Abstract
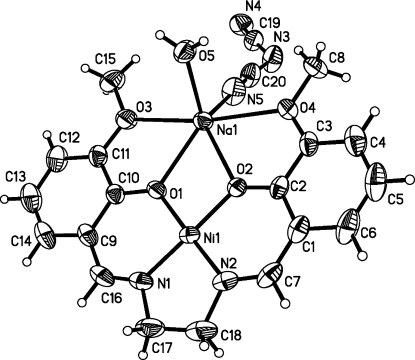
The molecule of the title compound, [NaNi(C18H18N2O4)(C2N3)(H2O)], is approximately planar, with a maximum deviation from the molecular plane of 0.770 (5) Å. The coordination environment of the Ni2+ ion is distorted square-planar and it is N2O2 coordinated by the 6,6′-dimethoxy-2,2′-[ethane-1,2-diylbis(nitrilomethylidyne)]diphenolate Schiff base ligand. The Na+ atom is chelated by the four O atoms of the Schiff base ligand, a water ligand and a dicyanamide anion. The structure displays intermolecular O—H⋯N hydrogen bonding.
Related literature
For chemical background, see: Ohba & Okawa (2000 ▶). For related structures, see: Correia et al. (2005 ▶); Costes et al.(2004 ▶).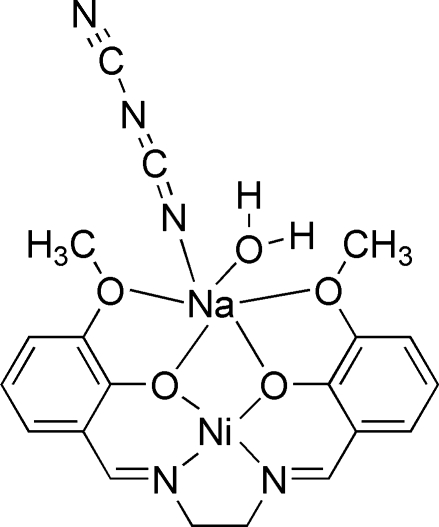
Experimental
Crystal data
[NaNi(C18H18N2O4)(C2N3)(H2O)]
M r = 492.11
Monoclinic,
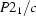
a = 7.4654 (14) Å
b = 22.745 (4) Å
c = 13.177 (3) Å
β = 101.282 (4)°
V = 2194.2 (8) Å3
Z = 4
Mo Kα radiation
μ = 0.95 mm−1
T = 293 K
0.14 × 0.13 × 0.11 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2003 ▶) T min = 0.879, T max = 0.903
10817 measured reflections
3864 independent reflections
2815 reflections with I > 2σ(I)
R int = 0.032
Refinement
R[F 2 > 2σ(F 2)] = 0.039
wR(F 2) = 0.099
S = 1.02
3864 reflections
291 parameters
54 restraints
H-atom parameters constrained
Δρmax = 0.33 e Å−3
Δρmin = −0.35 e Å−3
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: SAINT-Plus (Bruker, 2001 ▶); data reduction: SAINT-Plus; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S160053680901438X/hg2500sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680901438X/hg2500Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O5—H5A⋯N3i | 0.82 | 2.14 | 2.960 (4) | 175 |
| O5—H5B⋯N4ii | 0.82 | 2.03 | 2.852 (4) | 177 |
Symmetry codes: (i) 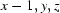 ; (ii)
; (ii) 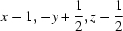 .
.
Acknowledgments
This work was supported by the Zhejiang Provincial Natural Science Foundation (grant No. Y4080395).
supplementary crystallographic information
Comment
The dicyanamide ligand N(CN)2, has attracted continuous attention in the past four years for the buildup of interesting extended architectures. Its versatile coordination behavior and its ability to organize solids into polymeric structures with a rich diversity of magnetic properties have attracted interest toward this research area (Ohba et al., 2000). N,N-disalicylideneethylenediamine type Schiff bases ligands present versatile steric, electronic and lipophilic properties (Correia et al. 2005). We report here the synthesis and crystal structure of the title compound. The molecular structure is shown in Fig. 1. The values of the geometric parameters in this compound are normal (Costes et al., 2004). NiII and NaI are connected via two bridging O atoms of the ligand. The six-coordinate Na atom adopts a distorted octahedral coordination geometry while the four-coordinate Ni gives a planar coordination geometry.
