Abstract
The title compound, C16H20N4O3S·0.25CH4O·0.25H2O, is a hydrate/methanol solvate of torasemide, a diuretic drug used in the treatment of hypertension. The asymmetric unit contains two torasemide molecules and half-occupied methanol and water molecules. It is isomorphous with the previously reported nonsolvated T–II form of torasemide. The water molecules contribute to the stability of the structure by participating in an extensive system of O—H⋯O hydrogen bonds; N—H⋯N and N—H⋯O hydrogen bonds are also present. Both asymmetric molecules of torasemide form inversion dimers in the crystal.
Related literature
For background on the medicinal properties and polymorphism of torasemide, see: Uchida et al. (1991 ▶); Broekhuysen et al. (1986 ▶); Ghys et al. (1985 ▶); Ishido & Senzaki (2008 ▶); Cosin & Diez (2002 ▶); Murray et al. (2001 ▶); Dupont et al. (1978 ▶); Danilovski et al. (2001 ▶).
Experimental
Crystal data
C16H20N4O3S·0.25CH4O·0.25H2O
M r = 360.94
Monoclinic,

a = 16.8477 (2) Å
b = 11.5951 (1) Å
c = 20.3256 (2) Å
β = 108.646 (1)°
V = 3762.21 (7) Å3
Z = 8
Cu Kα radiation
μ = 1.74 mm−1
T = 200 K
0.45 × 0.38 × 0.12 mm
Data collection
Oxford Diffraction Xcalibur PX Ultra CCD diffractometer
Absorption correction: multi-scan (ABSPACK; Oxford Diffraction, 2006 ▶) T min = 0.502, T max = 1.000 (expected range = 0.407–0.811)
50677 measured reflections
7402 independent reflections
6976 reflections with I > 2σ(I)
R int = 0.031
Refinement
R[F 2 > 2σ(F 2)] = 0.052
wR(F 2) = 0.156
S = 1.04
7402 reflections
496 parameters
6 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.95 e Å−3
Δρmin = −0.57 e Å−3
Data collection: CrysAlisPro CCD (Oxford Diffraction, 2006 ▶); cell refinement: CrysAlisPro CCD; data reduction: CrysAlisPro RED (Oxford Diffraction, 2006 ▶); program(s) used to solve structure: SIR97 (Altomare et al., 1999 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶) and PLATON (Spek, 2009 ▶); software used to prepare material for publication: SHELXL97 (Sheldrick, 2008 ▶), WinGX (Farrugia, 1999 ▶) and PARST (Nardelli, 1995 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S160053680901160X/hb2934sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680901160X/hb2934Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N⋯N3 | 0.83 (3) | 2.37 (3) | 2.986 (2) | 131 (2) |
| N1—H1N⋯N3i | 0.83 (3) | 2.38 (3) | 3.060 (2) | 139 (2) |
| N2—H2N⋯O5ii | 0.84 (3) | 2.14 (3) | 2.847 (2) | 142 (3) |
| N2—H2N⋯O6ii | 0.84 (3) | 2.21 (3) | 2.822 (2) | 130 (2) |
| N4—H4N⋯O1i | 0.90 (3) | 2.02 (3) | 2.919 (2) | 174 (3) |
| N5—H5N⋯N7 | 0.86 (3) | 2.21 (3) | 2.910 (2) | 138 (2) |
| N5—H5N⋯N7iii | 0.86 (3) | 2.47 (3) | 3.098 (2) | 130 (2) |
| N6—H6N⋯O2iv | 0.92 (3) | 2.53 (3) | 3.036 (2) | 115 (2) |
| N6—H6N⋯O3iv | 0.92 (3) | 1.79 (3) | 2.667 (2) | 158 (2) |
| N8—H8N⋯O4iii | 0.85 (3) | 2.21 (3) | 3.034 (3) | 164 (3) |
| O7—H7O⋯O8 | 0.923 (8) | 1.70 (5) | 2.480 (8) | 140 (7) |
| O7—H7O⋯O8v | 0.923 (8) | 2.50 (10) | 2.995 (9) | 114 (8) |
| O8—H82O⋯O6 | 0.75 (5) | 2.00 (6) | 2.735 (5) | 166 (11) |
| O8—H81O⋯O6v | 0.75 (5) | 2.02 (6) | 2.721 (5) | 158 (12) |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  .
.
Acknowledgments
The authors acknowledge financial support from the Italian Ministero dell’Istruzione, dell’Universitá e della Ricerca.
supplementary crystallographic information
Comment
Torasemide has been developed as a longlasting loop diuretic that combines the effects of both furosemide and spironolactone (Uchida et al., 1991; Broekhuysen et al., 1986; Ghys et al., 1985). It is used in the treatment of hypertension and of edema associated with congestive heart failure (Ishido & Senzaki, 2008). Torasemide has been reported to be more effective than furosemide in chronic heart failure, with a lower mortality (Cosin & Diez, 2002; Murray et al., 2001).
Three polymorphic forms have been reported up to now for torasemide, denoted T–I, T–II (Dupont et al., 1978) and T–N (Danilovski et al., 2001). We have obtained crystals of a solvate (I) which, besides being isomorphous with form T–II (with the a and c unit cell axes interchanged), contains water and methanol molecules interspersed in the lattice. The solvate molecules are disorderly arranged in proximity of a twofold rotation axis and one molecule of each type resides, with 0.50 occupancy, in the asymmetric unit of the monoclinic unit cell which comprises, in addition, two independent torasemide molecules (Fig. 1), as found for form T–II. Although data collection for I was carried out at a temperature of 200 K, only a 0.8% decrease in the cell volume was found with respect to the room–temperature value of T–II; this might be ascribed in part to the presence of the additional solvate molecules in the lattice of I. The 200 K data collection temperature represented a compromise between conditions of high thermal motion at room temperature, particularly affecting the terminal methyl groups of the chain, and crystal deterioration at lower temperatures. The conformations of the two independent molecules in the structure of I (respectively formed by carbon atoms C1 to C16 and C17 to C32, hereafter referred to as molecules A and B, in the above order, for consistency with previous notation) are similar to those of the A and B molecules of the T–II form, with an 8.4° largest difference in the value of the C22—N8—C23—C24 torsion angle (this corresponds to the C6—N4—C7—C9 chain of molecule B of the T–II form, according to Dupont's notation for the non–solvated form). The present values of the dihedral angles between the best planes through the phenyl and pyridyl rings in the two molecules, of 61.18 (7)° (A) and 78.71 (7)° (B), match those reported (61.2° and 79.1°) for the corresponding molecules of form T—II.
