Abstract
In the title compound, [Cu(C18H15P)2(H2O)]BF4, the CuI atom is coordinated by two P atoms from triphenylphosphine ligands and one water molecule in a distorted trigonal geometry. In the BF4 − anion, three F atoms are disordered over two sites around the B—F bond, the site-occupancy ratio being 0.67 (6):0.33 (6). The Cu⋯F distance of 2.602 (5) Å between the Cu atom and the ordered F atom may suggest a weak but genuine interaction. O—H⋯F and weak C—H⋯F hydrogen bonding is present in the crystal structure.
Related literature
For the applications of CuI complexes, see: Kirchhoff et al. (1985 ▶); Zhang et al. (2004 ▶); Moudam et al. (2007 ▶). For the tetrahedral coordination geometry of CuI complexes, see: Engelhardt et al. (1985 ▶); Barron et al. (1987 ▶). For the weak Cu⋯F interaction, see: Mao et al. (2003 ▶); Fu et al. (2004 ▶). For Cu—P and Cu—O bond distances, see: Meng et al. (2006 ▶).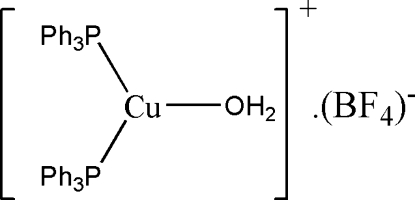
Experimental
Crystal data
[Cu(C18H15P)2(H2O)]BF4
M r = 692.91
Monoclinic,
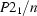
a = 13.9737 (14) Å
b = 12.4258 (11) Å
c = 19.4276 (18) Å
β = 94.521 (1)°
V = 3362.8 (5) Å3
Z = 4
Mo Kα radiation
μ = 0.79 mm−1
T = 298 K
0.48 × 0.19 × 0.16 mm
Data collection
Bruker SMART APEXII area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.702, T max = 0.883
17192 measured reflections
5914 independent reflections
3008 reflections with I > 2σ(I)
R int = 0.078
Refinement
R[F 2 > 2σ(F 2)] = 0.061
wR(F 2) = 0.202
S = 1.04
5914 reflections
434 parameters
1 restraint
H-atom parameters constrained
Δρmax = 0.93 e Å−3
Δρmin = −0.36 e Å−3
Data collection: APEX2 (Bruker, 2008 ▶); cell refinement: SAINT (Bruker, 2008 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809029559/xu2555sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809029559/xu2555Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Selected bond lengths (Å).
| Cu1—O1 | 2.105 (5) |
| Cu1—P1 | 2.2318 (18) |
| Cu1—P2 | 2.2478 (18) |
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1C⋯F2 | 0.85 | 1.87 | 2.71 (3) | 171 |
| O1—H1D⋯F3i | 0.85 | 1.98 | 2.82 (3) | 171 |
| C28—H28⋯F4ii | 0.93 | 2.51 | 3.25 (3) | 137 |
Symmetry codes: (i)  ; (ii)
; (ii) 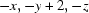 .
.
supplementary crystallographic information
Comment
Copper(I) complexes with phosphine ligand have attracted much attention because of their rich photophysical properties and potential applications in organic light-emitting diodes (OLEDs) (Kirchhoff et al., 1985; Zhang et al., 2004; Moudam et al., 2007). These complexes usually adopt tetrahedron coordination geometry (Engelhardt et al., 1985; Barron et al., 1987), three-coordinated copper(I) complexes with phosphine ligands is relatively little known. We reported here the title three-coordinated copper(I) complex.
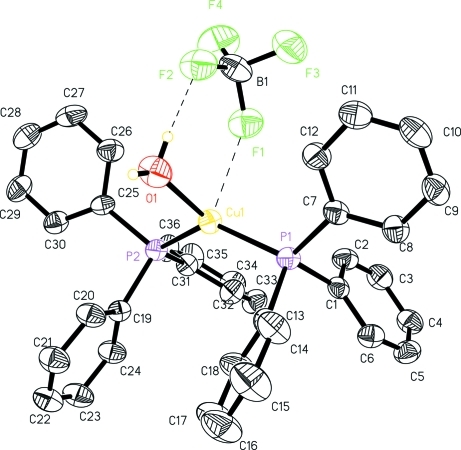
The molecular structure is depicted in Fig. 1. The copper(I) atom is three-coordinated in distorted trigonal geometry (Table 1) by two P atoms from two triphenylphosphine ligands and one water molecule. The Cu1—P and Cu1—O bond distances are comparable to those found in related complexes (Engelhardt et al., 1985; Barron et al., 1987; Meng et al., 2006). The coordination angles around the Cu1 atom are ranging from 104.80 (16)° to 133.89 (7)°. In the BF4 anion three F atoms are disordered over two sites around the B1—F1 bond. The Cu1···F1 distance of 2.602 (5) Å between the Cu1 atom and the ordered F1 atom may suggests a weak but genuine interaction, similar to the situation found in the related structures (Fu et al., 2004); Mao et al., 2003).
The O—H···F and weak C—H···F hydrogen bonding is present in the crystal structure (Table 2).