Experimental
A mixture of 6,6'-dimethoxy-2,2'-(ethane-1,2-diyldiiminodimethylene)diphenol (1 mmol) and nickel chloride (1 mmol) in methanol (15 ml) was stirred for 30 min and sodium dicyanamide (1 mmol) was added, stirred for another 15 min and then filtered. The resulting clear orange solution was vapor at room temperature for 7 d, after which large orange block-shaped crystals of the title complex suitable for X-ray diffraction analysis were obtained.
Refinement
The H atoms were fixed geometrically and were treated as riding on their parent C atoms, with C—H distances in the range of 0.93–0.97 Å and with Uiso(H) = 1.2Ueq(parent atom), or Uiso(H) = 1.5Ueq(Cmethyl).
Figures
Fig. 1.
The independent molecules of the title compound, showing 30% probability displacement ellipsoids and the atom-numbering scheme.
Crystal data
| [NaNi(C18H18N2O4)(C2N3)(H2O)] | F(000) = 1016 |
| Mr = 492.11 | Dx = 1.490 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 3120 reflections |
| a = 7.4654 (14) Å | θ = 2.5–24.6° |
| b = 22.745 (4) Å | µ = 0.95 mm−1 |
| c = 13.177 (3) Å | T = 293 K |
| β = 101.282 (4)° | Block, orange |
| V = 2194.2 (8) Å3 | 0.14 × 0.13 × 0.11 mm |
| Z = 4 |
Data collection
| Bruker SMART CCD area-detector diffractometer | 3864 independent reflections |
| Radiation source: fine-focus sealed tube | 2815 reflections with I > 2σ(I) |
| graphite | Rint = 0.032 |
| φ and ω scans | θmax = 25.0°, θmin = 1.8° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2003) | h = −8→8 |
| Tmin = 0.879, Tmax = 0.903 | k = −26→27 |
| 10817 measured reflections | l = −15→14 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.039 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.099 | H-atom parameters constrained |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.0455P)2 + 0.4335P] where P = (Fo2 + 2Fc2)/3 |
| 3864 reflections | (Δ/σ)max = 0.001 |
| 291 parameters | Δρmax = 0.33 e Å−3 |
| 54 restraints | Δρmin = −0.35 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Ni1 | 0.24893 (5) | 0.506776 (16) | 0.06029 (3) | 0.04684 (15) | |
| Na1 | 0.36614 (16) | 0.38280 (5) | 0.20328 (9) | 0.0527 (3) | |
| O1 | 0.2723 (3) | 0.48215 (8) | 0.19509 (16) | 0.0545 (5) | |
| O2 | 0.3315 (3) | 0.43203 (8) | 0.03988 (15) | 0.0497 (5) | |
| O3 | 0.3154 (4) | 0.42650 (10) | 0.36882 (17) | 0.0732 (7) | |
| O4 | 0.4284 (3) | 0.32315 (9) | 0.04958 (19) | 0.0655 (6) | |
| O5 | 0.1624 (3) | 0.30933 (10) | 0.21624 (17) | 0.0731 (7) | |
| H5A | 0.1005 | 0.3032 | 0.2601 | 0.088* | |
| H5B | 0.1305 | 0.2871 | 0.1665 | 0.088* | |