It should be noted that the present set of data, contrary to the older one for T–II, has allowed to assign unambiguously and refine the positions of all hydrogen atoms attached to the N atoms in the two molecules. In particular, the N—H amine bond formed by N1 in molecule A and by N5 in B, is almost parallel, in each case, to the plane of the pyridyl ring, yielding a substantially planar immediate environment of the nitrogen atom, whereas such bond is considerably displaced from the plane of the phenyl ring. This may rationalize the large difference, ca 0.09 Å, between the lengths of the two N—C bonds formed by N1 and, respectively, N5 in the two molecules. Indeed, as a consequence of the above N—H bond orientations, the nitrogen lone pair may favourably interact with antibonding orbitals of the pyridyl ring, but not with those of the phenyl ring, yielding shorter N1—C1 and N5—C17 bonds compared to the N1—C10 and N5—C26 ones (with the additional consequence that the two contiguous C—C bonds in each pyridyl ring are the longest of all bonds in the present rings).
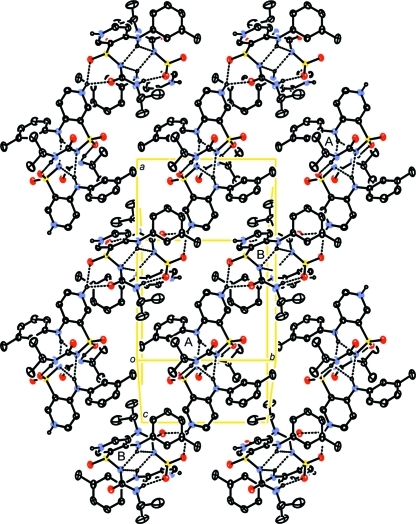
The crystal structure is stabilized by an extensive system of hydrogen bonds (Table 1), several of these in bifurcated mode. There are centrosymmetric dimers of molecule A, internally connnected by the N1···N3 (N1···N3 = 2.986 (2) Å, N1—H1N···N3 = 131 (2)°), N1···N3i (N1···N3i = 3.060 (2) Å, N1—H1N···N3i = 139 (2)°; symmetry code (i): - x, 1 - y, - z) and N4···O1i (N4···O1i = 2.919 (2) Å, N4—H4N···O1i = 174 (3)°) hydrogen bonds and centrosymmetric dimers of molecule B, linked by the N5···N7 (N5···N7 = 2.910 (2) Å, N5—H5N···N7 = 138 (2)°), N5···N7iii (N5···N7iii = 3.098 (2) Å, N5—H5N···N7iii = 130 (2)°; symmetry code (iii): 1 - x, - y, - z) and N8···O4iii (N8···O4iii = 3.034 (3) Å, N8—H8N···O4iii = 164 (3)° hydrogen bonds. Dimers of the above two types, connected through the N2···O5ii (N2···O5ii = 2.847 (2) Å, N2—H2N···O5ii = 142 (3)°; symmetry code (ii): 1 - x, 1 - y, - z) and N2···O6ii (N2···O6ii = 2.822 (2) Å, N2—H2N···O6ii = 130 (2)°) hydrogen bonds, form chains in an AABBAA fashion, which are stacked, with no intervening hydrogen bonds among them, on planes parallel to the ab cell face (Fig. 2). On contiguous planes of this set, spaced by c/2 intervals, the chains are alternatively parallel to the [110] and [1–10] directions. Furthermore, a three–dimensional network arises since dimeric groups from the above adjacent planes are connected through the N6···O2iv (N6···O2iv = 3.036 (2) Å, N6—H6N···O2iv = 115 (2)°; symmetry code (iv): 1/2 + x, 1 - y, 1/2 + z) and N6···O3iv (N6···O3iv = 2.667 (2) Å, N6—H6N···O3iv = 158 (2)°) linkages. Also the latter system of connections gives rise to planar arrays of AABBAA chains, these being parallel to the bc cell face (Fig. 3). Also in this case, there are alternative [01–1] and [011] chain orientations on adjacent planes, at a/2 intervals. It is remarkable that a similar arrangement (with one exception, see the accompanying paper) is found for the structure of the T–N polymorph, in spite of quite different cell settings. The (disordered) water molecule bridges between the chains (Fig. 4), through the O8···O6 (O8···O6 = 2.735 (5) Å, O8—H82O···O6 = 166 (11)°) and O8···O6v (O8···O6v = 2.721 (5) Å, O8—H81O···O6v = 158 (12)°; symmetry code (v): 3/2 - x, y, 1/2 - z) hydrogen bonds and each water fraction accepts one such bond, O7···O8 (O7···O8 = 2.480 (8) Å, O7—H7O···O8 = 140 (7)°), from the closest fraction of the methanol molecule.
Experimental
Samples of torasemide were kindly provided by SIMS (SIMS srl, Reggello Firenze, Italy). Crystals of (I), in the form of colourless prisms suitable for X-ray diffraction analysis, were obtained by slow evaporation from 2:1 methanol:butanol solutions. The presence of a small amount of water molecules in the structure may be rationalized considering that the operations were not performed in completely anhydrous conditions.
Refinement
H atoms bound to carbon atoms were in geometrically generated positions, riding. The coordinates of those bound to the N atoms of the torasemide molecules were refined freely, whereas those of H atoms of the solvent molecules were refined with geometric restraints. The constraint U(H) = 1.2Ueq(C,N) on hydrogen temperature factors was applied [U(H) = 1.5Ueq(C) for the H atoms of methyl groups and solvate molecules]. The N—H bond distances formed by refined hydrogen atoms were in the range 0.835 – 0.917 Å, the H2O O—H bonds refined to 0.75 (5) Å and the methanol O—H to 0.923 (8) Å.
Figures
Fig. 1.
A view of the content of the asymmetric unit of (I), where the sites of the solvate water and methanol molecules have 0.50 occupancy. Displacement ellipsoids are drawn at the 50% probability level.
Fig. 2.
A view of the crystal packing in the structure of (I), in proximity of the ab face. Hydrogen bonds are denoted by dashed lines. Only hydrogen atoms involved in the formation of hydrogen bonds are shown. Chains of hydrogen–bonded molecules, parallel to the [110] direction, are formed by centrosymmetric dimers of the two types of symmetry–independent molecules (denoted by the A and B letters, respectively). The solvent molecules, not shown in this drawing, lie between this layer and the parallel one at c = 1/2, where the chains of hydrogen–bonded molecules exhibit the alternative [1–10] orientation.
Fig. 3.
The arrangement of chains, parallel to the [01–1] direction, of hydrogen–bonded molecules in proximity of the bc face. A similar arrangement, however with the alternative [011] chain orientation, exists on the parallel flanking planes at a/2 distance from the one shown.
Fig. 4.