Experimental
[Cu(CH3CN)4]BF4 (0.031 g, 0.1 mmol) was added to a solution of triphenylphosphine (0.052 g, 0.2 mmol) in 30 ml dichloromethane with small amount of water under nitrogen atmosphere. The mixture was stirred at room temperature for 2 h to obtain the yellow solution. Crystallization by slow diffusion of diethyl ether into the dichloromethane solution yielded yellow crystals suitable for X-ray diffraction (yield: 47%). Analysis calculated for [Cu(H2O)(C18H15P)2].(BF4): C 62.40, H 4.66%; Found: C 62.08, H 4.93%.
Refinement
All H atoms were positioned geometrically and treated as riding (O—H = 0.65 Å and C—H = 0.93 Å, and refined in riding mode with Uiso(H) = 1.2Ueq(C,O). The F2, F3 and F4 atoms are disordered over two sites, site occupancy factors were refined to 0.67 (6) and 0.33 (6).
Figures
Fig. 1.
The molecular structure of the title compound. Displacement ellipsoids are drawn at the 30% probability level. The H atoms in benzene rings and the minor disorder component of the F2—F4 are omitted for clarity. The Cu···F weak interaction and O—H···F hydrogen bond are indicated by dashed lines.
Crystal data
| [Cu(C18H15P)2(H2O)]BF4 | F(000) = 1424 |
| Mr = 692.91 | Dx = 1.369 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 2641 reflections |
| a = 13.9737 (14) Å | θ = 2.2–21.6° |
| b = 12.4258 (11) Å | µ = 0.79 mm−1 |
| c = 19.4276 (18) Å | T = 298 K |
| β = 94.521 (1)° | Block, yellow |
| V = 3362.8 (5) Å3 | 0.48 × 0.19 × 0.16 mm |
| Z = 4 |
Data collection
| Bruker SMART APEXII area-detector diffractometer | 5914 independent reflections |
| Radiation source: fine-focus sealed tube | 3008 reflections with I > 2σ(I) |
| graphite | Rint = 0.078 |
| φ and ω scans | θmax = 25.0°, θmin = 1.7° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −16→15 |
| Tmin = 0.702, Tmax = 0.883 | k = −14→14 |
| 17192 measured reflections | l = −23→22 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.061 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.202 | H-atom parameters constrained |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0646P)2 + 7.3064P] where P = (Fo2 + 2Fc2)/3 |
| 5914 reflections | (Δ/σ)max = 0.001 |
| 434 parameters | Δρmax = 0.93 e Å−3 |
| 1 restraint | Δρmin = −0.36 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| Cu1 | 0.36105 (6) | 0.80395 (7) | 0.13634 (4) | 0.0517 (3) | |
| O1 | 0.2503 (4) | 0.7985 (4) | 0.2037 (2) | 0.0875 (17) | |
| H1C | 0.2344 | 0.8634 | 0.2107 | 0.105* | |
| H1D | 0.2314 | 0.7597 | 0.2361 | 0.105* | |
| P1 | 0.50924 (12) | 0.81613 (14) | 0.18770 (8) | 0.0460 (4) | |
| P2 | 0.29834 (12) | 0.72605 (14) | 0.03854 (8) | 0.0464 (4) | |
| C1 | 0.6043 (4) | 0.8457 (5) | 0.1314 (3) | 0.0463 (15) | |
| C2 | 0.5854 (5) | 0.9201 (6) | 0.0780 (3) | 0.0606 (19) | |
| H2 | 0.5247 | 0.9510 | 0.0715 | 0.073* | |
| C3 | 0.6543 (6) | 0.9483 (6) | 0.0352 (4) | 0.069 (2) | |
| H3 | 0.6399 | 0.9972 | −0.0003 | 0.082* | |