| N1 | 0.1665 (3) | 0.58076 (11) | 0.0844 (2) | 0.0584 (7) | |
| N2 | 0.2232 (3) | 0.52892 (13) | −0.0760 (2) | 0.0589 (7) | |
| N3 | 0.9189 (4) | 0.28821 (17) | 0.3648 (3) | 0.0917 (9) | |
| N4 | 1.0360 (5) | 0.26666 (15) | 0.5450 (3) | 0.0888 (10) | |
| N5 | 0.6431 (5) | 0.34246 (17) | 0.2954 (3) | 0.0934 (9) | |
| C1 | 0.3049 (4) | 0.43665 (18) | −0.1445 (3) | 0.0652 (10) | |
| C2 | 0.3415 (4) | 0.40716 (15) | −0.0485 (2) | 0.0511 (8) | |
| C3 | 0.3908 (4) | 0.34689 (15) | −0.0474 (3) | 0.0601 (9) | |
| C4 | 0.4007 (6) | 0.3183 (2) | −0.1377 (4) | 0.0885 (13) | |
| H4 | 0.4321 | 0.2787 | −0.1361 | 0.106* | |
| C5 | 0.3642 (7) | 0.3480 (3) | −0.2315 (4) | 0.1134 (17) | |
| H5 | 0.3708 | 0.3281 | −0.2923 | 0.136* | |
| C6 | 0.3189 (6) | 0.4060 (3) | −0.2353 (3) | 0.0998 (15) | |
| H6 | 0.2969 | 0.4256 | −0.2985 | 0.120* | |
| C7 | 0.2509 (5) | 0.49684 (19) | −0.1515 (3) | 0.0694 (11) | |
| H7 | 0.2342 | 0.5144 | −0.2164 | 0.083* | |
| C8 | 0.4977 (6) | 0.26446 (15) | 0.0610 (3) | 0.0903 (13) | |
| H8A | 0.4044 | 0.2375 | 0.0292 | 0.135* | |
| H8B | 0.5337 | 0.2553 | 0.1332 | 0.135* | |
| H8C | 0.6014 | 0.2611 | 0.0283 | 0.135* | |
| C9 | 0.1804 (5) | 0.57147 (14) | 0.2674 (3) | 0.0631 (9) | |
| C10 | 0.2389 (4) | 0.51231 (13) | 0.2744 (3) | 0.0524 (8) | |
| C11 | 0.2599 (5) | 0.48361 (15) | 0.3708 (3) | 0.0615 (9) | |
| C12 | 0.2255 (6) | 0.5127 (2) | 0.4565 (3) | 0.0842 (12) | |
| H12 | 0.2398 | 0.4933 | 0.5196 | 0.101* | |
| C13 | 0.1694 (7) | 0.5711 (2) | 0.4492 (4) | 0.0982 (14) | |
| H13 | 0.1469 | 0.5905 | 0.5076 | 0.118* | |
| C14 | 0.1473 (6) | 0.59969 (18) | 0.3580 (4) | 0.0854 (12) | |
| H14 | 0.1095 | 0.6387 | 0.3543 | 0.102* | |
| C15 | 0.3392 (6) | 0.39343 (19) | 0.4619 (3) | 0.0919 (13) | |
| H15A | 0.4253 | 0.4130 | 0.5147 | 0.138* | |
| H15B | 0.3841 | 0.3550 | 0.4502 | 0.138* | |
| H15C | 0.2241 | 0.3900 | 0.4836 | 0.138* | |
| C17 | 0.1111 (6) | 0.61685 (18) | −0.0096 (4) | 0.0889 (13) | |
| H16A | −0.0209 | 0.6203 | −0.0257 | 0.107* | |
| H16B | 0.1619 | 0.6560 | 0.0028 | 0.107* | |
| C18 | 0.1737 (7) | 0.59090 (18) | −0.0957 (4) | 0.0952 (14) | |
| H17A | 0.2791 | 0.6124 | −0.1086 | 0.114* | |
| H17B | 0.0782 | 0.5938 | −0.1571 | 0.114* | |
| C19 | 0.9741 (5) | 0.27798 (17) | 0.4624 (3) | 0.0681 (9) | |
| C20 | 0.7695 (6) | 0.31745 (18) | 0.3323 (3) | 0.0762 (9) | |
| C16 | 0.1480 (5) | 0.60153 (15) | 0.1717 (3) | 0.0691 (10) | |
| H20 | 0.1091 | 0.6404 | 0.1724 | 0.083* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Ni1 | 0.0435 (2) | 0.0440 (2) | 0.0519 (3) | −0.00269 (17) | 0.00649 (17) | 0.00924 (18) |
