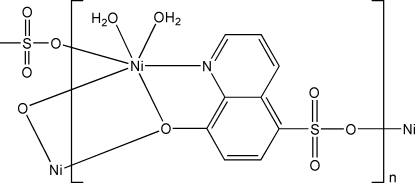
Experimental
Crystal data
[Ni(C9H5NO4S)(H2O)2]
M r = 317.94
Orthorhombic,
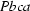
a = 9.2067 (8) Å
b = 15.0504 (13) Å
c = 16.1599 (14) Å
V = 2239.2 (3) Å3
Z = 8
Mo Kα radiation
μ = 1.94 mm−1
T = 293 K
0.28 × 0.22 × 0.18 mm
Data collection
Bruker SMART APEX CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2001 ▶) T min = 0.601, T max = 0.701
11973 measured reflections
2198 independent reflections
1874 reflections with I > 2σ(I)
R int = 0.037
Refinement
R[F 2 > 2σ(F 2)] = 0.031
wR(F 2) = 0.079
S = 1.02
2198 reflections
171 parameters
4 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.64 e Å−3
Δρmin = −0.27 e Å−3
Data collection: SMART (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶) and DIAMOND (Brandenburg, 1999 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809026105/hy2203sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809026105/hy2203Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Selected bond lengths (Å).
| Ni1—O1i | 2.0153 (17) |
| Ni1—O6W | 2.0285 (19) |
| Ni1—O1 | 2.0443 (16) |
| Ni1—N1 | 2.052 (2) |
| Ni1—O5W | 2.0936 (19) |
| Ni1—O2ii | 2.1437 (17) |
Symmetry codes: (i) 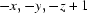 ; (ii)
; (ii) 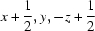 .
.
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O5W—H5WA⋯O3iii | 0.82 | 2.00 | 2.812 (2) | 171 |
| O5W—H5WB⋯O4iv | 0.80 (2) | 2.07 (2) | 2.866 (3) | 170 (2) |
| O6W—H6WA⋯O3iv | 0.82 | 1.93 | 2.687 (2) | 153 |
| O6W—H6WB⋯O4v | 0.78 (2) | 2.04 (2) | 2.787 (3) | 159 (3) |
Symmetry codes: (iii) 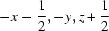 ; (iv)
; (iv) 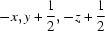 ; (v)
; (v) 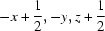 .
.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant No. 20571030).
supplementary crystallographic information
Comment
The metal complexes with organic ligands containing sulfonate group still remain largely unexplored. We have been investigating the formation of novel transition metal coordination polymers employing hydrothermal methods. During the course of the investigation employing 8-hydroxylquinoline-5-sulfonic acids (H2QS) as organic ligand, which has received a little attention (Ammor et al., 1992; Petit et al., 1993a,b; Rao et al., 2003; Wu et al., 2008; Xie et al., 1992), and nickel(II) as metal center, we isolated a new two-dimensional coordination polymer.