| C4 | 0.7436 (6) | 0.9051 (6) | 0.0442 (4) | 0.067 (2) | |
| H4 | 0.7902 | 0.9242 | 0.0149 | 0.081* | |
| C5 | 0.7649 (5) | 0.8330 (6) | 0.0971 (4) | 0.068 (2) | |
| H5 | 0.8265 | 0.8044 | 0.1038 | 0.082* | |
| C6 | 0.6956 (5) | 0.8025 (6) | 0.1404 (3) | 0.0583 (18) | |
| H6 | 0.7105 | 0.7530 | 0.1756 | 0.070* | |
| C7 | 0.5344 (5) | 0.9112 (5) | 0.2578 (3) | 0.0512 (17) | |
| C8 | 0.6274 (5) | 0.9386 (6) | 0.2829 (3) | 0.0610 (19) | |
| H8 | 0.6793 | 0.9041 | 0.2652 | 0.073* | |
| C9 | 0.6440 (6) | 1.0154 (6) | 0.3332 (4) | 0.070 (2) | |
| H9 | 0.7066 | 1.0337 | 0.3487 | 0.084* | |
| C10 | 0.5692 (7) | 1.0643 (7) | 0.3602 (4) | 0.077 (2) | |
| H10 | 0.5807 | 1.1151 | 0.3950 | 0.092* | |
| C11 | 0.4772 (6) | 1.0403 (7) | 0.3370 (4) | 0.083 (3) | |
| H11 | 0.4262 | 1.0753 | 0.3554 | 0.100* | |
| C12 | 0.4599 (5) | 0.9630 (6) | 0.2857 (3) | 0.067 (2) | |
| H12 | 0.3970 | 0.9462 | 0.2702 | 0.080* | |
| C13 | 0.5397 (5) | 0.6851 (5) | 0.2224 (3) | 0.0527 (17) | |
| C14 | 0.5584 (6) | 0.6645 (6) | 0.2916 (4) | 0.077 (2) | |
| H14 | 0.5589 | 0.7206 | 0.3234 | 0.092* | |
| C15 | 0.5764 (7) | 0.5595 (8) | 0.3144 (5) | 0.105 (3) | |
| H15 | 0.5880 | 0.5459 | 0.3614 | 0.126* | |
| C16 | 0.5774 (8) | 0.4772 (8) | 0.2688 (5) | 0.106 (3) | |
| H16 | 0.5910 | 0.4078 | 0.2845 | 0.127* | |
| C17 | 0.5585 (7) | 0.4957 (7) | 0.2005 (5) | 0.094 (3) | |
| H17 | 0.5586 | 0.4392 | 0.1691 | 0.113* | |
| C18 | 0.5394 (6) | 0.5986 (6) | 0.1781 (4) | 0.075 (2) | |
| H18 | 0.5257 | 0.6105 | 0.1311 | 0.090* | |
| C19 | 0.3094 (4) | 0.5817 (5) | 0.0445 (3) | 0.0479 (16) | |
| C20 | 0.2846 (5) | 0.5319 (6) | 0.1046 (4) | 0.067 (2) | |
| H20 | 0.2710 | 0.5738 | 0.1422 | 0.080* | |
| C21 | 0.2797 (6) | 0.4224 (7) | 0.1093 (4) | 0.077 (2) | |
| H21 | 0.2616 | 0.3908 | 0.1496 | 0.092* | |
| C22 | 0.3010 (6) | 0.3593 (7) | 0.0560 (4) | 0.077 (2) | |
| H22 | 0.2961 | 0.2849 | 0.0593 | 0.092* | |
| C23 | 0.3296 (6) | 0.4054 (7) | −0.0029 (4) | 0.081 (2) | |
| H23 | 0.3467 | 0.3624 | −0.0392 | 0.097* | |
| C24 | 0.3332 (5) | 0.5159 (6) | −0.0084 (4) | 0.066 (2) | |
| H24 | 0.3521 | 0.5466 | −0.0488 | 0.080* | |
| C25 | 0.1693 (5) | 0.7425 (6) | 0.0208 (3) | 0.0487 (16) | |
| C26 | 0.1270 (5) | 0.8370 (6) | 0.0406 (4) | 0.070 (2) | |
| H26 | 0.1638 | 0.8891 | 0.0645 | 0.084* | |
| C27 | 0.0280 (6) | 0.8544 (8) | 0.0245 (4) | 0.083 (3) | |
| H27 | −0.0007 | 0.9177 | 0.0380 | 0.099* | |
| C28 | −0.0256 (6) | 0.7771 (8) | −0.0113 (4) | 0.079 (2) | |
| H28 | −0.0904 | 0.7894 | −0.0235 | 0.095* | |
| C29 | 0.0150 (5) | 0.6832 (7) | −0.0291 (4) | 0.073 (2) | |
| H29 | −0.0223 | 0.6302 | −0.0520 | 0.088* | |
| C30 | 0.1115 (5) | 0.6663 (6) | −0.0133 (3) | 0.0610 (19) | |
| H30 | 0.1386 | 0.6016 | −0.0259 | 0.073* | |
| C31 | 0.3468 (5) | 0.7605 (5) | −0.0425 (3) | 0.0512 (17) | |
| C32 | 0.4439 (5) | 0.7700 (6) | −0.0454 (4) | 0.065 (2) | |
| H32 | 0.4840 | 0.7570 | −0.0057 | 0.078* | |
| C33 | 0.4846 (6) | 0.7984 (6) | −0.1059 (4) | 0.074 (2) | |
