Abstract
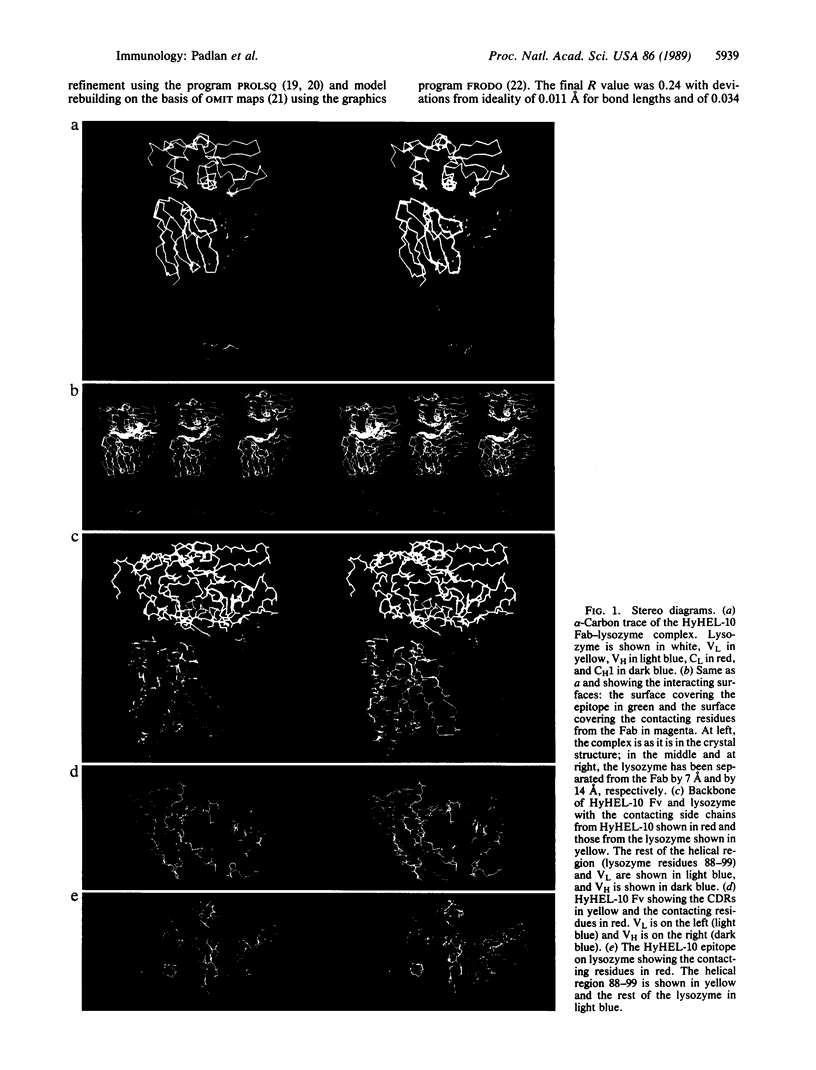
The crystal structure of the complex of the anti-lysozyme HyHEL-10 Fab and hen egg white lysozyme has been determined to a nominal resolution of 3.0 A. The antigenic determinant (epitope) on the lysozyme is discontinuous, consisting of residues from four different regions of the linear sequence. It consists of the exposed residues of an alpha-helix together with surrounding amino acids. The epitope crosses the active-site cleft and includes a tryptophan located within this cleft. The combining site of the antibody is mostly flat with a protuberance made up of two tyrosines that penetrate the cleft. All six complementarity-determining regions of the Fab contribute at least one residue to the binding; one residue from the framework is also in contact with the lysozyme. The contacting residues on the antibody contain a disproportionate number of aromatic side chains. The antibody-antigen contact mainly involves hydrogen bonds and van der Waals interactions; there is one ion-pair interaction but it is weak.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amit A. G., Mariuzza R. A., Phillips S. E., Poljak R. J. Three-dimensional structure of an antigen-antibody complex at 2.8 A resolution. Science. 1986 Aug 15;233(4765):747–753. doi: 10.1126/science.2426778. [DOI] [PubMed] [Google Scholar]
- Benjamin D. C., Berzofsky J. A., East I. J., Gurd F. R., Hannum C., Leach S. J., Margoliash E., Michael J. G., Miller A., Prager E. M. The antigenic structure of proteins: a reappraisal. Annu Rev Immunol. 1984;2:67–101. doi: 10.1146/annurev.iy.02.040184.000435. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Bjorkman P. J., Saper M. A., Samraoui B., Bennett W. S., Strominger J. L., Wiley D. C. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987 Oct 8;329(6139):506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- Case D. A., Karplus M. Dynamics of ligand binding to heme proteins. J Mol Biol. 1979 Aug 15;132(3):343–368. doi: 10.1016/0022-2836(79)90265-1. [DOI] [PubMed] [Google Scholar]
- Chothia C. Hydrophobic bonding and accessible surface area in proteins. Nature. 1974 Mar 22;248(446):338–339. doi: 10.1038/248338a0. [DOI] [PubMed] [Google Scholar]
- Colman P. M., Laver W. G., Varghese J. N., Baker A. T., Tulloch P. A., Air G. M., Webster R. G. Three-dimensional structure of a complex of antibody with influenza virus neuraminidase. 1987 Mar 26-Apr 1Nature. 326(6111):358–363. doi: 10.1038/326358a0. [DOI] [PubMed] [Google Scholar]