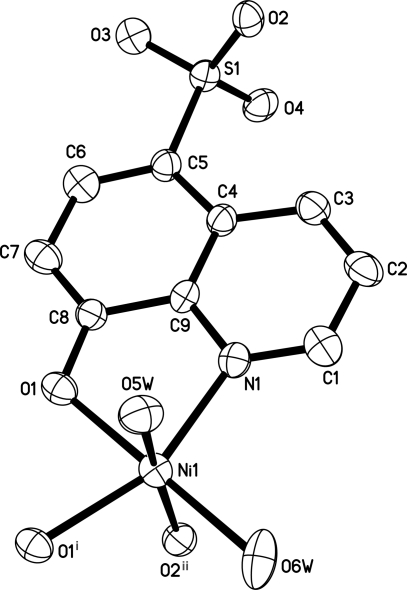
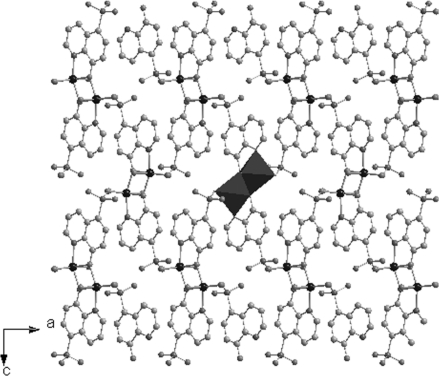
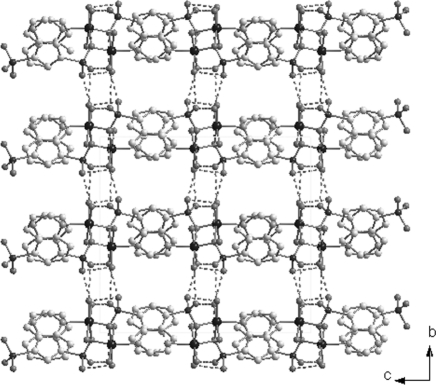
As shown in Fig. 1, the asymmetric unit of the title compound contains one NiII atom, one QS ligand, and two water molecules. The NiII atom adopts a distorted octahedral coordination geometry, defined by one N atom and two bridging olate O atoms from two QS ligands, one sulfonate O atom from a third ligand, and two water molecules (Table 1). Two crystallographically equivalent Ni atoms [Ni1 and Ni1i, symmetry code: (i) -x, -y, 1 - z] link to each other through two bridging atoms O1 and O1i, forming an edge-sharing dimer. These dimers are connected by the sulfonate groups of the QS ligands into an infinite two-dimensional coordination network with a (4,4) topology along the [0 1 0] direction, as shown in Fig. 2. These networks are further connected by hydrogen bonds between the coordinated water molecules and the uncoordinated O atoms of the sulfonate groups into a three-dimensional supramolecular structure (Fig. 3 and Table 2).
Experimental
A mixture of Ni(NO3)2.4H2O (0.250 g, 1 mmol) and H2QS (0.024 g, 0.1 mmol) was dissolved in 6 ml H2O with stirring about half an hour and pH = 6. The mixture was transferred to a 15 ml Teflon-lined stainless-steel hydrothermal autoclave and heated at 413 K for two weeks under autogenous pressure. The green block crystals were filtered off, washed with ethanol and dried at room temperature.
Refinement
H atoms on C atoms are positioned geometrically and refined as riding atoms, with C—H = 0.93 Å and Uiso = 1.2Ueq(C). H atoms of water molecules are located in a difference Fourier map. Two H atoms (H5WA and H6WA) were refined as riding atoms, with O—H = 0.82 Å and Uiso = 1.5Ueq(O), and the other two (H5WB and H6WB) were refined isotropically.
Figures
Fig. 1.
Coordination environment of the Ni atom in the title compound. Displacement ellipsoids are drawn at the 50% probability level. [Symmetry code: (i) -x,-y,-z + 1; (ii) x + 1/2, y, -z + 1/2.]
Fig. 2.
The two-dimensional coordination network of the title compound with a (4,4) topology, viewed along the [0 1 0] direction.
Fig. 3.
The three-dimensional supramolecular structure of the title compound, connected by hydrogen bonds between the coordinated water molecules and the uncoordinated sulfonate O atoms.