| H33 | 0.5509 | 0.8030 | −0.1069 | 0.089* | |
| C34 | 0.4255 (6) | 0.8195 (7) | −0.1639 (4) | 0.076 (2) | |
| H34 | 0.4519 | 0.8389 | −0.2046 | 0.091* | |
| C35 | 0.3292 (6) | 0.8125 (7) | −0.1625 (4) | 0.080 (2) | |
| H35 | 0.2896 | 0.8278 | −0.2021 | 0.096* | |
| C36 | 0.2894 (5) | 0.7826 (6) | −0.1026 (3) | 0.067 (2) | |
| H36 | 0.2231 | 0.7771 | −0.1023 | 0.080* | |
| B1 | 0.2480 (10) | 1.0703 (10) | 0.1638 (6) | 0.086 (3) | |
| F1 | 0.3118 (4) | 1.0059 (4) | 0.1338 (2) | 0.0998 (15) | |
| F2 | 0.206 (2) | 1.010 (2) | 0.2129 (18) | 0.108 (7) | 0.67 (6) |
| F3 | 0.3076 (17) | 1.150 (2) | 0.1979 (18) | 0.134 (9) | 0.67 (6) |
| F4 | 0.188 (2) | 1.115 (3) | 0.1190 (10) | 0.134 (10) | 0.67 (6) |
| F2' | 0.258 (5) | 1.174 (2) | 0.162 (3) | 0.138 (17) | 0.33 (6) |
| F3' | 0.155 (3) | 1.054 (5) | 0.123 (3) | 0.125 (15) | 0.33 (6) |
| F4' | 0.232 (5) | 1.050 (6) | 0.228 (2) | 0.110 (15) | 0.33 (6) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cu1 | 0.0530 (5) | 0.0605 (6) | 0.0401 (5) | 0.0008 (4) | −0.0054 (3) | −0.0045 (4) |
| O1 | 0.121 (4) | 0.076 (4) | 0.069 (3) | −0.003 (3) | 0.034 (3) | 0.015 (3) |
| P1 | 0.0515 (10) | 0.0508 (11) | 0.0342 (9) | 0.0055 (9) | −0.0063 (7) | −0.0053 (8) |
| P2 | 0.0489 (10) | 0.0510 (11) | 0.0378 (9) | 0.0044 (8) | −0.0063 (7) | −0.0019 (8) |
| C1 | 0.053 (4) | 0.044 (4) | 0.040 (4) | −0.001 (3) | −0.005 (3) | −0.009 (3) |
| C2 | 0.068 (5) | 0.055 (5) | 0.058 (4) | 0.013 (4) | −0.003 (4) | 0.000 (4) |
| C3 | 0.085 (6) | 0.058 (5) | 0.062 (5) | −0.002 (5) | 0.004 (4) | 0.008 (4) |
| C4 | 0.082 (6) | 0.059 (5) | 0.063 (5) | −0.002 (4) | 0.016 (4) | −0.004 (4) |
| C5 | 0.062 (5) | 0.075 (6) | 0.069 (5) | 0.012 (4) | 0.009 (4) | −0.012 (4) |
| C6 | 0.066 (5) | 0.059 (5) | 0.050 (4) | 0.010 (4) | 0.000 (4) | 0.003 (3) |
| C7 | 0.063 (4) | 0.052 (4) | 0.037 (4) | 0.004 (4) | −0.005 (3) | −0.007 (3) |
| C8 | 0.067 (5) | 0.067 (5) | 0.048 (4) | 0.001 (4) | −0.008 (3) | −0.011 (4) |
| C9 | 0.080 (5) | 0.072 (6) | 0.056 (5) | −0.007 (5) | −0.010 (4) | −0.012 (4) |
| C10 | 0.110 (7) | 0.069 (6) | 0.050 (5) | −0.006 (5) | 0.002 (5) | −0.019 (4) |
| C11 | 0.089 (6) | 0.089 (7) | 0.074 (6) | 0.004 (5) | 0.022 (5) | −0.032 (5) |
| C12 | 0.070 (5) | 0.075 (5) | 0.055 (4) | 0.002 (4) | 0.002 (4) | −0.020 (4) |
| C13 | 0.059 (4) | 0.055 (5) | 0.042 (4) | −0.001 (4) | −0.006 (3) | −0.002 (3) |
| C14 | 0.108 (6) | 0.060 (5) | 0.058 (5) | −0.002 (5) | −0.016 (4) | 0.003 (4) |
| C15 | 0.161 (10) | 0.080 (7) | 0.068 (6) | 0.000 (7) | −0.028 (6) | 0.024 (5) |
| C16 | 0.153 (9) | 0.062 (6) | 0.097 (8) | 0.014 (6) | −0.023 (7) | 0.016 (6) |
| C17 | 0.136 (8) | 0.060 (6) | 0.085 (7) | 0.013 (6) | −0.007 (6) | −0.004 (5) |
| C18 | 0.105 (6) | 0.059 (5) | 0.059 (5) | 0.016 (5) | −0.004 (4) | 0.002 (4) |
| C19 | 0.050 (4) | 0.053 (4) | 0.040 (4) | 0.006 (3) | −0.001 (3) | 0.000 (3) |
| C20 | 0.088 (6) | 0.057 (5) | 0.057 (5) | 0.009 (4) | 0.014 (4) | 0.000 (4) |