A view of the packing in the structure of (I) showing channels hosting the solvent molecules. Dimers of A or B molecules aligned along b, according to this view, are not connected by hydrogen bonds.
Crystal data
| C16H20N4O3S·0.25CH4O·0.25H2O | F(000) = 1528 |
| Mr = 360.94 | Dx = 1.274 Mg m−3 |
| Monoclinic, P2/n | Cu Kα radiation, λ = 1.54180 Å |
| Hall symbol: -P 2yac | Cell parameters from 37208 reflections |
| a = 16.8477 (2) Å | θ = 4.1–72.4° |
| b = 11.5951 (1) Å | µ = 1.74 mm−1 |
| c = 20.3256 (2) Å | T = 200 K |
| β = 108.646 (1)° | Prism, colorless |
| V = 3762.21 (7) Å3 | 0.45 × 0.38 × 0.12 mm |
| Z = 8 |
Data collection
| Oxford Diffraction Xcalibur PX Ultra CCD diffractometer | 7402 independent reflections |
| Radiation source: fine-focus sealed tube | 6976 reflections with I > 2σ(I) |
| Oxford Diffraction Enhance ULTRA assembly | Rint = 0.031 |
| Detector resolution: 8.1241 pixels mm-1 | θmax = 72.9°, θmin = 4.1° |
| ω scans | h = −17→20 |
| Absorption correction: multi-scan (ABSPACK; Oxford Diffraction, 2006) | k = −13→13 |
| Tmin = 0.502, Tmax = 1.000 | l = −25→25 |
| 50677 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.052 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.156 | w = 1/[σ2(Fo2) + (0.0978P)2 + 2.03P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.04 | (Δ/σ)max = 0.001 |
| 7402 reflections | Δρmax = 0.95 e Å−3 |
| 496 parameters | Δρmin = −0.57 e Å−3 |
| 6 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.00135 (17) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| C1 | 0.19533 (12) | 0.48508 (17) | −0.00966 (10) | 0.0373 (4) | |
| C2 | 0.27341 (13) | 0.43982 (19) | −0.01066 (12) | 0.0437 (5) | |
| H2 | 0.2910 | 0.3659 | 0.0087 | 0.052* | |
| C3 | 0.32285 (14) | 0.5010 (2) | −0.03890 (12) | 0.0485 (5) | |
| H3 | 0.3756 | 0.4706 | −0.0379 | 0.058* | |
| N2 | 0.29823 (12) | 0.60467 (17) | −0.06847 (10) | 0.0459 (4) | |
| H2N | 0.3334 (18) | 0.643 (2) | −0.0811 (15) | 0.055* | |
| C4 | 0.22400 (13) | 0.64928 (19) | −0.07146 (11) | 0.0420 (4) | |
| H4 | 0.2076 | 0.7215 | −0.0937 | 0.050* | |
| C5 | 0.17111 (12) | 0.59364 (17) | −0.04330 (10) | 0.0369 (4) | |
| S1 | 0.07610 (3) | 0.66425 (4) | −0.04649 (2) | 0.03510 (15) | |
| O1 | 0.08434 (9) | 0.69312 (13) | 0.02476 (7) | 0.0404 (3) | |
| O2 | 0.06917 (10) | 0.76225 (12) | −0.09190 (8) | 0.0426 (3) | |
| N3 | 0.00665 (10) | 0.56968 (14) | −0.07018 (8) | 0.0357 (3) | |
| C6 | −0.00564 (12) | 0.51828 (17) | −0.13380 (10) | 0.0365 (4) | |
| O3 | 0.02024 (10) | 0.55425 (13) | −0.18126 (8) | 0.0460 (4) | |
| N4 | −0.04911 (12) | 0.41930 (16) | −0.14092 (9) | 0.0445 (4) | |
| H4N | −0.0622 (17) | 0.389 (2) | −0.1047 (15) | 0.053* | |
| C7 | −0.05800 (17) | 0.3402 (2) | −0.19833 (12) | 0.0533 (6) | |
| H7 | −0.0722 | 0.3856 | −0.2424 | 0.064* | |
| C8 | −0.1274 (2) | 0.2567 (3) | −0.20331 (17) | 0.0795 (10) | |
| H81 | −0.1136 | 0.2106 | −0.1607 | 0.119* | |
| H82 | −0.1346 | 0.2058 | −0.2433 | 0.119* | |
| H83 | −0.1795 | 0.2992 | −0.2092 | 0.119* | |
| C9 | 0.0240 (2) | 0.2749 (3) | −0.1887 (2) | 0.0837 (10) | |
| H91 | 0.0699 | 0.3303 | −0.1823 | 0.126* | |
| H92 | 0.0186 | 0.2277 | −0.2299 | 0.126* | |
| H93 | 0.0361 | 0.2251 | −0.1477 | 0.126* | |
| N1 | 0.14707 (11) | 0.42831 (15) | 0.02100 (10) | 0.0403 (4) | |
| H1N | 0.0987 (18) | 0.452 (2) | 0.0161 (14) | 0.048* | |
| C10 | 0.17350 (13) | 0.32972 (18) | 0.06475 (11) | 0.0404 (4) | |
| C11 | 0.12689 (15) | 0.23004 (18) | 0.04998 (12) | 0.0445 (5) | |
| H11 | 0.0792 | 0.2260 | 0.0095 | 0.053* | |
| C12 | 0.14906 (17) | 0.1347 (2) | 0.09404 (14) | 0.0527 (6) | |
| C13 | 0.21970 (18) | 0.1432 (2) | 0.15223 (14) | 0.0602 (7) | |
| H13 | 0.2363 | 0.0787 | 0.1824 | 0.072* | |
| C14 | 0.26604 (18) | 0.2426 (3) | 0.16715 (15) | 0.0632 (7) | |
| H14 | 0.3140 | 0.2464 | 0.2074 | 0.076* | |
