Abstract
There are two crystallographically independent molecules in the asymmetric unit of the title compound, C15H16O5. In each, the 1,3-dioxane ring is in an envelope conformation with the C atom common to the cyclohexane ring forming the flap. The dihedral angles between the five essentially planar [maximum deviations from the least-squares planes of 0.049 (3) and 0.042 (3) Å] atoms of the 1,3-dioxane ring and the furan ring in the two molecules are 7.15 (1) and 6.80 (1)°. The crystal structure is stabilized by weak intermolecular C—H⋯O hydrogen bonds.
Related literature
For background to the applications of spiro compounds, see: Yaozhong et al. (1998 ▶); Lian et al. (2008 ▶); Wei et al. (2008 ▶). For the crystal structure of 3-(furan-2-ylmethylene)-1,5-dioxaspiro[5.5]undecane-2,4-dione, see: Zeng & Jian (2009 ▶).
Experimental
Crystal data
C15H16O5
M r = 276.28
Monoclinic,

a = 19.314 (4) Å
b = 6.8289 (14) Å
c = 20.468 (4) Å
β = 97.04 (3)°
V = 2679.2 (9) Å3
Z = 8
Mo Kα radiation
μ = 0.10 mm−1
T = 293 K
0.22 × 0.18 × 0.15 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.978, T max = 0.985
21978 measured reflections
6028 independent reflections
3716 reflections with I > 2σ(I)
R int = 0.049
Refinement
R[F 2 > 2σ(F 2)] = 0.060
wR(F 2) = 0.198
S = 1.02
6028 reflections
363 parameters
H-atom parameters constrained
Δρmax = 0.40 e Å−3
Δρmin = −0.36 e Å−3
Data collection: SMART (Bruker, 1997 ▶); cell refinement: SAINT (Bruker, 1997 ▶); data reduction: SAINT; program(s) used to refine structure: SHELXS97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S160053680902947X/lh2861sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680902947X/lh2861Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C4A—H4AA⋯O4Bi | 0.93 | 2.59 | 3.408 (3) | 147 |
| C1B—H1BA⋯O4Aii | 0.93 | 2.50 | 3.376 (3) | 157 |
| C12B—H12B⋯O4Bi | 0.97 | 2.53 | 3.420 (3) | 152 |
| C13A—H13A⋯O4Aii | 0.97 | 2.54 | 3.405 (3) | 149 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
supplementary crystallographic information
Comment
Spiro compounds are widely used in medicine, catalysis and optical material (Lian et al., 2008; Yaozhong et al., 1998; Wei et al., 2008) owing to their interesting conformational features. We report here the synthesis and structure of the title compound, (I), as part of our ongoing studies on new spiro compounds with potentially higher bioactivity and have recently determined the crystal structure of 3-(furan-2-ylmethylene)-1,5-dioxaspiro[5.5]undecane-2,4-dione, (Zeng & Jian, 2009).
