Abstract
The University of Minnesota Biocatalysis/Biodegradation Database (UM-BBD, http://umbbd.ahc.umn.edu/) provides curated information on microbial catabolic enzymes and their organization into metabolic pathways. Currently, it contains information on over 400 enzymes. In the last year the enzyme page was enhanced to contain more internal and external links; it also displays the different metabolic pathways in which each enzyme participates. In collaboration with the Nomenclature Commission of the International Union of Biochemistry and Molecular Biology, 35 UM-BBD enzymes were assigned complete EC codes during 2000. Bacterial oxygenases are heavily represented in the UM-BBD; they are known to have broad substrate specificity. A compilation of known reactions of naphthalene and toluene dioxygenases were recently added to the UM-BBD; 73 and 108 were listed respectively. In 2000 the UM-BBD is mirrored by two prestigious groups: the European Bioinformatics Institute and KEGG (the Kyoto Encyclopedia of Genes and Genomes). Collaborations with other groups are being developed. The increased emphasis on UM-BBD enzymes is important for predicting novel metabolic pathways that might exist in nature or could be engineered. It also is important for current efforts in microbial genome annotation.
INTRODUCTION
As the University of Minnesota Biocatalysis/Biodegradation Database (UM-BBD, http://umbbd.ahc.umn.edu/index.html) starts its sixth year, there are 30 complete and 127 on-going microbial genome sequencing projects (1, http://wit.integratedgenomics.com/gold/). Genomic sequence information is increasing exponentially, with a doubling time of less than 1 year. This information explosion has influenced the growth of the UM-BBD in the past year. We have strengthened our collaboration with the European Bioinformatics Institute, Kyoto University and the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology. UM-BBD enzyme information has played a major role in these collaborations and others. UM-BBD present and potential future status, and the increased emphasis on its enzyme information, is discussed in more detail below.
PRESENT STATUS
UM-BBD data content and methods, including data format, update and access, have been reported (2,3). By the end of 2000 it will have grown to contain over 100 pathways, 700 reactions, 600 compounds, 400 enzymes and nearly 300 microorganism entries. A goal of the UM-BBD is to document the breadth of reaction types catalyzed by microbes. Reaction types of interest are those in which a unique organic functional group is transformed or the bond between functional groups is cleaved. The list of organic functional groups contained in the UM-BBD has grown to 49 [J.Liu and J.Kang (2000) Organic Functional Groups, http://umbbd.ahc.umn.edu/search/FuncGrps.html], including bicycloaliphatic ring, tricycloaliphatic ring, unsaturated N-heterocyclic ring, epoxide, peroxide, oxime and cyanamide, are all transformed by one or more UM-BBD enzymes. A list of the more than 400 UM-BBD enzymes, ordered by EC number, is available (UM-BBD, 2000. List of All Enzymes, http://umbbd.ahc.umn.edu/cgi-bin/page.cgi?ptype=allenzymes). An excerpt from a representative enzyme page is shown in Figure 1.
Figure 1.
Excerpt from a UM-BBD enzyme page. This page for the enzyme haloalkane dehalogenase includes, among other information: (A) a link to the BRENDA database; (B) a dynamic search of the GenBank database; (C) a dynamic search of the GenPept database; and (D) a dynamic search of the PDB database. The complete enzyme page is available at http://umbbd.ahc.umn.edu/servlets/pageservlet?ptype=ep&enzymeID=e0003.
The UM-BBD enzyme page has greatly increased in importance in the past year. Before then, it duplicated a subset of the information found on the UM-BBD reaction page (excerpted in Fig. 2). However, the format of the reaction page restricted the amount of information that page could contain; adding links to it would detract from its focus on the reaction. UM-BBD users required additional enzyme information; the UM-BBD enzyme page expanded to meet this need and the UM-BBD increased this page’s visibility.
Figure 2.
Excerpt from a UM-BBD reaction page. This page for the reaction from 1,2-dichloroethane to 2-chloroethanol includes, among other information, (A) a link to a Medline abstract which contains information on the enzyme’s structure; (B) a link to the UM-BBD enzyme page for its enzyme, excerpt shown in Figure 1; and (C) a link to a generated pathway starting from this reaction. An example of the latter is shown in Figure 3. The complete reaction page is available at http://umbbd.ahc.umn.edu/servlets/pageservlet?ptype=r&reacID=r0001.
Enzyme pages are now more easily accessed with the addition of a link to them through the EC code on reaction pages (Fig. 2B). They are also now included in the list of compounds and reactions for each pathway; this list is linked to at the top and bottom of every pathway page.