- Cygler M., Anderson W. F. Application of the molecular replacement method to multidomain proteins. 1. Determination of the orientation of an immunoglobulin Fab fragment. Acta Crystallogr A. 1988 Jan 1;44(Pt 1):38–45. doi: 10.1107/s0108767387008365. [DOI] [PubMed] [Google Scholar]
- Davies D. R., Sheriff S., Padlan E. A. Antibody-antigen complexes. J Biol Chem. 1988 Aug 5;263(22):10541–10544. [PubMed] [Google Scholar]
- Diamond R. Real-space refinement of the structure of hen egg-white lysozyme. J Mol Biol. 1974 Jan 25;82(3):371–391. doi: 10.1016/0022-2836(74)90598-1. [DOI] [PubMed] [Google Scholar]
- Epp O., Lattman E. E., Schiffer M., Huber R., Palm W. The molecular structure of a dimer composed of the variable portions of the Bence-Jones protein REI refined at 2.0-A resolution. Biochemistry. 1975 Nov 4;14(22):4943–4952. doi: 10.1021/bi00693a025. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Shi J. P., Knill-Jones J., Lowe D. M., Wilkinson A. J., Blow D. M., Brick P., Carter P., Waye M. M., Winter G. Hydrogen bonding and biological specificity analysed by protein engineering. Nature. 1985 Mar 21;314(6008):235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
- Friguet B., Chaffotte A. F., Djavadi-Ohaniance L., Goldberg M. E. Measurements of the true affinity constant in solution of antigen-antibody complexes by enzyme-linked immunosorbent assay. J Immunol Methods. 1985 Mar 18;77(2):305–319. doi: 10.1016/0022-1759(85)90044-4. [DOI] [PubMed] [Google Scholar]
- Harper M., Lema F., Boulot G., Poljak R. J. Antigen specificity and cross-reactivity of monoclonal anti-lysozyme antibodies. Mol Immunol. 1987 Feb;24(2):97–108. doi: 10.1016/0161-5890(87)90081-2. [DOI] [PubMed] [Google Scholar]
- KAUZMANN W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- Marquart M., Deisenhofer J., Huber R., Palm W. Crystallographic refinement and atomic models of the intact immunoglobulin molecule Kol and its antigen-binding fragment at 3.0 A and 1.0 A resolution. J Mol Biol. 1980 Aug 25;141(4):369–391. doi: 10.1016/0022-2836(80)90252-1. [DOI] [PubMed] [Google Scholar]
- Phillips D. C. The three-dimensional structure of an enzyme molecule. Sci Am. 1966 Nov;215(5):78–90. doi: 10.1038/scientificamerican1166-78. [DOI] [PubMed] [Google Scholar]
- Satow Y., Cohen G. H., Padlan E. A., Davies D. R. Phosphocholine binding immunoglobulin Fab McPC603. An X-ray diffraction study at 2.7 A. J Mol Biol. 1986 Aug 20;190(4):593–604. doi: 10.1016/0022-2836(86)90245-7. [DOI] [PubMed] [Google Scholar]
- Saul F. A., Amzel L. M., Poljak R. J. Preliminary refinement and structural analysis of the Fab fragment from human immunoglobulin new at 2.0 A resolution. J Biol Chem. 1978 Jan 25;253(2):585–597. [PubMed] [Google Scholar]