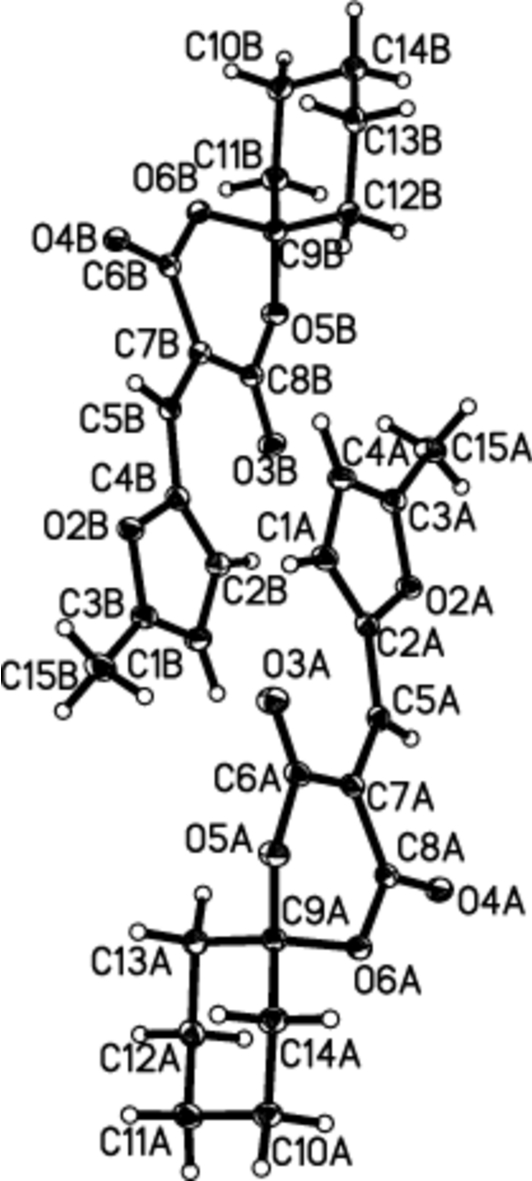
The asymmetric unit of (I) is shown in Fig. 1. In both independent molecules, the 1,3-dioxane ring is in an envelope conformation with atoms C9A and C9B forming the flap in each. The mean planes of the other five essentially planar atoms (O5A/O6A/C6A—C8A and O5B/O6B/C6B—C8B) form dihedral angles of 7.15 (1)° and 6.80 (1)° with the furan ring (O2A/C1A-C4A and O2B/C1B-C4B). The crystal structure is stabilized by weak intermolecular C—H···O hydrogen bonds (Table 1).
Experimental
A mixture of malonic acid (6.24 g, 0.06 mol) and acetic anhydride(9 ml) in conc. sulfuric acid (0.25 ml) was stirred with water at 303K, After dissolving, cyclohexanone (5.88 g, 0.06 mol) was added dropwise into solution for 1 h. The reaction was allowed to proceed for 4 h. The mixture was cooled and filtered, and then an ethanol solution of 5-methylfuran-2-carbaldehyde (6.60 g,0.06 mol) was added. The solution was then filtered and concentrated. Single crystals were obtained by evaporation of an acetone-petroleum aether (2:1 v/v) solution of (I) at room temperature over a period of one week.
Refinement
The H atoms were placed in calculated positions (C—H = 0.93–0.97 Å), and refined as riding with Uiso(H) = 1.2Ueq(C) or 1.5Ueq(methyl C).
Figures
Fig. 1.
The asymmetric unit of (I), drawn with 30% probability ellipsoids and spheres of arbritrary size for the H atoms.
Crystal data
| C15H16O5 | F(000) = 1168 |
| Mr = 276.28 | Dx = 1.370 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 3716 reflections |
| a = 19.314 (4) Å | θ = 3.1–27.5° |
| b = 6.8289 (14) Å | µ = 0.10 mm−1 |
| c = 20.468 (4) Å | T = 293 K |
| β = 97.04 (3)° | Block, yellow |
| V = 2679.2 (9) Å3 | 0.22 × 0.18 × 0.15 mm |
| Z = 8 |
Data collection
| Bruker SMART CCD area-detector diffractometer | 6028 independent reflections |
| Radiation source: fine-focus sealed tube | 3716 reflections with I > 2σ(I) |
| graphite | Rint = 0.049 |
| φ and ω scans | θmax = 27.5°, θmin = 3.1° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −25→24 |
| Tmin = 0.978, Tmax = 0.985 | k = −8→8 |
| 21978 measured reflections | l = −26→26 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.060 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.198 | H-atom parameters constrained |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.119P)2] where P = (Fo2 + 2Fc2)/3 |