| C15 | 0.24336 (16) | 0.3373 (2) | 0.12404 (13) | 0.0542 (6) | |
| H15 | 0.2750 | 0.4066 | 0.1347 | 0.065* | |
| C16 | 0.0971 (2) | 0.0273 (2) | 0.07844 (19) | 0.0770 (9) | |
| H161 | 0.1053 | −0.0119 | 0.0384 | 0.116* | |
| H162 | 0.0378 | 0.0474 | 0.0679 | 0.116* | |
| H163 | 0.1141 | −0.0241 | 0.1188 | 0.116* | |
| C17 | 0.46551 (12) | 0.07450 (16) | 0.14989 (10) | 0.0361 (4) | |
| C18 | 0.45704 (14) | 0.05823 (19) | 0.21650 (10) | 0.0427 (5) | |
| H18 | 0.4363 | −0.0129 | 0.2275 | 0.051* | |
| C19 | 0.47838 (14) | 0.14364 (19) | 0.26474 (11) | 0.0437 (5) | |
| H19 | 0.4733 | 0.1307 | 0.3094 | 0.052* | |
| N6 | 0.50654 (12) | 0.24638 (16) | 0.25054 (9) | 0.0418 (4) | |
| H6N | 0.5204 (16) | 0.304 (2) | 0.2829 (14) | 0.050* | |
| C20 | 0.51362 (13) | 0.26759 (17) | 0.18762 (11) | 0.0381 (4) | |
| H20 | 0.5332 | 0.3408 | 0.1785 | 0.046* | |
| C21 | 0.49306 (12) | 0.18543 (16) | 0.13642 (10) | 0.0344 (4) | |
| S2 | 0.50042 (3) | 0.21861 (4) | 0.05304 (2) | 0.03520 (15) | |
| O5 | 0.53461 (10) | 0.33358 (12) | 0.05809 (8) | 0.0430 (3) | |
| O4 | 0.41666 (9) | 0.20622 (13) | 0.00468 (8) | 0.0438 (3) | |
| N7 | 0.55423 (10) | 0.11763 (14) | 0.03788 (8) | 0.0360 (3) | |
| C22 | 0.63508 (13) | 0.10566 (17) | 0.08225 (10) | 0.0373 (4) | |
| O6 | 0.67497 (9) | 0.18090 (13) | 0.12360 (7) | 0.0432 (3) | |
| N8 | 0.66900 (14) | 0.00256 (19) | 0.07774 (11) | 0.0555 (5) | |
| H8N | 0.636 (2) | −0.047 (3) | 0.0531 (17) | 0.067* | |
| C23 | 0.75356 (18) | −0.0308 (3) | 0.11811 (16) | 0.0696 (8) | |
| H23 | 0.7776 | 0.0268 | 0.1559 | 0.084* | |
| C24 | 0.7492 (3) | −0.1528 (5) | 0.1507 (3) | 0.134 (2) | |
| H241 | 0.7311 | −0.1440 | 0.1917 | 0.201* | |
| H242 | 0.8046 | −0.1890 | 0.1642 | 0.201* | |
| H243 | 0.7089 | −0.2015 | 0.1164 | 0.201* | |
| C25 | 0.8076 (3) | −0.0405 (5) | 0.0741 (3) | 0.128 (2) | |
| H251 | 0.7865 | −0.1020 | 0.0398 | 0.192* | |
| H252 | 0.8649 | −0.0588 | 0.1031 | 0.192* | |
| H253 | 0.8074 | 0.0328 | 0.0500 | 0.192* | |
| N5 | 0.45039 (12) | −0.01004 (14) | 0.10282 (9) | 0.0404 (4) | |
| H5N | 0.4718 (16) | −0.004 (2) | 0.0697 (14) | 0.048* | |
| C26 | 0.41125 (13) | −0.11707 (16) | 0.10831 (10) | 0.0370 (4) | |
| C27 | 0.45708 (15) | −0.21778 (18) | 0.11739 (11) | 0.0426 (5) | |
| H27 | 0.5151 | −0.2154 | 0.1226 | 0.051* | |
| C28 | 0.41752 (17) | −0.32354 (19) | 0.11896 (11) | 0.0497 (5) | |
| C29 | 0.33272 (17) | −0.3224 (2) | 0.11144 (12) | 0.0525 (6) | |
| H29 | 0.3052 | −0.3934 | 0.1131 | 0.063* | |
| C30 | 0.28749 (16) | −0.2224 (2) | 0.10169 (13) | 0.0521 (6) | |
| H30 | 0.2293 | −0.2246 | 0.0959 | 0.063* | |
| C31 | 0.32645 (14) | −0.11830 (19) | 0.10030 (11) | 0.0440 (5) | |
| H31 | 0.2956 | −0.0483 | 0.0939 | 0.053* | |
| C32 | 0.4664 (2) | −0.4336 (2) | 0.12887 (16) | 0.0704 (8) | |
| H321 | 0.4287 | −0.4990 | 0.1264 | 0.106* | |
| H322 | 0.5100 | −0.4328 | 0.1744 | 0.106* | |
| H323 | 0.4925 | −0.4410 | 0.0923 | 0.106* | |
| O8 | 0.7099 (4) | 0.2571 (5) | 0.2572 (2) | 0.0972 (18) | 0.50 |
| H81O | 0.740 (6) | 0.221 (10) | 0.284 (4) | 0.146* | 0.50 |
| H82O | 0.706 (7) | 0.228 (10) | 0.223 (4) | 0.146* | 0.50 |
| O7 | 0.6701 (3) | 0.4556 (5) | 0.2137 (3) | 0.0920 (14) | 0.50 |
| H7O | 0.679 (6) | 0.397 (2) | 0.2459 (11) | 0.138* | 0.50 |
| C33 | 0.7049 (4) | 0.5514 (6) | 0.2484 (5) | 0.087 (2) | 0.50 |
| H331 | 0.6809 | 0.5664 | 0.2856 | 0.131* | 0.50 |
| H332 | 0.6935 | 0.6171 | 0.2164 | 0.131* | 0.50 |
| H333 | 0.7656 | 0.5405 | 0.2685 | 0.131* | 0.50 |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0406 (10) | 0.0365 (10) | 0.0361 (9) | −0.0021 (8) | 0.0140 (8) | 0.0001 (8) |
| C2 | 0.0443 (11) | 0.0418 (11) | 0.0480 (11) | 0.0013 (8) | 0.0192 (9) | −0.0009 (9) |
| C3 | 0.0451 (11) | 0.0534 (13) | 0.0518 (12) | −0.0019 (10) | 0.0221 (10) | −0.0072 (10) |
| N2 | 0.0467 (10) | 0.0478 (10) | 0.0502 (10) | −0.0092 (8) | 0.0254 (8) | −0.0023 (8) |