- Segal D. M., Padlan E. A., Cohen G. H., Rudikoff S., Potter M., Davies D. R. The three-dimensional structure of a phosphorylcholine-binding mouse immunoglobulin Fab and the nature of the antigen binding site. Proc Natl Acad Sci U S A. 1974 Nov;71(11):4298–4302. doi: 10.1073/pnas.71.11.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheriff S., Hendrickson W. A., Smith J. L. Structure of myohemerythrin in the azidomet state at 1.7/1.3 A resolution. J Mol Biol. 1987 Sep 20;197(2):273–296. doi: 10.1016/0022-2836(87)90124-0. [DOI] [PubMed] [Google Scholar]
- Sheriff S., Silverton E. W., Padlan E. A., Cohen G. H., Smith-Gill S. J., Finzel B. C., Davies D. R. Three-dimensional structure of an antibody-antigen complex. Proc Natl Acad Sci U S A. 1987 Nov;84(22):8075–8079. doi: 10.1073/pnas.84.22.8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverton E. W., Padlan E. A., Davies D. R., Smith-Gill S., Potter M. Crystalline monoclonal antibody Fabs complexed to hen egg white lysozyme. J Mol Biol. 1984 Dec 15;180(3):761–765. doi: 10.1016/0022-2836(84)90037-8. [DOI] [PubMed] [Google Scholar]
- Smith-Gill S. J., Lavoie T. B., Mainhart C. R. Antigenic regions defined by monoclonal antibodies correspond to structural domains of avian lysozyme. J Immunol. 1984 Jul;133(1):384–393. [PubMed] [Google Scholar]
- Smith-Gill S. J., Mainhart C., Lavoie T. B., Feldmann R. J., Drohan W., Brooks B. R. A three-dimensional model of an anti-lysozyme antibody. J Mol Biol. 1987 Apr 20;194(4):713–724. doi: 10.1016/0022-2836(87)90249-x. [DOI] [PubMed] [Google Scholar]
- Smith-Gill S. J., Rupley J. A., Pincus M. R., Carty R. P., Scheraga H. A. Experimental identification of a theoretically predicted "left-sided" binding mode for (GlcNAc)6 in the active site of lysozyme. Biochemistry. 1984 Feb 28;23(5):993–997. doi: 10.1021/bi00300a030. [DOI] [PubMed] [Google Scholar]
- Suh S. W., Bhat T. N., Navia M. A., Cohen G. H., Rao D. N., Rudikoff S., Davies D. R. The galactan-binding immunoglobulin Fab J539: an X-ray diffraction study at 2.6-A resolution. Proteins. 1986 Sep;1(1):74–80. doi: 10.1002/prot.340010112. [DOI] [PubMed] [Google Scholar]
- Sussman J. L. Constrained-restrained least-squares (CORELS) refinement of proteins and nucleic acids. Methods Enzymol. 1985;115:271–303. doi: 10.1016/0076-6879(85)15022-6. [DOI] [PubMed] [Google Scholar]
- Verhoeyen M., Milstein C., Winter G. Reshaping human antibodies: grafting an antilysozyme activity. Science. 1988 Mar 25;239(4847):1534–1536. doi: 10.1126/science.2451287. [DOI] [PubMed] [Google Scholar]