| 6028 reflections | (Δ/σ)max = 0.001 |
| 363 parameters | Δρmax = 0.40 e Å−3 |
| 0 restraints | Δρmin = −0.36 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O2A | 0.77282 (7) | 0.9079 (2) | 0.18291 (7) | 0.0240 (4) | |
| O3A | 0.85666 (7) | 1.0679 (3) | 0.38985 (7) | 0.0302 (4) | |
| O4A | 1.01152 (7) | 1.0476 (3) | 0.22791 (7) | 0.0298 (4) | |
| O5A | 0.97036 (7) | 1.1039 (2) | 0.41380 (7) | 0.0266 (4) | |
| O6A | 1.04784 (7) | 1.0940 (2) | 0.33323 (7) | 0.0250 (4) | |
| C1A | 0.74943 (10) | 0.9542 (4) | 0.28658 (10) | 0.0246 (5) | |
| H1AA | 0.7543 | 0.9805 | 0.3315 | 0.030* | |
| C2A | 0.80226 (10) | 0.9507 (3) | 0.24675 (9) | 0.0226 (5) | |
| C3A | 0.70337 (10) | 0.8848 (3) | 0.18383 (10) | 0.0241 (5) | |
| C4A | 0.68697 (10) | 0.9107 (4) | 0.24655 (11) | 0.0264 (5) | |
| H4AA | 0.6428 | 0.9012 | 0.2600 | 0.032* | |
| C5A | 0.87528 (10) | 0.9763 (3) | 0.25215 (9) | 0.0223 (5) | |
| H5AA | 0.8931 | 0.9559 | 0.2125 | 0.027* | |
| C6A | 0.91304 (10) | 1.0628 (3) | 0.37011 (10) | 0.0234 (5) | |
| C7A | 0.92562 (10) | 1.0240 (3) | 0.30207 (9) | 0.0227 (5) | |
| C8A | 0.99672 (10) | 1.0523 (4) | 0.28365 (10) | 0.0244 (5) | |
| C9A | 1.03783 (10) | 1.0380 (3) | 0.39939 (9) | 0.0225 (5) | |
| C10A | 1.16592 (10) | 1.0837 (4) | 0.43581 (10) | 0.0294 (5) | |
| H10A | 1.1762 | 1.1199 | 0.3922 | 0.035* | |
| H10B | 1.1991 | 1.1501 | 0.4677 | 0.035* | |
| C11A | 1.17412 (10) | 0.8647 (4) | 0.44459 (10) | 0.0295 (5) | |
| H11A | 1.1698 | 0.8308 | 0.4899 | 0.035* | |
| H11B | 1.2203 | 0.8263 | 0.4355 | 0.035* | |
| C12A | 1.11918 (10) | 0.7519 (4) | 0.39880 (10) | 0.0265 (5) | |
| H12C | 1.1237 | 0.6130 | 0.4082 | 0.032* | |
| H12D | 1.1273 | 0.7724 | 0.3534 | 0.032* | |
| C13A | 1.04505 (10) | 0.8192 (4) | 0.40769 (10) | 0.0252 (5) | |
| H13A | 1.0117 | 0.7541 | 0.3755 | 0.030* | |
| H13B | 1.0346 | 0.7824 | 0.4512 | 0.030* | |
| C14A | 1.09154 (10) | 1.1511 (4) | 0.44459 (10) | 0.0275 (5) | |
| H14C | 1.0834 | 1.1312 | 0.4899 | 0.033* | |
| H14D | 1.0868 | 1.2899 | 0.4349 | 0.033* | |
| C15A | 0.66067 (11) | 0.8365 (4) | 0.12100 (11) | 0.0323 (5) | |
| H15A | 0.6659 | 0.9375 | 0.0893 | 0.049* | |
| H15B | 0.6125 | 0.8269 | 0.1280 | 0.049* | |
| H15C | 0.6758 | 0.7137 | 0.1049 | 0.049* | |
| O2B | 0.72619 (7) | 0.4699 (2) | 0.32748 (7) | 0.0260 (4) | |
| O3B | 0.65167 (7) | 0.3142 (3) | 0.11772 (7) | 0.0311 (4) | |
| O4B | 0.48850 (7) | 0.3305 (3) | 0.27231 (7) | 0.0312 (4) | |