| Na1 | 0.0576 (7) | 0.0475 (7) | 0.0521 (7) | 0.0048 (6) | 0.0090 (6) | 0.0046 (5) |
| O1 | 0.0737 (15) | 0.0407 (11) | 0.0502 (13) | 0.0055 (10) | 0.0146 (11) | −0.0009 (10) |
| O2 | 0.0576 (13) | 0.0493 (12) | 0.0426 (12) | 0.0010 (10) | 0.0109 (10) | 0.0014 (9) |
| O3 | 0.115 (2) | 0.0601 (15) | 0.0482 (14) | 0.0084 (14) | 0.0235 (13) | 0.0051 (11) |
| O4 | 0.0800 (16) | 0.0474 (13) | 0.0746 (17) | −0.0003 (11) | 0.0284 (13) | −0.0082 (12) |
| O5 | 0.0885 (17) | 0.0713 (15) | 0.0650 (16) | −0.0195 (13) | 0.0290 (13) | −0.0105 (12) |
| N1 | 0.0475 (16) | 0.0432 (15) | 0.081 (2) | −0.0028 (12) | 0.0046 (14) | 0.0120 (15) |
| N2 | 0.0461 (16) | 0.0653 (18) | 0.0625 (19) | −0.0061 (13) | 0.0036 (14) | 0.0215 (15) |
| N3 | 0.0765 (19) | 0.131 (2) | 0.0687 (18) | 0.0310 (17) | 0.0164 (15) | 0.0124 (18) |
| N4 | 0.099 (2) | 0.085 (2) | 0.076 (2) | 0.0171 (19) | 0.0042 (18) | 0.0172 (18) |
| N5 | 0.0782 (19) | 0.125 (2) | 0.0725 (19) | 0.0268 (18) | 0.0037 (16) | 0.0108 (17) |
| C1 | 0.046 (2) | 0.100 (3) | 0.049 (2) | −0.0013 (19) | 0.0089 (15) | −0.001 (2) |
| C2 | 0.0405 (17) | 0.070 (2) | 0.0432 (19) | −0.0088 (16) | 0.0099 (14) | −0.0071 (16) |
| C3 | 0.056 (2) | 0.067 (2) | 0.060 (2) | −0.0097 (17) | 0.0196 (17) | −0.0206 (19) |
| C4 | 0.088 (3) | 0.097 (3) | 0.083 (3) | −0.003 (2) | 0.023 (2) | −0.034 (3) |
| C5 | 0.119 (4) | 0.159 (5) | 0.066 (3) | 0.015 (4) | 0.027 (3) | −0.038 (3) |
| C6 | 0.097 (3) | 0.160 (5) | 0.044 (2) | 0.008 (3) | 0.018 (2) | −0.009 (3) |
| C7 | 0.051 (2) | 0.108 (3) | 0.048 (2) | −0.007 (2) | 0.0077 (16) | 0.021 (2) |
| C8 | 0.111 (3) | 0.045 (2) | 0.124 (4) | 0.000 (2) | 0.045 (3) | −0.009 (2) |
| C9 | 0.057 (2) | 0.052 (2) | 0.079 (3) | 0.0015 (16) | 0.0093 (18) | −0.0180 (19) |
| C10 | 0.0513 (19) | 0.0493 (18) | 0.057 (2) | −0.0025 (15) | 0.0122 (15) | −0.0081 (16) |
| C11 | 0.066 (2) | 0.065 (2) | 0.056 (2) | −0.0009 (18) | 0.0165 (17) | −0.0114 (18) |
| C12 | 0.097 (3) | 0.100 (3) | 0.057 (3) | 0.000 (3) | 0.019 (2) | −0.018 (2) |
| C13 | 0.111 (4) | 0.099 (4) | 0.088 (4) | 0.006 (3) | 0.027 (3) | −0.043 (3) |
| C14 | 0.083 (3) | 0.071 (3) | 0.102 (3) | 0.015 (2) | 0.016 (2) | −0.033 (3) |
| C15 | 0.124 (4) | 0.096 (3) | 0.058 (2) | 0.001 (3) | 0.025 (2) | 0.022 (2) |
| C17 | 0.079 (3) | 0.075 (3) | 0.113 (4) | 0.017 (2) | 0.018 (3) | 0.047 (3) |
| C18 | 0.108 (3) | 0.080 (3) | 0.094 (3) | 0.005 (3) | 0.011 (3) | 0.041 (3) |
| C19 | 0.0625 (19) | 0.080 (2) | 0.0614 (18) | 0.0125 (16) | 0.0121 (16) | 0.0107 (18) |
| C20 | 0.068 (2) | 0.105 (2) | 0.0550 (18) | 0.0168 (19) | 0.0113 (16) | 0.0098 (18) |