The number of static enzyme links has increased. For example, a link to the BRENDA Comprehensive Enzyme Information System [D.Schomburg (2000) http://www.brenda.uni-koeln.de/] was added to all pages for enzymes which had been assigned a four-digit EC code (Fig. 1A).
The ability to search remote databases was also expanded. From its very beginning the UM-BBD has included dynamic searches of the GenBank database of nucleic acid sequences, for UM-BBD enzymes whose sequences were present in GenBank (Fig. 1B). With the increase in genomic data mentioned in the Introduction, larger DNA fragments are deposited and users have a harder time locating the region of interest. Thus we added dynamic searches of the NCBI GenPept (4, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=protein), Figure 1C.
Enzyme structure information was initially included through links to Medline abstracts reporting enzyme structures (Fig. 2A). However, only one structure could be indicated in this way. With the proliferation of structure information, we now include a dynamic link to the PDB Protein Structure Database (5, http://www.rcsb.org/pdb/) when such structures exist (Fig. 1D).
Some of these features, such as the link to BRENDA, require assignment of a four-digit EC code. In 1997, a collaboration began between Keith Tipton, designated member of the Nomenclature Commission of the International Union of Biochemistry and Molecular Biology (NC-IUBMB) with responsibility for enzyme classification and nomenclature, Lynda Ellis, co-director of the UM-BBD, and Toni Kazic, director of the KLOTHO database, funded by the NIH (PI, Toni Kazic). As part of this collaboration, 35 UM-BBD enzymes (listed in Supplementary Material) were assigned four-digit EC codes, systematic names and other attributes by the NC-IUBMB in 2000, and many more will gain this information in 2001. As they are approved, these enzyme classification details are made available to the scientific community for comment (http://www.chem.qmw.ac.uk/iubmb/enzyme/newenz.html) prior to incorporation into the enzyme database (http://www.chem.qmw.ac.uk/iubmb/enzyme/). Nomenclature Commission staff report that UM-BBD organization, primary reaction references and dynamic reference searches greatly facilitate their task of classifying its enzymes (S.Boyce, personal communication, June 2000).
Over the past year we continued to develop pages that document the biocatalytic versatility of UM-BBD enzymes. Building on the previous list of 73 reactions catalyzed by the enzyme naphthalene 1,2-dioxygenase, EC 1.14.12.12 [J.Liu (1999) Reactions of Naphthalene 1,2-Dioxygenase. http://umbbd.ahc.umn.edu/ naph/ndo.html], we compiled a list of 108 reactions catalyzed by toluene dioxygenase, EC 1.14.12.11. The types and numbers of substrates transformed by this versatile enzyme are shown in Table 1. Such lists document the broad substrate specificity often found for enzymes involved in biodegradation and their wide biocatalytic potential.
Table 1. Type (and number) of substrates transformed by toluene 1,2-dioxygenase, EC 1.14.12.11, from Ps.putida F1a.
| Monocyclic aromatics (69) |
| Dioxygenation (60) |
| Monooxygenation (5) |
| Sulfoxidation (4) |
| Fused aromatics (3) |
| Linked aromatics (7) |
| Aliphatic olefins (17) |
| Allylic methyl group monooxygenation (8) |
| Monooxygenation with allylic rearrangement (1) |
| Dioxygenation (8) |
| Other substrates (12) |
aTaken from a complete list of 108 reactions, J.Liu and A.Negrete (2000) Toluene Dioxygenase Reactions. http://www.labmed.umn.edu/umbbd/tol/tdo.html
Future plans include establishing and maintaining mirror sites at geographically dispersed locations; improving interface with other databases; and new directions in pathway visualization and prediction.
MIRROR SITES
The past year saw the first UM-BBD mirror site, hosted by the European Bioinformatics Institute on their SRS server [T.Etzold, G.Verde, D.Kreil and P.Carter (1999) http://srs.ebi.ac.uk/]. This year, UM-BBD pathways began to be duplicated in KEGG, the Kyoto Encyclopedia of Genes and Genomes (6, http://www. genome.ad.jp/kegg/kegg.html). Additional mirror sites may be set up in the future. Our two present mirrors each integrate the UM-BBD with other databases that they host. We are working with others to increase the availability of our information.