| O5B | 0.53852 (7) | 0.2773 (3) | 0.08899 (7) | 0.0289 (4) | |
| O6B | 0.45715 (7) | 0.2830 (3) | 0.16647 (7) | 0.0269 (4) | |
| C1B | 0.81544 (10) | 0.4690 (4) | 0.26779 (11) | 0.0289 (5) | |
| H1BA | 0.8604 | 0.4789 | 0.2562 | 0.035* | |
| C2B | 0.75465 (10) | 0.4255 (4) | 0.22541 (11) | 0.0262 (5) | |
| H2BA | 0.7519 | 0.3999 | 0.1805 | 0.031* | |
| C3B | 0.79632 (10) | 0.4941 (4) | 0.32922 (11) | 0.0269 (5) | |
| C4B | 0.69981 (10) | 0.4275 (3) | 0.26259 (10) | 0.0242 (5) | |
| C5B | 0.62670 (10) | 0.4016 (3) | 0.25433 (10) | 0.0229 (5) | |
| H5BA | 0.6071 | 0.4197 | 0.2932 | 0.027* | |
| C6B | 0.50626 (10) | 0.3260 (4) | 0.21756 (10) | 0.0255 (5) | |
| C7B | 0.57830 (10) | 0.3558 (3) | 0.20207 (10) | 0.0238 (5) | |
| C8B | 0.59395 (10) | 0.3186 (4) | 0.13503 (10) | 0.0249 (5) | |
| C9B | 0.46957 (10) | 0.3378 (4) | 0.10066 (10) | 0.0251 (5) | |
| C10B | 0.34343 (11) | 0.2776 (4) | 0.06026 (11) | 0.0326 (6) | |
| H10C | 0.3122 | 0.2052 | 0.0282 | 0.039* | |
| H10D | 0.3319 | 0.2447 | 0.1037 | 0.039* | |
| C11B | 0.41892 (11) | 0.2164 (4) | 0.05488 (11) | 0.0309 (5) | |
| H11C | 0.4249 | 0.0789 | 0.0661 | 0.037* | |
| H11D | 0.4288 | 0.2335 | 0.0099 | 0.037* | |
| C12B | 0.45988 (10) | 0.5554 (4) | 0.09071 (10) | 0.0263 (5) | |
| H12A | 0.4725 | 0.5916 | 0.0479 | 0.032* | |
| H12B | 0.4905 | 0.6249 | 0.1240 | 0.032* | |
| C13B | 0.38411 (10) | 0.6153 (4) | 0.09512 (10) | 0.0285 (5) | |
| H13C | 0.3738 | 0.5974 | 0.1399 | 0.034* | |
| H13D | 0.3783 | 0.7530 | 0.0842 | 0.034* | |
| C14B | 0.33275 (11) | 0.4947 (4) | 0.04849 (10) | 0.0331 (6) | |
| H14A | 0.2854 | 0.5296 | 0.0550 | 0.040* | |
| H14B | 0.3390 | 0.5252 | 0.0033 | 0.040* | |
| C15B | 0.83547 (11) | 0.5452 (4) | 0.39357 (12) | 0.0354 (6) | |
| H15D | 0.8248 | 0.4525 | 0.4262 | 0.053* | |
| H15E | 0.8225 | 0.6742 | 0.4061 | 0.053* | |
| H15F | 0.8846 | 0.5421 | 0.3902 | 0.053* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O2A | 0.0205 (6) | 0.0310 (10) | 0.0199 (7) | −0.0014 (6) | −0.0002 (6) | −0.0009 (6) |
| O3A | 0.0241 (7) | 0.0446 (12) | 0.0225 (7) | 0.0018 (7) | 0.0052 (6) | −0.0025 (7) |
| O4A | 0.0222 (7) | 0.0473 (12) | 0.0202 (7) | −0.0007 (7) | 0.0042 (6) | 0.0054 (7) |
| O5A | 0.0221 (7) | 0.0372 (11) | 0.0205 (7) | 0.0017 (6) | 0.0022 (6) | −0.0029 (6) |
| O6A | 0.0209 (6) | 0.0344 (10) | 0.0191 (7) | −0.0023 (6) | 0.0007 (6) | 0.0042 (6) |
| C1A | 0.0231 (9) | 0.0292 (14) | 0.0216 (10) | 0.0022 (9) | 0.0028 (8) | −0.0001 (8) |