| C21 | 0.099 (6) | 0.064 (6) | 0.069 (5) | 0.007 (5) | 0.016 (5) | 0.011 (4) |
| C22 | 0.096 (6) | 0.054 (5) | 0.080 (6) | 0.010 (5) | 0.002 (5) | 0.004 (5) |
| C23 | 0.107 (7) | 0.062 (6) | 0.073 (6) | 0.014 (5) | 0.013 (5) | −0.010 (4) |
| C24 | 0.084 (5) | 0.060 (5) | 0.056 (5) | 0.011 (4) | 0.009 (4) | 0.001 (4) |
| C25 | 0.052 (4) | 0.054 (4) | 0.040 (4) | 0.013 (3) | 0.002 (3) | 0.004 (3) |
| C26 | 0.070 (5) | 0.066 (5) | 0.072 (5) | 0.013 (4) | −0.001 (4) | 0.000 (4) |
| C27 | 0.077 (6) | 0.082 (6) | 0.091 (6) | 0.035 (5) | 0.014 (5) | 0.012 (5) |
| C28 | 0.060 (5) | 0.106 (8) | 0.071 (6) | 0.017 (5) | −0.004 (4) | 0.009 (5) |
| C29 | 0.052 (4) | 0.102 (7) | 0.063 (5) | 0.002 (5) | −0.007 (4) | −0.007 (5) |
| C30 | 0.054 (4) | 0.074 (5) | 0.053 (4) | 0.009 (4) | −0.006 (3) | −0.007 (4) |
| C31 | 0.057 (4) | 0.054 (4) | 0.042 (4) | 0.000 (3) | −0.002 (3) | 0.002 (3) |
| C32 | 0.063 (5) | 0.075 (5) | 0.056 (5) | 0.003 (4) | 0.002 (4) | 0.009 (4) |
| C33 | 0.067 (5) | 0.081 (6) | 0.075 (5) | 0.004 (4) | 0.019 (4) | 0.013 (5) |
| C34 | 0.091 (6) | 0.081 (6) | 0.058 (5) | −0.002 (5) | 0.018 (5) | 0.008 (4) |
| C35 | 0.088 (6) | 0.099 (7) | 0.051 (5) | −0.011 (5) | −0.005 (4) | 0.018 (4) |
| C36 | 0.063 (5) | 0.086 (6) | 0.050 (4) | −0.009 (4) | −0.001 (4) | 0.012 (4) |
| B1 | 0.123 (10) | 0.058 (8) | 0.075 (8) | 0.007 (8) | 0.003 (8) | 0.002 (6) |
| F1 | 0.111 (4) | 0.107 (4) | 0.085 (3) | 0.013 (3) | 0.026 (3) | 0.007 (3) |
| F2 | 0.131 (14) | 0.094 (13) | 0.108 (16) | 0.015 (9) | 0.058 (12) | 0.023 (9) |
| F3 | 0.134 (12) | 0.113 (11) | 0.151 (18) | −0.001 (10) | −0.009 (11) | −0.046 (11) |
| F4 | 0.139 (14) | 0.14 (2) | 0.119 (9) | 0.048 (15) | −0.014 (9) | 0.047 (13) |
| F2' | 0.18 (4) | 0.097 (19) | 0.14 (3) | −0.014 (19) | −0.01 (3) | 0.015 (17) |
| F3' | 0.14 (2) | 0.10 (3) | 0.13 (2) | 0.02 (2) | −0.016 (18) | −0.024 (19) |
| F4' | 0.15 (3) | 0.12 (4) | 0.060 (15) | 0.02 (3) | 0.013 (16) | 0.013 (18) |
Geometric parameters (Å, °)
| Cu1—O1 | 2.105 (5) | C17—H17 | 0.9300 |
| Cu1—P1 | 2.2318 (18) | C18—H18 | 0.9300 |
| Cu1—P2 | 2.2478 (18) | C19—C24 | 1.375 (9) |
| O1—H1C | 0.8500 | C19—C20 | 1.389 (9) |
| O1—H1D | 0.8500 | C20—C21 | 1.366 (10) |
| P1—C13 | 1.800 (7) | C20—H20 | 0.9300 |
| P1—C7 | 1.816 (6) | C21—C22 | 1.349 (10) |
| P1—C1 | 1.823 (6) | C21—H21 | 0.9300 |
| P2—C19 | 1.803 (7) | C22—C23 | 1.368 (10) |
| P2—C31 | 1.814 (6) | C22—H22 | 0.9300 |
| P2—C25 | 1.820 (6) | C23—C24 | 1.377 (10) |
| C1—C6 | 1.383 (8) | C23—H23 | 0.9300 |
| C1—C2 | 1.400 (9) | C24—H24 | 0.9300 |
| C2—C3 | 1.366 (9) | C25—C30 | 1.380 (9) |
| C2—H2 | 0.9300 | C25—C26 | 1.382 (9) |
| C3—C4 | 1.357 (10) | C26—C27 | 1.410 (10) |
| C3—H3 | 0.9300 | C26—H26 | 0.9300 |
| C4—C5 | 1.378 (10) | C27—C28 | 1.373 (11) |
| C4—H4 | 0.9300 | C27—H27 | 0.9300 |
| C5—C6 | 1.385 (9) | C28—C29 | 1.355 (11) |
| C5—H5 | 0.9300 | C28—H28 | 0.9300 |