INTERACTIONS WITH OTHER DATABASES
The collaboration of Keith Tipton, Lynda Ellis and Toni Kazic, mentioned earlier, is developing The Agora, a distributed computational environment for biochemical information. This includes information on enzymes, compounds and reactions. The environment will permit reliable sharing of curatorial functions and queries among independent participating databases, to permit the scientific community to deposit, review and query biochemical information, while at the same time allowing each database to preserve its native semantics, data model and query language. The UM-BBD is one of the founding databases in this collaboration (7).
We are also collaborating with Terri Attwood, curator of PRINTS, a compendium of groups of conserved motifs (fingerprints) used to characterize a protein family (8, http://bioinf.man.ac.uk/dbbrowser/prints/). She is developing fingerprints to characterize the protein families that contain selected UM-BBD enzymes.
The Biodegradative Strain Database [BSD, J.Urbance, J.Cole, and J.Tiedje (2000) http://bsd.cme.msu.edu/], an on-line database of information on described, biodegradative microbial strains, debuted on the web in searchable form in 2000. The BSD presently links to UM-BBD information and reciprocal links will be implemented in the coming year.
PATHWAY VISUALIZATION AND PREDICTION
While all curated UM-BBD pathway diagrams are handcrafted to insure a clear and aesthetically pleasing rendering, the option to dynamically generate pathways, which can start at any UM-BBD reaction, is also offered. One such generated pathway is shown in Figure 3. In collaboration with Markus Eiglsperger in the Department of Computer Science at the University of Tuebingen, Germany, tools are being developed to better visualize these dynamically generated pathways. A prototype is shown in Figure 4.
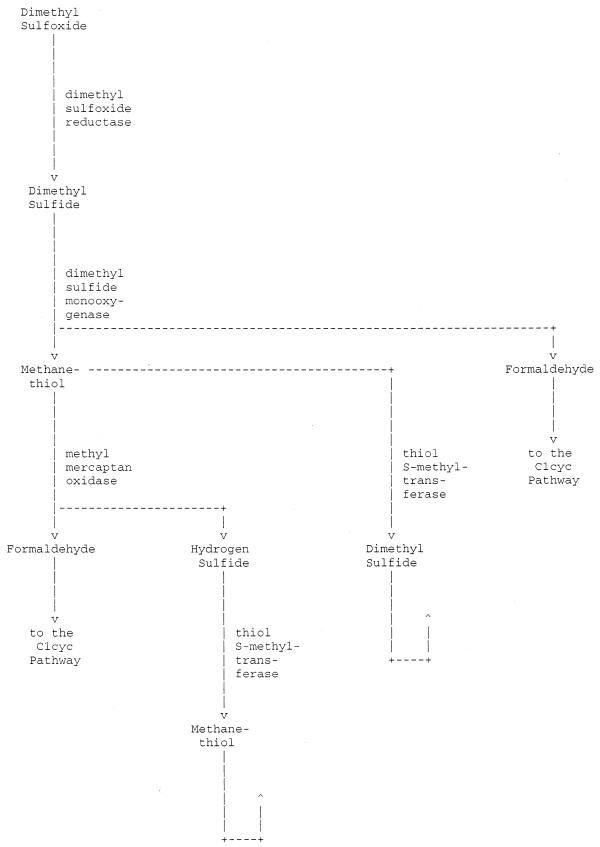
Figure 3.
Excerpt from a UM-BBD generated metabolic pathway. This pathway was generated starting from the reaction from dimethyl sulfoxide to dimethyl sulfide. In the original all enzyme and compound names are hyperlinked to UM-BBD reaction and compound pages, respectively. The complete generated pathway page is available at http://umbbd.ahc.umn.edu/cgi-bin/page.cgi?ptype=p&reacID=r0207.
Figure 4.
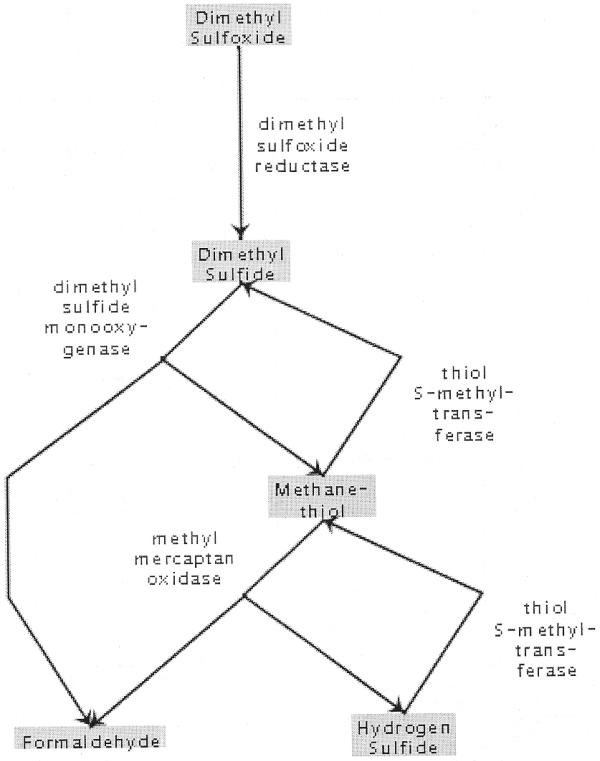
Prototype improved visualization of UM-BBD generated pathways. This is an excerpt that displays the same generated pathway shown in Figure 3. Compared to Figure 3, the prototype is more compact, more attractive and loops are displayed more intuitively.