| C2A | 0.0232 (9) | 0.0244 (13) | 0.0195 (9) | 0.0016 (8) | −0.0001 (8) | 0.0003 (8) |
| C3A | 0.0216 (9) | 0.0237 (13) | 0.0270 (10) | 0.0008 (8) | 0.0020 (9) | −0.0017 (8) |
| C4A | 0.0215 (9) | 0.0277 (14) | 0.0304 (11) | 0.0005 (9) | 0.0039 (9) | 0.0004 (9) |
| C5A | 0.0250 (9) | 0.0241 (13) | 0.0178 (9) | 0.0024 (8) | 0.0030 (8) | 0.0016 (8) |
| C6A | 0.0215 (9) | 0.0287 (14) | 0.0195 (9) | 0.0008 (8) | 0.0003 (8) | −0.0006 (8) |
| C7A | 0.0201 (9) | 0.0290 (14) | 0.0188 (9) | 0.0032 (8) | 0.0019 (8) | 0.0027 (8) |
| C8A | 0.0223 (9) | 0.0298 (14) | 0.0203 (10) | −0.0012 (9) | −0.0002 (8) | 0.0053 (8) |
| C9A | 0.0202 (9) | 0.0284 (13) | 0.0190 (9) | 0.0002 (8) | 0.0032 (8) | 0.0018 (8) |
| C10A | 0.0243 (10) | 0.0415 (16) | 0.0211 (10) | −0.0078 (9) | −0.0018 (9) | 0.0002 (9) |
| C11A | 0.0231 (9) | 0.0432 (16) | 0.0220 (10) | 0.0022 (9) | 0.0018 (9) | 0.0016 (9) |
| C12A | 0.0279 (10) | 0.0283 (14) | 0.0231 (10) | 0.0012 (9) | 0.0018 (9) | 0.0022 (9) |
| C13A | 0.0242 (9) | 0.0326 (14) | 0.0185 (9) | −0.0029 (9) | 0.0019 (8) | 0.0020 (8) |
| C14A | 0.0258 (10) | 0.0317 (15) | 0.0242 (10) | −0.0063 (9) | −0.0002 (9) | −0.0022 (9) |
| C15A | 0.0273 (10) | 0.0378 (16) | 0.0300 (11) | −0.0017 (10) | −0.0047 (9) | −0.0052 (10) |
| O2B | 0.0213 (7) | 0.0312 (10) | 0.0250 (7) | −0.0001 (6) | 0.0008 (6) | 0.0000 (6) |
| O3B | 0.0247 (7) | 0.0426 (12) | 0.0272 (8) | 0.0018 (7) | 0.0076 (6) | −0.0011 (7) |
| O4B | 0.0226 (7) | 0.0496 (12) | 0.0216 (7) | 0.0011 (7) | 0.0039 (6) | 0.0069 (7) |
| O5B | 0.0224 (7) | 0.0407 (11) | 0.0238 (7) | 0.0009 (7) | 0.0031 (6) | −0.0016 (7) |
| O6B | 0.0214 (7) | 0.0393 (11) | 0.0196 (7) | −0.0029 (6) | 0.0010 (6) | 0.0053 (6) |
| C1B | 0.0222 (10) | 0.0278 (14) | 0.0367 (12) | 0.0006 (9) | 0.0044 (9) | 0.0021 (10) |
| C2B | 0.0236 (9) | 0.0275 (14) | 0.0278 (11) | 0.0017 (9) | 0.0046 (9) | 0.0022 (9) |
| C3B | 0.0220 (9) | 0.0237 (13) | 0.0341 (11) | 0.0007 (8) | 0.0001 (9) | 0.0000 (9) |
| C4B | 0.0247 (10) | 0.0246 (13) | 0.0230 (10) | 0.0015 (8) | 0.0014 (9) | 0.0023 (8) |
| C5B | 0.0237 (9) | 0.0246 (13) | 0.0212 (9) | 0.0041 (8) | 0.0062 (8) | 0.0026 (8) |
| C6B | 0.0231 (9) | 0.0293 (14) | 0.0234 (10) | 0.0007 (9) | 0.0002 (9) | 0.0049 (9) |
| C7B | 0.0210 (9) | 0.0270 (13) | 0.0236 (10) | 0.0017 (8) | 0.0035 (8) | 0.0040 (8) |
| C8B | 0.0220 (9) | 0.0282 (14) | 0.0245 (10) | 0.0011 (8) | 0.0031 (9) | 0.0019 (9) |
| C9B | 0.0215 (9) | 0.0338 (14) | 0.0199 (9) | −0.0026 (9) | 0.0023 (8) | 0.0009 (9) |