| C6—H6 | 0.9300 | C29—C30 | 1.375 (9) |
| C7—C12 | 1.373 (9) | C29—H29 | 0.9300 |
| C7—C8 | 1.394 (9) | C30—H30 | 0.9300 |
| C8—C9 | 1.374 (9) | C31—C32 | 1.367 (9) |
| C8—H8 | 0.9300 | C31—C36 | 1.390 (9) |
| C9—C10 | 1.350 (10) | C32—C33 | 1.391 (9) |
| C9—H9 | 0.9300 | C32—H32 | 0.9300 |
| C10—C11 | 1.362 (10) | C33—C34 | 1.369 (10) |
| C10—H10 | 0.9300 | C33—H33 | 0.9300 |
| C11—C12 | 1.391 (10) | C34—C35 | 1.351 (10) |
| C11—H11 | 0.9300 | C34—H34 | 0.9300 |
| C12—H12 | 0.9300 | C35—C36 | 1.381 (9) |
| C13—C14 | 1.374 (9) | C35—H35 | 0.9300 |
| C13—C18 | 1.377 (9) | C36—H36 | 0.9300 |
| C14—C15 | 1.393 (11) | B1—F4 | 1.285 (19) |
| C14—H14 | 0.9300 | B1—F2' | 1.30 (3) |
| C15—C16 | 1.354 (12) | B1—F4' | 1.31 (5) |
| C15—H15 | 0.9300 | B1—F1 | 1.363 (12) |
| C16—C17 | 1.352 (11) | B1—F2 | 1.38 (3) |
| C16—H16 | 0.9300 | B1—F3 | 1.42 (2) |
| C17—C18 | 1.370 (10) | B1—F3' | 1.48 (4) |
| O1—Cu1—P1 | 115.18 (16) | C17—C18—C13 | 122.6 (7) |
| O1—Cu1—P2 | 104.80 (16) | C17—C18—H18 | 118.7 |
| P1—Cu1—P2 | 133.89 (7) | C13—C18—H18 | 118.7 |
| Cu1—O1—H1C | 106.5 | C24—C19—C20 | 117.0 (7) |
| Cu1—O1—H1D | 139.8 | C24—C19—P2 | 124.6 (5) |
| H1C—O1—H1D | 108.6 | C20—C19—P2 | 118.1 (5) |
| C13—P1—C7 | 106.4 (3) | C21—C20—C19 | 121.1 (7) |
| C13—P1—C1 | 104.2 (3) | C21—C20—H20 | 119.4 |
| C7—P1—C1 | 102.2 (3) | C19—C20—H20 | 119.4 |
| C13—P1—Cu1 | 106.8 (2) | C22—C21—C20 | 120.8 (8) |
| C7—P1—Cu1 | 119.8 (2) | C22—C21—H21 | 119.6 |
| C1—P1—Cu1 | 116.1 (2) | C20—C21—H21 | 119.6 |
| C19—P2—C31 | 104.8 (3) | C21—C22—C23 | 119.7 (8) |
| C19—P2—C25 | 101.7 (3) | C21—C22—H22 | 120.2 |
| C31—P2—C25 | 103.9 (3) | C23—C22—H22 | 120.2 |
| C19—P2—Cu1 | 110.4 (2) | C22—C23—C24 | 119.8 (8) |
| C31—P2—Cu1 | 119.0 (2) | C22—C23—H23 | 120.1 |
| C25—P2—Cu1 | 115.2 (2) | C24—C23—H23 | 120.1 |
| C6—C1—C2 | 118.0 (6) | C19—C24—C23 | 121.5 (7) |
| C6—C1—P1 | 123.7 (5) | C19—C24—H24 | 119.2 |
| C2—C1—P1 | 118.3 (5) | C23—C24—H24 | 119.2 |
| C3—C2—C1 | 121.3 (7) | C30—C25—C26 | 118.0 (6) |
| C3—C2—H2 | 119.4 | C30—C25—P2 | 123.2 (5) |
| C1—C2—H2 | 119.4 | C26—C25—P2 | 118.8 (6) |
| C4—C3—C2 | 120.3 (7) | C25—C26—C27 | 120.2 (8) |
| C4—C3—H3 | 119.8 | C25—C26—H26 | 119.9 |
| C2—C3—H3 | 119.8 | C27—C26—H26 | 119.9 |
| C3—C4—C5 | 119.8 (7) | C28—C27—C26 | 119.4 (8) |
| C3—C4—H4 | 120.1 | C28—C27—H27 | 120.3 |
| C5—C4—H4 | 120.1 | C26—C27—H27 | 120.3 |
| C4—C5—C6 | 120.7 (7) | C29—C28—C27 | 120.6 (8) |
| C4—C5—H5 | 119.7 | C29—C28—H28 | 119.7 |
| C6—C5—H5 | 119.7 | C27—C28—H28 | 119.7 |
| C1—C6—C5 | 119.9 (6) | C28—C29—C30 | 119.8 (8) |
| C1—C6—H6 | 120.0 | C28—C29—H29 | 120.1 |
| C5—C6—H6 | 120.0 | C30—C29—H29 | 120.1 |
| C12—C7—C8 | 117.6 (6) | C29—C30—C25 | 121.9 (7) |
| C12—C7—P1 | 119.6 (5) | C29—C30—H30 | 119.0 |
| C8—C7—P1 | 122.8 (5) | C25—C30—H30 | 119.0 |
| C9—C8—C7 | 121.3 (7) | C32—C31—C36 | 117.2 (6) |
| C9—C8—H8 | 119.4 | C32—C31—P2 | 119.7 (5) |
| C7—C8—H8 | 119.4 | C36—C31—P2 | 123.0 (5) |