Crystal data
| [Ni(C9H5NO4S)(H2O)2] | F(000) = 1296 |
| Mr = 317.94 | Dx = 1.886 Mg m−3Dm = 1.886 Mg m−3Dm measured by not measured |
| Orthorhombic, Pbca | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ac 2ab | Cell parameters from 2198 reflections |
| a = 9.2067 (8) Å | θ = 2.5–28.1° |
| b = 15.0504 (13) Å | µ = 1.94 mm−1 |
| c = 16.1599 (14) Å | T = 293 K |
| V = 2239.2 (3) Å3 | Block, green |
| Z = 8 | 0.28 × 0.22 × 0.18 mm |
Data collection
| Bruker SMART APEX CCD diffractometer | 2198 independent reflections |
| Radiation source: fine-focus sealed tube | 1874 reflections with I > 2σ(I) |
| graphite | Rint = 0.037 |
| φ and ω scans | θmax = 26.0°, θmin = 2.5° |
| Absorption correction: multi-scan (SADABS; Bruker, 2001) | h = −10→11 |
| Tmin = 0.601, Tmax = 0.701 | k = −18→17 |
| 11973 measured reflections | l = −18→19 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.031 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.079 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.0484P)2] where P = (Fo2 + 2Fc2)/3 |
| 2198 reflections | (Δ/σ)max = 0.001 |
| 171 parameters | Δρmax = 0.64 e Å−3 |
| 4 restraints | Δρmin = −0.27 e Å−3 |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Ni1 | 0.10943 (3) | 0.058524 (19) | 0.446648 (17) | 0.02363 (12) | |
| O5W | −0.0374 (2) | 0.16474 (12) | 0.44545 (10) | 0.0326 (4) | |
| H5WA | −0.0848 | 0.1645 | 0.4884 | 0.049* | |
| O1 | 0.04736 (19) | 0.03393 (11) | 0.56593 (9) | 0.0264 (4) | |
| N1 | 0.0998 (2) | 0.05336 (12) | 0.31986 (12) | 0.0250 (5) | |
| C8 | −0.0750 (3) | −0.05668 (14) | 0.35675 (14) | 0.0232 (5) | |
| C9 | 0.0015 (3) | −0.00829 (15) | 0.29298 (13) | 0.0231 (5) | |
| C4 | −0.0248 (3) | −0.02414 (16) | 0.20803 (13) | 0.0251 (5) | |
| C2 | 0.1498 (3) | 0.09088 (18) | 0.17944 (15) | 0.0338 (6) | |
| H2A | 0.2007 | 0.1263 | 0.1423 | 0.041* | |
| C3 | 0.0542 (3) | 0.02903 (17) | 0.15126 (14) | 0.0300 (6) | |
| H3B | 0.0406 | 0.0216 | 0.0947 | 0.036* | |
| C5 | −0.1275 (3) | −0.09119 (16) | 0.18792 (14) | 0.0256 (5) | |
| C1 | 0.1711 (3) | 0.10083 (16) | 0.26430 (15) | 0.0311 (6) | |
| H1B | 0.2383 | 0.1426 | 0.2826 | 0.037* | |
| C6 | −0.1969 (3) | −0.13796 (16) | 0.24900 (15) | 0.0310 (6) | |
| H6A | −0.2631 | −0.1819 | 0.2342 | 0.037* | |
| C7 | −0.1711 (3) | −0.12159 (15) | 0.33312 (15) | 0.0314 (6) | |
| H7A | −0.2193 | −0.1549 | 0.3731 | 0.038* | |
| O6W | 0.2752 (2) | 0.14637 (13) | 0.45841 (13) | 0.0415 (5) | |
| H6WA | 0.2501 | 0.1948 | 0.4401 | 0.062* | |
| H5WB | −0.011 (3) | 0.2155 (12) | 0.4428 (14) | 0.029 (7)* | |
| H6WB | 0.347 (3) | 0.134 (2) | 0.4814 (19) | 0.060 (11)* | |
| S1 | −0.16782 (7) | −0.11580 (4) | 0.08365 (4) | 0.02460 (16) | |
| O2 | −0.21464 (19) | −0.03403 (11) | 0.04239 (9) | 0.0293 (4) | |
| O4 | −0.03579 (19) | −0.15043 (12) | 0.04607 (10) | 0.0352 (4) | |