| C10B | 0.0266 (10) | 0.0460 (17) | 0.0240 (10) | −0.0102 (10) | −0.0013 (9) | 0.0016 (10) |
| C11B | 0.0289 (10) | 0.0371 (16) | 0.0257 (10) | −0.0044 (10) | −0.0002 (9) | −0.0022 (10) |
| C12B | 0.0265 (10) | 0.0335 (14) | 0.0191 (9) | −0.0038 (9) | 0.0030 (9) | 0.0020 (9) |
| C13B | 0.0293 (10) | 0.0317 (15) | 0.0242 (10) | 0.0022 (9) | 0.0026 (9) | 0.0049 (9) |
| C14B | 0.0256 (10) | 0.0517 (18) | 0.0209 (10) | −0.0012 (10) | −0.0013 (9) | 0.0070 (10) |
| C15B | 0.0294 (11) | 0.0390 (17) | 0.0352 (12) | −0.0029 (10) | −0.0059 (10) | −0.0007 (11) |
Geometric parameters (Å, °)
| O2A—C3A | 1.353 (2) | O2B—C3B | 1.361 (2) |
| O2A—C2A | 1.391 (2) | O2B—C4B | 1.393 (2) |
| O3A—C6A | 1.207 (2) | O3B—C8B | 1.211 (2) |
| O4A—C8A | 1.210 (2) | O4B—C6B | 1.212 (2) |
| O5A—C6A | 1.364 (2) | O5B—C8B | 1.366 (2) |
| O5A—C9A | 1.443 (2) | O5B—C9B | 1.442 (2) |
| O6A—C8A | 1.357 (2) | O6B—C6B | 1.355 (2) |
| O6A—C9A | 1.443 (2) | O6B—C9B | 1.446 (2) |
| C1A—C2A | 1.382 (3) | C1B—C3B | 1.364 (3) |
| C1A—C4A | 1.404 (3) | C1B—C2B | 1.403 (3) |
| C1A—H1AA | 0.9300 | C1B—H1BA | 0.9300 |
| C2A—C5A | 1.412 (3) | C2B—C4B | 1.378 (3) |
| C3A—C4A | 1.371 (3) | C2B—H2BA | 0.9300 |
| C3A—C15A | 1.478 (3) | C3B—C15B | 1.478 (3) |
| C4A—H4AA | 0.9300 | C4B—C5B | 1.412 (3) |
| C5A—C7A | 1.361 (3) | C5B—C7B | 1.368 (3) |
| C5A—H5AA | 0.9300 | C5B—H5BA | 0.9300 |
| C6A—C7A | 1.467 (3) | C6B—C7B | 1.478 (3) |
| C7A—C8A | 1.481 (3) | C7B—C8B | 1.463 (3) |
| C9A—C13A | 1.508 (3) | C9B—C12B | 1.508 (3) |
| C9A—C14A | 1.514 (3) | C9B—C11B | 1.516 (3) |
| C10A—C11A | 1.512 (4) | C10B—C14B | 1.512 (4) |
| C10A—C14A | 1.540 (3) | C10B—C11B | 1.534 (3) |
| C10A—H10A | 0.9700 | C10B—H10C | 0.9700 |
| C10A—H10B | 0.9700 | C10B—H10D | 0.9700 |
| C11A—C12A | 1.534 (3) | C11B—H11C | 0.9700 |
| C11A—H11A | 0.9700 | C11B—H11D | 0.9700 |
| C11A—H11B | 0.9700 | C12B—C13B | 1.533 (3) |
| C12A—C13A | 1.536 (3) | C12B—H12A | 0.9700 |
| C12A—H12C | 0.9700 | C12B—H12B | 0.9700 |
| C12A—H12D | 0.9700 | C13B—C14B | 1.530 (3) |
| C13A—H13A | 0.9700 | C13B—H13C | 0.9700 |
| C13A—H13B | 0.9700 | C13B—H13D | 0.9700 |
| C14A—H14C | 0.9700 | C14B—H14A | 0.9700 |
| C14A—H14D | 0.9700 | C14B—H14B | 0.9700 |
| C15A—H15A | 0.9600 | C15B—H15D | 0.9600 |
| C15A—H15B | 0.9600 | C15B—H15E | 0.9600 |
| C15A—H15C | 0.9600 | C15B—H15F | 0.9600 |
| C3A—O2A—C2A | 107.65 (15) | C3B—O2B—C4B | 107.26 (16) |