| C4 | 0.0476 (11) | 0.0412 (11) | 0.0407 (10) | −0.0066 (9) | 0.0188 (9) | −0.0011 (8) |
| C5 | 0.0421 (10) | 0.0360 (10) | 0.0341 (9) | −0.0047 (8) | 0.0142 (8) | 0.0008 (7) |
| S1 | 0.0427 (3) | 0.0318 (3) | 0.0331 (3) | −0.00044 (17) | 0.01538 (19) | 0.00419 (17) |
| O1 | 0.0506 (8) | 0.0383 (7) | 0.0344 (7) | −0.0024 (6) | 0.0165 (6) | 0.0000 (6) |
| O2 | 0.0553 (8) | 0.0328 (7) | 0.0412 (8) | −0.0009 (6) | 0.0174 (6) | 0.0084 (6) |
| N3 | 0.0400 (8) | 0.0349 (8) | 0.0341 (8) | −0.0001 (6) | 0.0146 (6) | 0.0034 (6) |
| C6 | 0.0414 (10) | 0.0349 (9) | 0.0343 (9) | 0.0022 (8) | 0.0139 (8) | 0.0068 (7) |
| O3 | 0.0672 (10) | 0.0375 (8) | 0.0396 (7) | −0.0043 (7) | 0.0258 (7) | 0.0047 (6) |
| N4 | 0.0593 (11) | 0.0420 (10) | 0.0351 (9) | −0.0114 (8) | 0.0191 (8) | 0.0007 (7) |
| C7 | 0.0709 (15) | 0.0504 (13) | 0.0381 (11) | −0.0137 (11) | 0.0166 (10) | −0.0022 (9) |
| C8 | 0.102 (2) | 0.077 (2) | 0.0585 (16) | −0.0408 (19) | 0.0252 (16) | −0.0156 (15) |
| C9 | 0.093 (2) | 0.074 (2) | 0.081 (2) | 0.0160 (17) | 0.0221 (18) | −0.0136 (17) |
| N1 | 0.0387 (9) | 0.0367 (9) | 0.0487 (10) | 0.0042 (7) | 0.0183 (7) | 0.0114 (7) |
| C10 | 0.0435 (10) | 0.0377 (10) | 0.0435 (11) | 0.0053 (8) | 0.0185 (9) | 0.0081 (8) |
| C11 | 0.0512 (12) | 0.0373 (11) | 0.0488 (12) | 0.0033 (9) | 0.0212 (10) | 0.0044 (9) |
| C12 | 0.0706 (15) | 0.0356 (11) | 0.0616 (14) | 0.0065 (10) | 0.0345 (12) | 0.0070 (10) |
| C13 | 0.0782 (17) | 0.0478 (13) | 0.0590 (15) | 0.0194 (12) | 0.0279 (13) | 0.0203 (11) |
| C14 | 0.0624 (15) | 0.0654 (16) | 0.0553 (14) | 0.0104 (13) | 0.0098 (12) | 0.0166 (12) |
| C15 | 0.0526 (13) | 0.0508 (14) | 0.0545 (14) | −0.0008 (10) | 0.0103 (10) | 0.0092 (10) |
| C16 | 0.107 (2) | 0.0411 (14) | 0.089 (2) | −0.0082 (14) | 0.0399 (19) | 0.0062 (14) |
| C17 | 0.0435 (10) | 0.0321 (9) | 0.0348 (9) | −0.0025 (8) | 0.0156 (8) | −0.0036 (7) |
| C18 | 0.0576 (12) | 0.0384 (10) | 0.0365 (10) | −0.0092 (9) | 0.0214 (9) | −0.0039 (8) |
| C19 | 0.0543 (12) | 0.0451 (11) | 0.0353 (10) | −0.0058 (9) | 0.0192 (9) | −0.0050 (8) |
| N6 | 0.0520 (10) | 0.0380 (9) | 0.0374 (9) | −0.0038 (8) | 0.0172 (7) | −0.0111 (7) |
| C20 | 0.0434 (10) | 0.0323 (9) | 0.0404 (10) | −0.0012 (8) | 0.0159 (8) | −0.0047 (8) |
| C21 | 0.0401 (9) | 0.0297 (9) | 0.0359 (9) | −0.0021 (7) | 0.0159 (8) | −0.0042 (7) |
| S2 | 0.0431 (3) | 0.0301 (3) | 0.0348 (3) | −0.00235 (17) | 0.0158 (2) | −0.00089 (16) |
| O5 | 0.0552 (8) | 0.0292 (7) | 0.0493 (8) | −0.0046 (6) | 0.0234 (7) | 0.0004 (6) |
| O4 | 0.0443 (8) | 0.0432 (8) | 0.0416 (8) | −0.0003 (6) | 0.0104 (6) | 0.0008 (6) |
| N7 | 0.0446 (9) | 0.0332 (8) | 0.0332 (8) | −0.0022 (7) | 0.0166 (7) | −0.0046 (6) |
| C22 | 0.0461 (10) | 0.0366 (10) | 0.0335 (9) | −0.0023 (8) | 0.0187 (8) | −0.0019 (7) |
| O6 | 0.0512 (8) | 0.0379 (7) | 0.0385 (7) | −0.0064 (6) | 0.0113 (6) | −0.0033 (6) |
| N8 | 0.0555 (12) | 0.0475 (11) | 0.0560 (12) | 0.0099 (9) | 0.0071 (9) | −0.0164 (9) |
| C23 | 0.0596 (15) | 0.0695 (18) | 0.0684 (17) | 0.0205 (13) | 0.0045 (13) | −0.0180 (14) |
| C24 | 0.101 (3) | 0.176 (5) | 0.126 (4) | 0.047 (3) | 0.037 (3) | 0.084 (4) |
| C25 | 0.087 (3) | 0.182 (5) | 0.128 (4) | 0.060 (3) | 0.054 (3) | 0.057 (4) |
| N5 | 0.0600 (11) | 0.0299 (8) | 0.0378 (9) | −0.0102 (7) | 0.0250 (8) | −0.0059 (7) |
| C26 | 0.0514 (11) | 0.0313 (9) | 0.0307 (9) | −0.0070 (8) | 0.0163 (8) | −0.0040 (7) |
| C27 | 0.0544 (12) | 0.0383 (11) | 0.0369 (10) | 0.0019 (9) | 0.0172 (9) | −0.0022 (8) |
| C28 | 0.0794 (16) | 0.0329 (11) | 0.0373 (11) | 0.0006 (10) | 0.0192 (10) | −0.0040 (8) |
| C29 | 0.0742 (16) | 0.0402 (12) | 0.0450 (12) | −0.0181 (11) | 0.0218 (11) | −0.0050 (9) |
| C30 | 0.0544 (13) | 0.0528 (14) | 0.0514 (13) | −0.0147 (10) | 0.0201 (10) | −0.0059 (10) |
| C31 | 0.0507 (12) | 0.0417 (11) | 0.0412 (11) | −0.0013 (9) | 0.0168 (9) | −0.0015 (8) |