| O3 | −0.28431 (19) | −0.18166 (11) | 0.08585 (10) | 0.0313 (4) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Ni1 | 0.0259 (2) | 0.0217 (2) | 0.02325 (19) | −0.00303 (12) | −0.00136 (12) | 0.00040 (11) |
| O5W | 0.0340 (11) | 0.0262 (10) | 0.0375 (10) | 0.0004 (8) | 0.0078 (8) | 0.0026 (8) |
| O1 | 0.0293 (10) | 0.0275 (9) | 0.0225 (8) | −0.0058 (8) | 0.0005 (7) | −0.0008 (7) |
| N1 | 0.0235 (11) | 0.0235 (11) | 0.0279 (11) | −0.0013 (8) | −0.0027 (8) | −0.0001 (8) |
| C8 | 0.0250 (13) | 0.0217 (12) | 0.0230 (12) | 0.0009 (9) | 0.0008 (9) | 0.0002 (9) |
| C9 | 0.0191 (11) | 0.0232 (12) | 0.0270 (12) | 0.0017 (9) | −0.0022 (9) | −0.0010 (9) |
| C4 | 0.0246 (13) | 0.0263 (12) | 0.0245 (12) | 0.0038 (10) | −0.0011 (10) | −0.0017 (9) |
| C2 | 0.0348 (15) | 0.0385 (15) | 0.0282 (13) | −0.0071 (12) | 0.0025 (11) | 0.0077 (12) |
| C3 | 0.0305 (14) | 0.0348 (14) | 0.0246 (12) | −0.0007 (11) | −0.0002 (10) | 0.0009 (10) |
| C5 | 0.0259 (13) | 0.0259 (12) | 0.0250 (12) | 0.0028 (10) | −0.0012 (10) | −0.0020 (10) |
| C1 | 0.0317 (14) | 0.0299 (14) | 0.0317 (13) | −0.0106 (11) | −0.0051 (11) | 0.0019 (10) |
| C6 | 0.0338 (14) | 0.0265 (13) | 0.0327 (13) | −0.0065 (11) | −0.0032 (11) | −0.0038 (11) |
| C7 | 0.0358 (15) | 0.0308 (14) | 0.0276 (13) | −0.0072 (11) | 0.0013 (11) | 0.0017 (10) |
| O6W | 0.0318 (11) | 0.0256 (10) | 0.0670 (13) | −0.0053 (8) | −0.0201 (10) | 0.0100 (9) |
| S1 | 0.0260 (3) | 0.0232 (3) | 0.0246 (3) | 0.0021 (2) | −0.0014 (2) | −0.0041 (2) |
| O2 | 0.0330 (10) | 0.0263 (9) | 0.0284 (9) | 0.0054 (8) | −0.0022 (7) | 0.0002 (7) |
| O4 | 0.0320 (10) | 0.0358 (11) | 0.0377 (10) | 0.0081 (8) | 0.0043 (8) | −0.0047 (8) |
| O3 | 0.0353 (10) | 0.0276 (9) | 0.0310 (9) | −0.0038 (8) | −0.0036 (7) | −0.0052 (7) |
Geometric parameters (Å, °)
| Ni1—O1i | 2.0153 (17) | C4—C5 | 1.421 (3) |
| Ni1—O6W | 2.0285 (19) | C2—C3 | 1.360 (4) |
| Ni1—O1 | 2.0443 (16) | C2—C1 | 1.393 (3) |
| Ni1—N1 | 2.052 (2) | C2—H2A | 0.9300 |
| Ni1—O5W | 2.0936 (19) | C3—H3B | 0.9300 |
| Ni1—O2ii | 2.1437 (17) | C5—C6 | 1.371 (3) |
| O5W—H5WA | 0.8200 | C5—S1 | 1.765 (2) |
| O5W—H5WB | 0.804 (17) | C1—H1B | 0.9300 |
| O1—C8i | 1.320 (3) | C6—C7 | 1.402 (3) |
| N1—C1 | 1.322 (3) | C6—H6A | 0.9300 |
| N1—C9 | 1.367 (3) | C7—H7A | 0.9300 |
| C8—O1i | 1.320 (3) | O6W—H6WA | 0.8200 |
| C8—C7 | 1.372 (3) | O6W—H6WB | 0.784 (17) |
| C8—C9 | 1.445 (3) | S1—O4 | 1.4553 (18) |
| C9—C4 | 1.414 (3) | S1—O3 | 1.4608 (17) |
| C4—C3 | 1.418 (3) | S1—O2 | 1.4645 (17) |
| O1i—Ni1—O6W | 176.93 (8) | C9—C4—C5 | 117.1 (2) |
| O1i—Ni1—O1 | 76.69 (7) | C3—C4—C5 | 126.5 (2) |
| O6W—Ni1—O1 | 103.89 (8) | C3—C2—C1 | 119.6 (2) |
| O1i—Ni1—N1 | 80.92 (7) | C3—C2—H2A | 120.2 |
| O6W—Ni1—N1 | 98.67 (8) | C1—C2—H2A | 120.2 |
| O1—Ni1—N1 | 157.28 (7) | C2—C3—C4 | 120.1 (2) |