| C6A—O5A—C9A | 118.84 (15) | C8B—O5B—C9B | 119.22 (16) |
| C8A—O6A—C9A | 118.53 (15) | C6B—O6B—C9B | 118.84 (15) |
| C2A—C1A—C4A | 107.20 (18) | C3B—C1B—C2B | 107.22 (18) |
| C2A—C1A—H1AA | 126.4 | C3B—C1B—H1BA | 126.4 |
| C4A—C1A—H1AA | 126.4 | C2B—C1B—H1BA | 126.4 |
| C1A—C2A—O2A | 108.14 (16) | C4B—C2B—C1B | 107.33 (19) |
| C1A—C2A—C5A | 138.96 (19) | C4B—C2B—H2BA | 126.3 |
| O2A—C2A—C5A | 112.90 (16) | C1B—C2B—H2BA | 126.3 |
| O2A—C3A—C4A | 109.96 (18) | O2B—C3B—C1B | 109.97 (19) |
| O2A—C3A—C15A | 117.54 (17) | O2B—C3B—C15B | 116.81 (18) |
| C4A—C3A—C15A | 132.49 (18) | C1B—C3B—C15B | 133.19 (19) |
| C3A—C4A—C1A | 107.04 (18) | C2B—C4B—O2B | 108.21 (17) |
| C3A—C4A—H4AA | 126.5 | C2B—C4B—C5B | 139.2 (2) |
| C1A—C4A—H4AA | 126.5 | O2B—C4B—C5B | 112.55 (17) |
| C7A—C5A—C2A | 134.69 (18) | C7B—C5B—C4B | 134.40 (18) |
| C7A—C5A—H5AA | 112.7 | C7B—C5B—H5BA | 112.8 |
| C2A—C5A—H5AA | 112.7 | C4B—C5B—H5BA | 112.8 |
| O3A—C6A—O5A | 117.86 (18) | O4B—C6B—O6B | 117.93 (17) |
| O3A—C6A—C7A | 125.73 (18) | O4B—C6B—C7B | 125.12 (19) |
| O5A—C6A—C7A | 116.36 (16) | O6B—C6B—C7B | 116.91 (17) |
| C5A—C7A—C6A | 124.74 (17) | C5B—C7B—C8B | 124.97 (17) |
| C5A—C7A—C8A | 116.08 (18) | C5B—C7B—C6B | 115.77 (18) |
| C6A—C7A—C8A | 119.01 (18) | C8B—C7B—C6B | 119.03 (18) |
| O4A—C8A—O6A | 118.46 (17) | O3B—C8B—O5B | 117.70 (18) |
| O4A—C8A—C7A | 124.77 (19) | O3B—C8B—C7B | 125.61 (19) |
| O6A—C8A—C7A | 116.72 (17) | O5B—C8B—C7B | 116.63 (16) |
| O5A—C9A—O6A | 109.72 (16) | O5B—C9B—O6B | 110.01 (16) |
| O5A—C9A—C13A | 111.06 (16) | O5B—C9B—C12B | 111.21 (17) |
| O6A—C9A—C13A | 110.32 (17) | O6B—C9B—C12B | 110.41 (17) |
| O5A—C9A—C14A | 106.51 (17) | O5B—C9B—C11B | 106.25 (17) |
| O6A—C9A—C14A | 106.11 (16) | O6B—C9B—C11B | 105.43 (17) |
| C13A—C9A—C14A | 112.94 (18) | C12B—C9B—C11B | 113.30 (18) |
| C11A—C10A—C14A | 111.55 (18) | C14B—C10B—C11B | 111.61 (19) |
| C11A—C10A—H10A | 109.3 | C14B—C10B—H10C | 109.3 |
| C14A—C10A—H10A | 109.3 | C11B—C10B—H10C | 109.3 |
| C11A—C10A—H10B | 109.3 | C14B—C10B—H10D | 109.3 |
| C14A—C10A—H10B | 109.3 | C11B—C10B—H10D | 109.3 |
| H10A—C10A—H10B | 108.0 | H10C—C10B—H10D | 108.0 |
| C10A—C11A—C12A | 111.68 (18) | C9B—C11B—C10B | 110.70 (19) |
| C10A—C11A—H11A | 109.3 | C9B—C11B—H11C | 109.5 |
| C12A—C11A—H11A | 109.3 | C10B—C11B—H11C | 109.5 |
| C10A—C11A—H11B | 109.3 | C9B—C11B—H11D | 109.5 |
| C12A—C11A—H11B | 109.3 | C10B—C11B—H11D | 109.5 |
| H11A—C11A—H11B | 107.9 | H11C—C11B—H11D | 108.1 |