| C10—C9—C8 | 119.8 (7) | C31—C32—C33 | 122.0 (7) |
| C10—C9—H9 | 120.1 | C31—C32—H32 | 119.0 |
| C8—C9—H9 | 120.1 | C33—C32—H32 | 119.0 |
| C9—C10—C11 | 120.9 (7) | C34—C33—C32 | 118.9 (7) |
| C9—C10—H10 | 119.6 | C34—C33—H33 | 120.5 |
| C11—C10—H10 | 119.6 | C32—C33—H33 | 120.5 |
| C10—C11—C12 | 119.6 (7) | C35—C34—C33 | 120.6 (7) |
| C10—C11—H11 | 120.2 | C35—C34—H34 | 119.7 |
| C12—C11—H11 | 120.2 | C33—C34—H34 | 119.7 |
| C7—C12—C11 | 120.8 (7) | C34—C35—C36 | 120.1 (7) |
| C7—C12—H12 | 119.6 | C34—C35—H35 | 120.0 |
| C11—C12—H12 | 119.6 | C36—C35—H35 | 120.0 |
| C14—C13—C18 | 117.2 (7) | C35—C36—C31 | 121.2 (7) |
| C14—C13—P1 | 123.8 (6) | C35—C36—H36 | 119.4 |
| C18—C13—P1 | 118.9 (5) | C31—C36—H36 | 119.4 |
| C13—C14—C15 | 120.0 (8) | F4—B1—F1 | 112.3 (13) |
| C13—C14—H14 | 120.0 | F4—B1—F2 | 114.1 (18) |
| C15—C14—H14 | 120.0 | F1—B1—F2 | 107.7 (14) |
| C16—C15—C14 | 120.7 (8) | F4—B1—F3 | 110.3 (14) |
| C16—C15—H15 | 119.6 | F1—B1—F3 | 103.1 (12) |
| C14—C15—H15 | 119.6 | F2—B1—F3 | 108.8 (13) |
| C17—C16—C15 | 120.2 (9) | F2'—B1—F4' | 104 (3) |
| C17—C16—H16 | 119.9 | F2'—B1—F3' | 103 (2) |
| C15—C16—H16 | 119.9 | F4'—B1—F3' | 106 (3) |
| C16—C17—C18 | 119.2 (8) | F1—B1—F3' | 105.6 (16) |
| C16—C17—H17 | 120.4 | F2'—B1—F1 | 120 (2) |
| C18—C17—H17 | 120.4 | F4'—B1—F1 | 117 (3) |
| O1—Cu1—P1—C13 | 75.2 (3) | C13—C14—C15—C16 | 1.1 (15) |
| P2—Cu1—P1—C13 | −72.4 (2) | C14—C15—C16—C17 | −1.5 (17) |
| O1—Cu1—P1—C7 | −45.7 (3) | C15—C16—C17—C18 | 0.6 (17) |
| P2—Cu1—P1—C7 | 166.8 (2) | C16—C17—C18—C13 | 0.8 (15) |
| O1—Cu1—P1—C1 | −169.2 (3) | C14—C13—C18—C17 | −1.3 (12) |
| P2—Cu1—P1—C1 | 43.3 (3) | P1—C13—C18—C17 | −177.8 (7) |
| O1—Cu1—P2—C19 | −83.8 (3) | C31—P2—C19—C24 | −10.6 (7) |
| P1—Cu1—P2—C19 | 66.0 (2) | C25—P2—C19—C24 | 97.4 (6) |
| O1—Cu1—P2—C31 | 155.1 (3) | Cu1—P2—C19—C24 | −139.8 (5) |
| P1—Cu1—P2—C31 | −55.1 (3) | C31—P2—C19—C20 | 175.1 (5) |
| O1—Cu1—P2—C25 | 30.6 (3) | C25—P2—C19—C20 | −76.9 (6) |
| P1—Cu1—P2—C25 | −179.5 (2) | Cu1—P2—C19—C20 | 45.8 (6) |
| C13—P1—C1—C6 | −26.4 (6) | C24—C19—C20—C21 | −3.0 (11) |
| C7—P1—C1—C6 | 84.2 (6) | P2—C19—C20—C21 | 171.7 (6) |
| Cu1—P1—C1—C6 | −143.5 (5) | C19—C20—C21—C22 | 1.4 (13) |
| C13—P1—C1—C2 | 156.7 (5) | C20—C21—C22—C23 | 1.4 (13) |
| C7—P1—C1—C2 | −92.7 (5) | C21—C22—C23—C24 | −2.4 (13) |
| Cu1—P1—C1—C2 | 39.6 (6) | C20—C19—C24—C23 | 2.0 (11) |
| C6—C1—C2—C3 | 1.0 (10) | P2—C19—C24—C23 | −172.4 (6) |
| P1—C1—C2—C3 | 178.1 (6) | C22—C23—C24—C19 | 0.7 (12) |
| C1—C2—C3—C4 | −0.8 (11) | C19—P2—C25—C30 | −28.7 (6) |
| C2—C3—C4—C5 | −0.3 (12) | C31—P2—C25—C30 | 80.0 (6) |
| C3—C4—C5—C6 | 1.1 (11) | Cu1—P2—C25—C30 | −148.0 (5) |
| C2—C1—C6—C5 | −0.2 (10) | C19—P2—C25—C26 | 153.1 (5) |
| P1—C1—C6—C5 | −177.1 (5) | C31—P2—C25—C26 | −98.3 (6) |
| C4—C5—C6—C1 | −0.8 (11) | Cu1—P2—C25—C26 | 33.7 (6) |
| C13—P1—C7—C12 | −111.1 (6) | C30—C25—C26—C27 | −1.5 (10) |
| C1—P1—C7—C12 | 139.9 (6) | P2—C25—C26—C27 | 176.8 (6) |