| C16 | 0.061 (2) | 0.0391 (18) | 0.102 (3) | 0.0030 (16) | 0.004 (2) | −0.006 (2) |
Geometric parameters (Å, °)
| Ni1—O1 | 1.838 (2) | C3—C4 | 1.371 (5) |
| Ni1—N2 | 1.839 (3) | C4—C5 | 1.387 (6) |
| Ni1—N1 | 1.840 (3) | C4—H4 | 0.9300 |
| Ni1—O2 | 1.8457 (19) | C5—C6 | 1.361 (7) |
| Na1—O5 | 2.288 (2) | C5—H5 | 0.9300 |
| Na1—O1 | 2.362 (2) | C6—H6 | 0.9300 |
| Na1—N5 | 2.368 (4) | C7—H7 | 0.9300 |
| Na1—O2 | 2.395 (2) | C8—H8A | 0.9600 |
| Na1—O3 | 2.492 (3) | C8—H8B | 0.9600 |
| Na1—O4 | 2.555 (2) | C8—H8C | 0.9600 |
| O1—C10 | 1.314 (4) | C9—C10 | 1.412 (4) |
| O2—C2 | 1.310 (3) | C9—C16 | 1.413 (5) |
| O3—C11 | 1.365 (4) | C9—C14 | 1.419 (5) |
| O3—C15 | 1.420 (4) | C10—C11 | 1.410 (5) |
| O4—C3 | 1.365 (4) | C11—C12 | 1.375 (5) |
| O4—C8 | 1.429 (4) | C12—C13 | 1.389 (6) |
| O5—H5A | 0.8200 | C12—H12 | 0.9300 |
| O5—H5B | 0.8246 | C13—C14 | 1.349 (6) |
| N1—C16 | 1.276 (4) | C13—H13 | 0.9300 |
| N1—C17 | 1.476 (4) | C14—H14 | 0.9300 |
| N2—C7 | 1.282 (4) | C15—H15A | 0.9600 |
| N2—C18 | 1.468 (5) | C15—H15B | 0.9600 |
| N3—C19 | 1.292 (5) | C15—H15C | 0.9600 |
| N3—C20 | 1.297 (5) | C17—C18 | 1.436 (6) |
| N4—C19 | 1.127 (4) | C17—H16A | 0.9700 |
| N5—C20 | 1.127 (4) | C17—H16B | 0.9700 |
| C1—C6 | 1.405 (5) | C18—H17A | 0.9700 |
| C1—C2 | 1.411 (5) | C18—H17B | 0.9700 |
| C1—C7 | 1.425 (5) | C16—H20 | 0.9300 |
| C2—C3 | 1.419 (5) | ||
| O1—Ni1—N2 | 178.09 (11) | C6—C5—C4 | 120.6 (4) |
| O1—Ni1—N1 | 94.79 (11) | C6—C5—H5 | 119.7 |
| N2—Ni1—N1 | 86.76 (14) | C4—C5—H5 | 119.7 |
| O1—Ni1—O2 | 83.62 (9) | C5—C6—C1 | 120.5 (4) |
| N2—Ni1—O2 | 94.82 (11) | C5—C6—H6 | 119.7 |
| N1—Ni1—O2 | 178.41 (11) | C1—C6—H6 | 119.7 |
| O5—Na1—O1 | 120.42 (9) | N2—C7—C1 | 125.7 (3) |
| O5—Na1—N5 | 101.85 (12) | N2—C7—H7 | 117.1 |
| O1—Na1—N5 | 127.99 (12) | C1—C7—H7 | 117.1 |
| O5—Na1—O2 | 116.86 (9) | O4—C8—H8A | 109.5 |
| O1—Na1—O2 | 62.15 (7) | O4—C8—H8B | 109.5 |
| N5—Na1—O2 | 124.88 (11) | H8A—C8—H8B | 109.5 |
| O5—Na1—O3 | 90.51 (9) | O4—C8—H8C | 109.5 |
| O1—Na1—O3 | 64.15 (8) | H8A—C8—H8C | 109.5 |
| N5—Na1—O3 | 88.43 (11) | H8B—C8—H8C | 109.5 |
| O2—Na1—O3 | 126.30 (8) | C10—C9—C16 | 121.1 (3) |
| O5—Na1—O4 | 84.15 (8) | C10—C9—C14 | 118.6 (4) |
| O1—Na1—O4 | 124.77 (9) | C16—C9—C14 | 120.2 (4) |
| N5—Na1—O4 | 85.68 (11) | O1—C10—C11 | 118.0 (3) |
| O2—Na1—O4 | 62.63 (7) | O1—C10—C9 | 123.4 (3) |
| O3—Na1—O4 | 171.07 (9) | C11—C10—C9 | 118.6 (3) |
| C10—O1—Ni1 | 127.8 (2) | O3—C11—C12 | 125.5 (4) |
| C10—O1—Na1 | 124.3 (2) | O3—C11—C10 | 113.8 (3) |
| Ni1—O1—Na1 | 107.87 (9) | C12—C11—C10 | 120.7 (4) |