Better pathway visualization will assist users in connecting enzyme-catalyzed reactions into productive pathways. The UM-BBD presents enzyme reactions broadly; not all reactions in depicted pathways necessarily exist in any one given organism. Nonetheless, its pathways represent plausible metabolism; and metabolic reconstruction has taken on much greater significance with the current focus on genome annotation. This also has importance for enhancing the ability to predict the metabolism of newly synthesized compounds for environmental purposes or for metabolic engineering.
Prediction is important because, with over 12 million organic compounds currently known, the UM-BBD will never contain biodegradation pathways for every one (3). We are developing a framework in which biodegradation pathways for a target compound are evaluated by mapping the chemical functional groups of that target compound against the capabilities of organisms to generate enzymes that operate on these functional groups. The UM-BBD serves as the main data source in this collaboration (9).
CONCLUSIONS
The UM-BBD’s emphasis on enzymes, their multiplicity of substrates and inclusion into different pathways is important for predicting novel metabolic pathways that might exist in nature or could be engineered. It also is important for current efforts in microbial genome annotation.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Robert Andrews, Minoru Kanihesa, Markus Eiglsperger, John Urbance, Jeffrey Varner, Toni Kazic, Terri Attwood, Keith Tipton and Sinèad Boyce for helpful discussions. Supported in part by NIH R01GM56529, NSF Postdoctoral Fellowship 9974209 in Bioinformatics to C.D.H. and an NLM Postdoctoral Traineeship in Bioinformatics (LM07041) to E.M.B.
References
- 1.Kyrpides N. (1999) Genomes OnLine Database (GOLD 1.0): a monitor of complete and ongoing genome projects world-wide. Bioinformatics, 15, 773–774. [DOI] [PubMed] [Google Scholar]
- 2.Ellis L.B.M., Hershberger,C.D. and Wackett,L.P. (1999) The University of Minnesota Biocatalysis/Biodegradation Database: specialized metabolism for functional genomics. Nucleic Acids Res., 27, 373–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellis L.B.M., Hershberger,C.D. and Wackett,L.P. (2000) The University of Minnesota Biocatalysis/Biodegradation Database: microorganisms, genomics and prediction. Nucleic Acids Res., 28, 377–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wheeler D.L., Chappey,C., Lash,A.E., Leipe,D.D., Madden,T.L., Schuler,G.D., Tatusova,T.A. and Rapp,B.A. (2000) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 28, 10–14. Updated article in this issue: Nucleic Acids Res. (2001), 29, 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. Updated article in this issue: Nucleic Acids Res. (2001), 29, 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanehisa M. and Goto,S. (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res., 28, 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bugrim A., Boyce,S., Yao,G., Fabrizio,F., McDonald,A., Slomczynski,J., Ouyang,J., Feng,B., Wise,W., Tipton,K., Ellis,L. and Kazic,T. (2000) The Agora—An Environment for Distributed Deposit, Review, and Analysis of Biochemical Information. Final Program, Intelligent Systems in Molecular Biology 2000, San Diego, CA, August 19–23, 2000, pp. 33.
- 8.Attwood T.K., Croning,M.D.R., Flower,D.R., Lewis,A.P., Mabey,J.E., Scordis,P., Selley,J. and Wright,W. (2000) PRINTS-S: the database formerly known as PRINTS. Nucleic Acids Res., 28, 225–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J., Vigouroux,M., Hershberger,C.D., Ellis,L.B.M., Wackett,L.P. and Varner,J.D. (2000) Heuristic-based Prediction of Specialized Metabolism. Final Program, Intelligent Systems in Molecular Biology 2000, San Diego, CA, August 19–23, 2000, p. 22.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.