| C32 | 0.103 (2) | 0.0414 (13) | 0.0661 (17) | 0.0128 (14) | 0.0255 (16) | 0.0007 (12) |
| O8 | 0.109 (4) | 0.119 (4) | 0.042 (2) | 0.056 (3) | −0.006 (2) | −0.015 (3) |
| O7 | 0.080 (3) | 0.090 (3) | 0.100 (4) | −0.010 (3) | 0.020 (3) | −0.007 (3) |
| C33 | 0.049 (3) | 0.066 (4) | 0.134 (6) | 0.007 (3) | 0.013 (3) | −0.012 (4) |
Geometric parameters (Å, °)
| C1—N1 | 1.344 (3) | C18—H18 | 0.9500 |
| C1—C2 | 1.422 (3) | C19—N6 | 1.347 (3) |
| C1—C5 | 1.428 (3) | C19—H19 | 0.9500 |
| C2—C3 | 1.354 (3) | N6—C20 | 1.345 (3) |
| C2—H2 | 0.9500 | N6—H6N | 0.92 (3) |
| C3—N2 | 1.348 (3) | C20—C21 | 1.371 (3) |
| C3—H3 | 0.9500 | C20—H20 | 0.9500 |
| N2—C4 | 1.337 (3) | C21—S2 | 1.781 (2) |
| N2—H2N | 0.84 (3) | S2—O5 | 1.4428 (14) |
| C4—C5 | 1.365 (3) | S2—O4 | 1.4462 (15) |
| C4—H4 | 0.9500 | S2—N7 | 1.5704 (16) |
| C5—S1 | 1.781 (2) | N7—C22 | 1.379 (3) |
| S1—O2 | 1.4449 (14) | C22—O6 | 1.248 (2) |
| S1—O1 | 1.4491 (14) | C22—N8 | 1.341 (3) |
| S1—N3 | 1.5630 (17) | N8—C23 | 1.451 (3) |
| N3—C6 | 1.378 (3) | N8—H8N | 0.85 (3) |
| C6—O3 | 1.250 (2) | C23—C25 | 1.471 (5) |
| C6—N4 | 1.344 (3) | C23—C24 | 1.574 (6) |
| N4—C7 | 1.454 (3) | C23—H23 | 1.0000 |
| N4—H4N | 0.90 (3) | C24—H241 | 0.9800 |
| C7—C8 | 1.496 (4) | C24—H242 | 0.9800 |
| C7—C9 | 1.533 (4) | C24—H243 | 0.9800 |
| C7—H7 | 1.0000 | C25—H251 | 0.9800 |
| C8—H81 | 0.9800 | C25—H252 | 0.9800 |
| C8—H82 | 0.9800 | C25—H253 | 0.9800 |
| C8—H83 | 0.9800 | N5—C26 | 1.426 (2) |
| C9—H91 | 0.9800 | N5—H5N | 0.86 (3) |
| C9—H92 | 0.9800 | C26—C27 | 1.379 (3) |
| C9—H93 | 0.9800 | C26—C31 | 1.385 (3) |
| N1—C10 | 1.429 (3) | C27—C28 | 1.401 (3) |
| N1—H1N | 0.83 (3) | C27—H27 | 0.9500 |
| C10—C11 | 1.375 (3) | C28—C29 | 1.387 (4) |
| C10—C15 | 1.392 (3) | C28—C32 | 1.497 (3) |
| C11—C12 | 1.396 (3) | C29—C30 | 1.367 (4) |
| C11—H11 | 0.9500 | C29—H29 | 0.9500 |
| C12—C13 | 1.387 (4) | C30—C31 | 1.379 (3) |
| C12—C16 | 1.498 (4) | C30—H30 | 0.9500 |
| C13—C14 | 1.371 (4) | C31—H31 | 0.9500 |
| C13—H13 | 0.9500 | C32—H321 | 0.9800 |
| C14—C15 | 1.380 (4) | C32—H322 | 0.9800 |
| C14—H14 | 0.9500 | C32—H323 | 0.9800 |
| C15—H15 | 0.9500 | O8—H81O | 0.75 (5) |
| C16—H161 | 0.9800 | O8—H82O | 0.75 (5) |
| C16—H162 | 0.9800 | O7—C33 | 1.346 (8) |
| C16—H163 | 0.9800 | O7—H7O | 0.923 (8) |
| C17—N5 | 1.336 (2) | C33—H331 | 0.9800 |
| C17—C18 | 1.419 (3) | C33—H332 | 0.9800 |
| C17—C21 | 1.423 (3) | C33—H333 | 0.9800 |
| C18—C19 | 1.359 (3) | ||
| N1—C1—C2 | 121.69 (19) | C19—C18—H18 | 119.7 |
| N1—C1—C5 | 122.07 (18) | C17—C18—H18 | 119.7 |
| C2—C1—C5 | 116.25 (18) | N6—C19—C18 | 121.33 (19) |
| C3—C2—C1 | 120.7 (2) | N6—C19—H19 | 119.3 |
| C3—C2—H2 | 119.7 | C18—C19—H19 | 119.3 |
| C1—C2—H2 | 119.7 | C20—N6—C19 | 120.66 (18) |
| N2—C3—C2 | 120.9 (2) | C20—N6—H6N | 117.9 (17) |
| N2—C3—H3 | 119.6 | C19—N6—H6N | 121.4 (17) |
| C2—C3—H3 | 119.6 | N6—C20—C21 | 121.13 (19) |
| C4—N2—C3 | 121.00 (19) | N6—C20—H20 | 119.4 |
| C4—N2—H2N | 121 (2) | C21—C20—H20 | 119.4 |
| C3—N2—H2N | 117.2 (19) | C20—C21—C17 | 120.05 (18) |
| N2—C4—C5 | 121.6 (2) | C20—C21—S2 | 119.65 (15) |
| N2—C4—H4 | 119.2 | C17—C21—S2 | 120.30 (14) |
| C5—C4—H4 | 119.2 | O5—S2—O4 | 114.89 (9) |
| C4—C5—C1 | 119.49 (19) | O5—S2—N7 | 117.42 (9) |
| C4—C5—S1 | 117.85 (16) | O4—S2—N7 | 106.96 (9) |
| C1—C5—S1 | 122.59 (15) | O5—S2—C21 | 106.24 (9) |
| O2—S1—O1 | 114.78 (9) | O4—S2—C21 | 106.00 (9) |
| O2—S1—N3 | 117.08 (9) | N7—S2—C21 | 104.24 (9) |
| O1—S1—N3 | 107.04 (9) | C22—N7—S2 | 117.27 (13) |
| O2—S1—C5 | 105.84 (9) | O6—C22—N8 | 121.3 (2) |
| O1—S1—C5 | 105.79 (9) | O6—C22—N7 | 125.21 (18) |
| N3—S1—C5 | 105.32 (9) | N8—C22—N7 | 113.48 (18) |
| C6—N3—S1 | 117.75 (14) | C22—N8—C23 | 124.2 (2) |
| O3—C6—N4 | 120.94 (19) | C22—N8—H8N | 116 (2) |
| O3—C6—N3 | 126.50 (19) | C23—N8—H8N | 119 (2) |
| N4—C6—N3 | 112.55 (17) | N8—C23—C25 | 111.3 (3) |