| C2—O2—Ni1 | 127.3 (2) | C11—C12—C13 | 120.4 (4) |
| C2—O2—Na1 | 125.61 (19) | C11—C12—H12 | 119.8 |
| Ni1—O2—Na1 | 106.26 (9) | C13—C12—H12 | 119.8 |
| C11—O3—C15 | 118.3 (3) | C14—C13—C12 | 120.3 (4) |
| C11—O3—Na1 | 119.68 (19) | C14—C13—H13 | 119.8 |
| C15—O3—Na1 | 122.0 (2) | C12—C13—H13 | 119.8 |
| C3—O4—C8 | 118.2 (3) | C13—C14—C9 | 121.3 (4) |
| C3—O4—Na1 | 119.73 (19) | C13—C14—H14 | 119.3 |
| C8—O4—Na1 | 122.1 (2) | C9—C14—H14 | 119.3 |
| Na1—O5—H5A | 130.6 | O3—C15—H15A | 109.5 |
| Na1—O5—H5B | 118.8 | O3—C15—H15B | 109.5 |
| H5A—O5—H5B | 110.0 | H15A—C15—H15B | 109.5 |
| C16—N1—C17 | 119.2 (3) | O3—C15—H15C | 109.5 |
| C16—N1—Ni1 | 126.4 (2) | H15A—C15—H15C | 109.5 |
| C17—N1—Ni1 | 114.3 (3) | H15B—C15—H15C | 109.5 |
| C7—N2—C18 | 118.9 (3) | C18—C17—N1 | 110.7 (3) |
| C7—N2—Ni1 | 126.8 (3) | C18—C17—H16A | 109.5 |
| C18—N2—Ni1 | 114.2 (3) | N1—C17—H16A | 109.5 |
| C19—N3—C20 | 120.5 (3) | C18—C17—H16B | 109.5 |
| C20—N5—Na1 | 171.8 (4) | N1—C17—H16B | 109.5 |
| C6—C1—C2 | 119.6 (4) | H16A—C17—H16B | 108.1 |
| C6—C1—C7 | 119.2 (4) | C17—C18—N2 | 111.4 (3) |
| C2—C1—C7 | 121.1 (3) | C17—C18—H17A | 109.4 |
| O2—C2—C1 | 123.9 (3) | N2—C18—H17A | 109.4 |
| O2—C2—C3 | 117.9 (3) | C17—C18—H17B | 109.4 |
| C1—C2—C3 | 118.2 (3) | N2—C18—H17B | 109.4 |
| O4—C3—C4 | 126.1 (4) | H17A—C18—H17B | 108.0 |
| O4—C3—C2 | 113.4 (3) | N4—C19—N3 | 173.6 (4) |
| C4—C3—C2 | 120.4 (4) | N5—C20—N3 | 173.8 (4) |
| C3—C4—C5 | 120.5 (4) | N1—C16—C9 | 126.5 (3) |
| C3—C4—H4 | 119.7 | N1—C16—H20 | 116.7 |
| C5—C4—H4 | 119.7 | C9—C16—H20 | 116.7 |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O5—H5A···N3i | 0.82 | 2.14 | 2.960 (4) | 175 |
| O5—H5B···N4ii | 0.82 | 2.03 | 2.852 (4) | 177 |
Symmetry codes: (i) x−1, y, z; (ii) x−1, −y+1/2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HG2500).
References
- Bruker (2001). SAINT-Plus Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2004). APEX2 Bruker AXS Inc., Madison, Wisconsin, USA.
- Correia, I., Duarte, M. T., Piedade, M. F. M., Jackush, T., Kiss, T., Castro, M. M., Geraldes, C. A., Carlos, F. G. C. & Avecilla, F. (2005). Eur. J. Inorg. Chem. pp. 732–744.
- Costes, J.-P., Novitchi, G., Shova, S., Dahan, F., Donnadieu, B. & Tuchagues, J.-P. (2004). Inorg. Chem.43, 7792–7799. [DOI] [PubMed]
- Ohba, M. & Okawa, H. (2000). Coord. Chem. Rev.198, 313–328.
- Sheldrick, G. M. (2003). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S160053680901438X/hg2500sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680901438X/hg2500Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report