| Cu1—P1—C7—C12 | 10.0 (7) | C25—C26—C27—C28 | −0.4 (12) |
| C13—P1—C7—C8 | 72.0 (6) | C26—C27—C28—C29 | 2.3 (13) |
| C1—P1—C7—C8 | −37.0 (6) | C27—C28—C29—C30 | −2.2 (12) |
| Cu1—P1—C7—C8 | −166.9 (5) | C28—C29—C30—C25 | 0.2 (11) |
| C12—C7—C8—C9 | −0.7 (10) | C26—C25—C30—C29 | 1.6 (10) |
| P1—C7—C8—C9 | 176.2 (6) | P2—C25—C30—C29 | −176.6 (5) |
| C7—C8—C9—C10 | 1.3 (11) | C19—P2—C31—C32 | −81.9 (6) |
| C8—C9—C10—C11 | −1.5 (13) | C25—P2—C31—C32 | 171.8 (6) |
| C9—C10—C11—C12 | 1.1 (13) | Cu1—P2—C31—C32 | 42.0 (7) |
| C8—C7—C12—C11 | 0.3 (11) | C19—P2—C31—C36 | 100.8 (6) |
| P1—C7—C12—C11 | −176.7 (6) | C25—P2—C31—C36 | −5.5 (7) |
| C10—C11—C12—C7 | −0.5 (13) | Cu1—P2—C31—C36 | −135.3 (6) |
| C7—P1—C13—C14 | 12.0 (7) | C36—C31—C32—C33 | −1.2 (11) |
| C1—P1—C13—C14 | 119.6 (6) | P2—C31—C32—C33 | −178.7 (6) |
| Cu1—P1—C13—C14 | −117.1 (6) | C31—C32—C33—C34 | 1.3 (12) |
| C7—P1—C13—C18 | −171.7 (6) | C32—C33—C34—C35 | −0.3 (13) |
| C1—P1—C13—C18 | −64.2 (6) | C33—C34—C35—C36 | −0.7 (13) |
| Cu1—P1—C13—C18 | 59.2 (6) | C34—C35—C36—C31 | 0.8 (13) |
| C18—C13—C14—C15 | 0.3 (12) | C32—C31—C36—C35 | 0.1 (11) |
| P1—C13—C14—C15 | 176.6 (7) | P2—C31—C36—C35 | 177.5 (6) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1C···F2 | 0.85 | 1.87 | 2.71 (3) | 171 |
| O1—H1D···F3i | 0.85 | 1.98 | 2.82 (3) | 171 |
| C28—H28···F4ii | 0.93 | 2.51 | 3.25 (3) | 137 |
Symmetry codes: (i) −x+1/2, y−1/2, −z+1/2; (ii) −x, −y+2, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: XU2555).
References
- Barron, P. F., Dyason, J. C., Healy, P. C., Engelhardt, L. M., Pakawatchai, C., Patrick, V. A. & White, A. H. (1987). J. Chem. Soc. Dalton Trans. pp. 1099–1106.
- Bruker (2008). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Engelhardt, L. M., Pakawatchai, C., White, A. H. & Healy, P. C. (1985). J. Chem. Soc. Dalton Trans. pp. 125–133.
- Fu, W.-F., Gan, X., Che, C.-M., Cao, Q.-Y., Zhou, Z.-Y. & Zhu, N.-Y. (2004). Chem. Eur. J.10, 2228–2236. [DOI] [PubMed]
- Kirchhoff, J. R., McMillin, D. R., Robinson, W. R., Powell, D. R., McKenzie, A. T. & Chen, S. (1985). Inorg. Chem.24, 3928–2933.
- Mao, Z., Chao, H.-Y., Hui, Z., Che, C.-M., Fu, W.-F., Cheung, K.-K. & Zhu, N. (2003). Chem. Eur. J.9, 2885–2997. [DOI] [PubMed]
- Meng, X.-T., Li, Q.-S., Xu, F.-B., Song, H.-B., Anson, C. E. & Zhang, Z.-Z. (2006). Inorg. Chem.45, 7986–7987. [DOI] [PubMed]
- Moudam, O., Kaeser, A., Delavaux-Nicot, B., Duhayon, C., Holler, M., Accorsi, G., Armaroli, N., Seguy, I., Navarro, J., Destruel, P. & Nierengarten, J. (2007). Chem. Commun. pp. 3077–3079. [DOI] [PubMed]
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Zhang, Q., Zhou, Q., Cheng, Y., Wang, L., Ma, D., Jing, X. & Wang, F. (2004). Adv. Mater.16, 432–436.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809029559/xu2555sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809029559/xu2555Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report