| C6—N4—C7 | 122.53 (18) | N8—C23—C24 | 107.9 (3) |
| C6—N4—H4N | 120.4 (18) | C25—C23—C24 | 108.0 (4) |
| C7—N4—H4N | 115.1 (18) | N8—C23—H23 | 109.9 |
| N4—C7—C8 | 109.9 (2) | C25—C23—H23 | 109.9 |
| N4—C7—C9 | 110.6 (2) | C24—C23—H23 | 109.9 |
| C8—C7—C9 | 110.0 (3) | C23—C24—H241 | 109.5 |
| N4—C7—H7 | 108.8 | C23—C24—H242 | 109.5 |
| C8—C7—H7 | 108.8 | H241—C24—H242 | 109.5 |
| C9—C7—H7 | 108.8 | C23—C24—H243 | 109.5 |
| C7—C8—H81 | 109.5 | H241—C24—H243 | 109.5 |
| C7—C8—H82 | 109.5 | H242—C24—H243 | 109.5 |
| H81—C8—H82 | 109.5 | C23—C25—H251 | 109.5 |
| C7—C8—H83 | 109.5 | C23—C25—H252 | 109.5 |
| H81—C8—H83 | 109.5 | H251—C25—H252 | 109.5 |
| H82—C8—H83 | 109.5 | C23—C25—H253 | 109.5 |
| C7—C9—H91 | 109.5 | H251—C25—H253 | 109.5 |
| C7—C9—H92 | 109.5 | H252—C25—H253 | 109.5 |
| H91—C9—H92 | 109.5 | C17—N5—C26 | 124.70 (17) |
| C7—C9—H93 | 109.5 | C17—N5—H5N | 117.8 (18) |
| H91—C9—H93 | 109.5 | C26—N5—H5N | 116.9 (18) |
| H92—C9—H93 | 109.5 | C27—C26—C31 | 121.27 (19) |
| C1—N1—C10 | 124.46 (18) | C27—C26—N5 | 119.60 (19) |
| C1—N1—H1N | 119.7 (19) | C31—C26—N5 | 119.01 (19) |
| C10—N1—H1N | 115.8 (19) | C26—C27—C28 | 119.6 (2) |
| C11—C10—C15 | 120.5 (2) | C26—C27—H27 | 120.2 |
| C11—C10—N1 | 119.59 (19) | C28—C27—H27 | 120.2 |
| C15—C10—N1 | 119.82 (19) | C29—C28—C27 | 118.0 (2) |
| C10—C11—C12 | 120.5 (2) | C29—C28—C32 | 121.7 (2) |
| C10—C11—H11 | 119.7 | C27—C28—C32 | 120.3 (2) |
| C12—C11—H11 | 119.7 | C30—C29—C28 | 122.0 (2) |
| C13—C12—C11 | 118.2 (2) | C30—C29—H29 | 119.0 |
| C13—C12—C16 | 121.5 (2) | C28—C29—H29 | 119.0 |
| C11—C12—C16 | 120.3 (3) | C29—C30—C31 | 119.9 (2) |
| C14—C13—C12 | 121.4 (2) | C29—C30—H30 | 120.0 |
| C14—C13—H13 | 119.3 | C31—C30—H30 | 120.0 |
| C12—C13—H13 | 119.3 | C30—C31—C26 | 119.1 (2) |
| C13—C14—C15 | 120.4 (3) | C30—C31—H31 | 120.4 |
| C13—C14—H14 | 119.8 | C26—C31—H31 | 120.4 |
| C15—C14—H14 | 119.8 | C28—C32—H321 | 109.5 |
| C14—C15—C10 | 119.1 (2) | C28—C32—H322 | 109.5 |
| C14—C15—H15 | 120.5 | H321—C32—H322 | 109.5 |
| C10—C15—H15 | 120.5 | C28—C32—H323 | 109.5 |
| C12—C16—H161 | 109.5 | H321—C32—H323 | 109.5 |
| C12—C16—H162 | 109.5 | H322—C32—H323 | 109.5 |
| H161—C16—H162 | 109.5 | H81O—O8—H82O | 105.0 (10) |
| C12—C16—H163 | 109.5 | C33—O7—H7O | 107.0 (7) |
| H161—C16—H163 | 109.5 | O7—C33—H331 | 109.5 |
| H162—C16—H163 | 109.5 | O7—C33—H332 | 109.5 |
| N5—C17—C18 | 122.11 (18) | H331—C33—H332 | 109.5 |
| N5—C17—C21 | 121.63 (17) | O7—C33—H333 | 109.5 |
| C18—C17—C21 | 116.25 (17) | H331—C33—H333 | 109.5 |
| C19—C18—C17 | 120.50 (19) | H332—C33—H333 | 109.5 |
| N1—C1—C2—C3 | −176.8 (2) | N5—C17—C18—C19 | 175.5 (2) |
| C5—C1—C2—C3 | 3.5 (3) | C21—C17—C18—C19 | −3.0 (3) |
| C1—C2—C3—N2 | −1.9 (3) | C17—C18—C19—N6 | 1.2 (4) |
| C2—C3—N2—C4 | −0.8 (3) | C18—C19—N6—C20 | 0.6 (3) |
| C3—N2—C4—C5 | 1.7 (3) | C19—N6—C20—C21 | −0.5 (3) |
| N2—C4—C5—C1 | 0.1 (3) | N6—C20—C21—C17 | −1.5 (3) |
| N2—C4—C5—S1 | 176.95 (16) | N6—C20—C21—S2 | 178.14 (16) |
| N1—C1—C5—C4 | 177.7 (2) | N5—C17—C21—C20 | −175.4 (2) |
| C2—C1—C5—C4 | −2.6 (3) | C18—C17—C21—C20 | 3.1 (3) |
| N1—C1—C5—S1 | 1.0 (3) | N5—C17—C21—S2 | 5.0 (3) |
| C2—C1—C5—S1 | −179.31 (15) | C18—C17—C21—S2 | −176.49 (16) |
| C4—C5—S1—O2 | 10.41 (19) | C20—C21—S2—O5 | 3.78 (19) |
| C1—C5—S1—O2 | −172.81 (16) | C17—C21—S2—O5 | −176.60 (16) |
| C4—C5—S1—O1 | −111.79 (17) | C20—C21—S2—O4 | −118.87 (17) |
| C1—C5—S1—O1 | 64.99 (18) | C17—C21—S2—O4 | 60.75 (18) |
| C4—C5—S1—N3 | 135.05 (17) | C20—C21—S2—N7 | 128.44 (17) |
| C1—C5—S1—N3 | −48.17 (18) | C17—C21—S2—N7 | −51.94 (18) |
| O2—S1—N3—C6 | 57.06 (17) | O5—S2—N7—C22 | 54.98 (17) |
| O1—S1—N3—C6 | −172.47 (14) | O4—S2—N7—C22 | −174.19 (14) |
| C5—S1—N3—C6 | −60.20 (16) | C21—S2—N7—C22 | −62.19 (16) |
| S1—N3—C6—O3 | −15.8 (3) | S2—N7—C22—O6 | −16.3 (3) |
| S1—N3—C6—N4 | 163.11 (15) | S2—N7—C22—N8 | 163.34 (16) |