| C11A—C12A—C13A | 111.25 (18) | C9B—C12B—C13B | 111.01 (18) |
| C11A—C12A—H12C | 109.4 | C9B—C12B—H12A | 109.4 |
| C13A—C12A—H12C | 109.4 | C13B—C12B—H12A | 109.4 |
| C11A—C12A—H12D | 109.4 | C9B—C12B—H12B | 109.4 |
| C13A—C12A—H12D | 109.4 | C13B—C12B—H12B | 109.4 |
| H12C—C12A—H12D | 108.0 | H12A—C12B—H12B | 108.0 |
| C9A—C13A—C12A | 110.95 (17) | C14B—C13B—C12B | 111.74 (19) |
| C9A—C13A—H13A | 109.4 | C14B—C13B—H13C | 109.3 |
| C12A—C13A—H13A | 109.4 | C12B—C13B—H13C | 109.3 |
| C9A—C13A—H13B | 109.4 | C14B—C13B—H13D | 109.3 |
| C12A—C13A—H13B | 109.4 | C12B—C13B—H13D | 109.3 |
| H13A—C13A—H13B | 108.0 | H13C—C13B—H13D | 107.9 |
| C9A—C14A—C10A | 110.81 (18) | C10B—C14B—C13B | 111.28 (18) |
| C9A—C14A—H14C | 109.5 | C10B—C14B—H14A | 109.4 |
| C10A—C14A—H14C | 109.5 | C13B—C14B—H14A | 109.4 |
| C9A—C14A—H14D | 109.5 | C10B—C14B—H14B | 109.4 |
| C10A—C14A—H14D | 109.5 | C13B—C14B—H14B | 109.4 |
| H14C—C14A—H14D | 108.1 | H14A—C14B—H14B | 108.0 |
| C3A—C15A—H15A | 109.5 | C3B—C15B—H15D | 109.5 |
| C3A—C15A—H15B | 109.5 | C3B—C15B—H15E | 109.5 |
| H15A—C15A—H15B | 109.5 | H15D—C15B—H15E | 109.5 |
| C3A—C15A—H15C | 109.5 | C3B—C15B—H15F | 109.5 |
| H15A—C15A—H15C | 109.5 | H15D—C15B—H15F | 109.5 |
| H15B—C15A—H15C | 109.5 | H15E—C15B—H15F | 109.5 |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C1A—H1AA···O3A | 0.93 | 2.26 | 2.878 (3) | 123 |
| C4A—H4AA···O4Bi | 0.93 | 2.59 | 3.408 (3) | 147 |
| C1B—H1BA···O4Aii | 0.93 | 2.50 | 3.376 (3) | 157 |
| C2B—H2BA···O3B | 0.93 | 2.26 | 2.884 (3) | 124 |
| C12B—H12B···O4Bi | 0.97 | 2.53 | 3.420 (3) | 152 |
| C13A—H13A···O4Aii | 0.97 | 2.54 | 3.405 (3) | 149 |
Symmetry codes: (i) −x+1, y+1/2, −z+1/2; (ii) −x+2, y−1/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: LH2861).
References
- Bruker (1997). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Lian, Y., Guo, J. J., Liu, X. M. & Wei, R. B. (2008). Chem. Res. Chin. Univ.24, 441–444.
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Wei, R. B., Liu, B., Guo, J. J., Liu, Y. & Zhang, D. W. (2008). Chin. J. Org. Chem 28, 1501–1514.
- Yaozhong, J., Song, X., Zhi, J., Jingen, D., Aiqiao, M. & Chan, A. S. C. (1998). Tetrahedron Assymetry, 9, 3185–3189.
- Zeng, W.-L. & Jian, F. (2009). Acta Cryst. E65, o1875. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S160053680902947X/lh2861sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680902947X/lh2861Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report