| O1i—Ni1—O5W | 93.64 (8) | C2—C3—H3B | 119.9 |
| O6W—Ni1—O5W | 89.40 (8) | C4—C3—H3B | 119.9 |
| O1—Ni1—O5W | 88.06 (7) | C6—C5—C4 | 120.7 (2) |
| N1—Ni1—O5W | 89.54 (7) | C6—C5—S1 | 118.81 (19) |
| O1i—Ni1—O2 | 59.09 (4) | C4—C5—S1 | 120.49 (18) |
| O6W—Ni1—O2 | 120.98 (6) | N1—C1—C2 | 122.7 (2) |
| O1—Ni1—O2 | 133.91 (5) | N1—C1—H1B | 118.6 |
| N1—Ni1—O2ii | 95.19 (7) | C2—C1—H1B | 118.6 |
| O5W—Ni1—O2ii | 170.00 (7) | C5—C6—C7 | 121.9 (2) |
| Ni1—O5W—H5WA | 109.5 | C5—C6—H6A | 119.0 |
| Ni1—O5W—H5WB | 122 (2) | C7—C6—H6A | 119.0 |
| H5WA—O5W—H5WB | 102.2 | C8—C7—C6 | 120.3 (2) |
| C8i—O1—Ni1i | 114.35 (14) | C8—C7—H7A | 119.9 |
| C8i—O1—Ni1 | 142.34 (15) | C6—C7—H7A | 119.9 |
| Ni1i—O1—Ni1 | 103.31 (7) | Ni1—O6W—H6WA | 109.5 |
| C1—N1—C9 | 118.7 (2) | Ni1—O6W—H6WB | 122 (2) |
| C1—N1—Ni1 | 129.50 (16) | H6WA—O6W—H6WB | 128.7 |
| C9—N1—Ni1 | 111.81 (15) | O4—S1—O3 | 112.36 (11) |
| O1i—C8—C7 | 124.9 (2) | O4—S1—O2 | 110.91 (10) |
| O1i—C8—C9 | 116.8 (2) | O3—S1—O2 | 111.42 (10) |
| C7—C8—C9 | 118.3 (2) | O4—S1—C5 | 107.33 (11) |
| N1—C9—C4 | 122.4 (2) | O3—S1—C5 | 105.87 (10) |
| N1—C9—C8 | 115.98 (19) | O2—S1—C5 | 108.68 (10) |
| C4—C9—C8 | 121.6 (2) | S1—O2—Ni1iii | 136.95 (11) |
| C9—C4—C3 | 116.4 (2) |
Symmetry codes: (i) −x, −y, −z+1; (ii) x+1/2, y, −z+1/2; (iii) x−1/2, y, −z+1/2.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O5W—H5WA···O3iv | 0.82 | 2.00 | 2.812 (2) | 171 |
| O5W—H5WB···O4v | 0.80 (2) | 2.07 (2) | 2.866 (3) | 170 (2) |
| O6W—H6WA···O3v | 0.82 | 1.93 | 2.687 (2) | 153 |
| O6W—H6WB···O4vi | 0.78 (2) | 2.04 (2) | 2.787 (3) | 159 (3) |
Symmetry codes: (iv) −x−1/2, −y, z+1/2; (v) −x, y+1/2, −z+1/2; (vi) −x+1/2, −y, z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HY2203).
References
- Ammor, S., Coquerel, G., Perez, G. & Robert, F. (1992). Eur. J. Solid State Inorg. Chem.29, 131–139.
- Brandenburg, K. (1999). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Bruker (2001). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2007). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Petit, S., Ammor, S., Coquerel, G., Mayer, G., Perez, G. & Dance, J.-M. (1993a). Eur. J. Solid State Inorg. Chem.39, 497–507.
- Petit, S., Coquerel, G., Perez, G., Louer, D. & Louer, M. (1993b). New J. Chem.17, 187–192.
- Rao, H.-Y., Tao, J. & Ng, S. W. (2003). Acta Cryst. E59, m859–m860.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Wu, H., Dong, X. W., Liu, H.-Y., Ma, J.-F., Li, S.-L., Yang, J., Liu, Y.-Y. & Su, Z.-M. (2008). Dalton Trans. pp. 5331–5341. [DOI] [PubMed]
- Xie, Z. X., Liu, W., Liu, H. F. & Zheng, L. S. (1992). Chin. J. Struct. Chem.11, 139–142.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809026105/hy2203sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809026105/hy2203Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report