| O3—C6—N4—C7 | 9.4 (3) | O6—C22—N8—C23 | −1.3 (4) |
| N3—C6—N4—C7 | −169.6 (2) | N7—C22—N8—C23 | 179.1 (2) |
| C6—N4—C7—C8 | −164.5 (2) | C22—N8—C23—C25 | −110.2 (4) |
| C6—N4—C7—C9 | 73.9 (3) | C22—N8—C23—C24 | 131.5 (3) |
| C2—C1—N1—C10 | 10.9 (3) | C18—C17—N5—C26 | 11.2 (3) |
| C5—C1—N1—C10 | −169.43 (19) | C21—C17—N5—C26 | −170.33 (19) |
| C1—N1—C10—C11 | −125.7 (2) | C17—N5—C26—C27 | −112.5 (2) |
| C1—N1—C10—C15 | 57.9 (3) | C17—N5—C26—C31 | 71.3 (3) |
| C15—C10—C11—C12 | −0.2 (3) | C31—C26—C27—C28 | −0.4 (3) |
| N1—C10—C11—C12 | −176.6 (2) | N5—C26—C27—C28 | −176.50 (18) |
| C10—C11—C12—C13 | −0.9 (3) | C26—C27—C28—C29 | −0.2 (3) |
| C10—C11—C12—C16 | 178.8 (2) | C26—C27—C28—C32 | −179.9 (2) |
| C11—C12—C13—C14 | 1.1 (4) | C27—C28—C29—C30 | 1.0 (3) |
| C16—C12—C13—C14 | −178.5 (3) | C32—C28—C29—C30 | −179.3 (2) |
| C12—C13—C14—C15 | −0.3 (4) | C28—C29—C30—C31 | −1.1 (4) |
| C13—C14—C15—C10 | −0.9 (4) | C29—C30—C31—C26 | 0.5 (3) |
| C11—C10—C15—C14 | 1.1 (4) | C27—C26—C31—C30 | 0.2 (3) |
| N1—C10—C15—C14 | 177.5 (2) | N5—C26—C31—C30 | 176.38 (19) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···N3 | 0.83 (3) | 2.37 (3) | 2.986 (2) | 131 (2) |
| N1—H1N···N3i | 0.83 (3) | 2.38 (3) | 3.060 (2) | 139 (2) |
| N2—H2N···O5ii | 0.84 (3) | 2.14 (3) | 2.847 (2) | 142 (3) |
| N2—H2N···O6ii | 0.84 (3) | 2.21 (3) | 2.822 (2) | 130 (2) |
| N4—H4N···O1i | 0.90 (3) | 2.02 (3) | 2.919 (2) | 174 (3) |
| N5—H5N···N7 | 0.86 (3) | 2.21 (3) | 2.910 (2) | 138 (2) |
| N5—H5N···N7iii | 0.86 (3) | 2.47 (3) | 3.098 (2) | 130 (2) |
| N6—H6N···O2iv | 0.92 (3) | 2.53 (3) | 3.036 (2) | 115 (2) |
| N6—H6N···O3iv | 0.92 (3) | 1.79 (3) | 2.667 (2) | 158 (2) |
| N8—H8N···O4iii | 0.85 (3) | 2.21 (3) | 3.034 (3) | 164 (3) |
| O7—H7O···O8 | 0.92 (1) | 1.70 (5) | 2.480 (8) | 140 (7) |
| O7—H7O···O8v | 0.92 (1) | 2.50 (10) | 2.995 (9) | 114 (8) |
| O8—H82O···O6 | 0.75 (5) | 2.00 (6) | 2.735 (5) | 166 (11) |
| O8—H81O···O6v | 0.75 (5) | 2.02 (6) | 2.721 (5) | 158 (12) |
Symmetry codes: (i) −x, −y+1, −z; (ii) −x+1, −y+1, −z; (iii) −x+1, −y, −z; (iv) x+1/2, −y+1, z+1/2; (v) −x+3/2, y, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB2934).
References
- Altomare, A., Burla, M. C., Camalli, M., Cascarano, G. L., Giacovazzo, C., Guagliardi, A., Moliterni, A. G. G., Polidori, G. & Spagna, R. (1999). J. Appl. Cryst.32, 115–119.
- Broekhuysen, J., Deger, F., Douchamps, J., Ducarne, H. & Herchuelz, A. (1986). Eur. J. Clin. Pharmacol.31 Suppl, 29–34. [DOI] [PubMed]
- Cosin, J. & Diez, J. (2002). Eur. J. Heart Failure, 4, 507–513.
- Danilovski, A., Filić, D., Orešić, M. & Dumić, M. (2001). Croat. Chim. Acta, 74, 103–120.
- Dupont, L., Campsteyn, H., Lamotte, J. & Vermeire, M. (1978). Acta Cryst. B34, 2659–2662.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Ghys, A., Denef, J., de Suray, J. M., Gerin, M., Georges, A., Delarge, J. & Willems, J. (1985). Arzneim. Forschung.35, 1520–1526. [PubMed]
- Ishido, H. & Senzaki, H. (2008). Cardiovas. Hematolog. Disorders Drug Targets, 8, 127–132. [DOI] [PubMed]
- Murray, M. D., Deer, M. M., Ferguson, J. A., Dexter, P. R., Bennett, S. J., Perkins, S. M., Smith, F. E., Lane, K. A., Adams, L. D., Tierney, W. M. & Brater, D. C. (2001). Am. J. Med.111, 513–520. [DOI] [PubMed]
- Nardelli, M. (1995). J. Appl. Cryst.28, 659.
- Oxford Diffraction (2006). CrysAlisPro CCD, CrysAlisPro RED and ABSPACK in CrysAlisPro RED Oxford Diffraction Ltd, Abingdon, England.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Uchida, T., Yamanaga, K., Nishikawa, M., Ohtaki, Y., Kido, H. & Watanabe, M. (1991). Eur. J. Pharmacol.205, 145–150. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S160053680901160X/hb2934sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053680901160X/hb2934Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report