Abstract
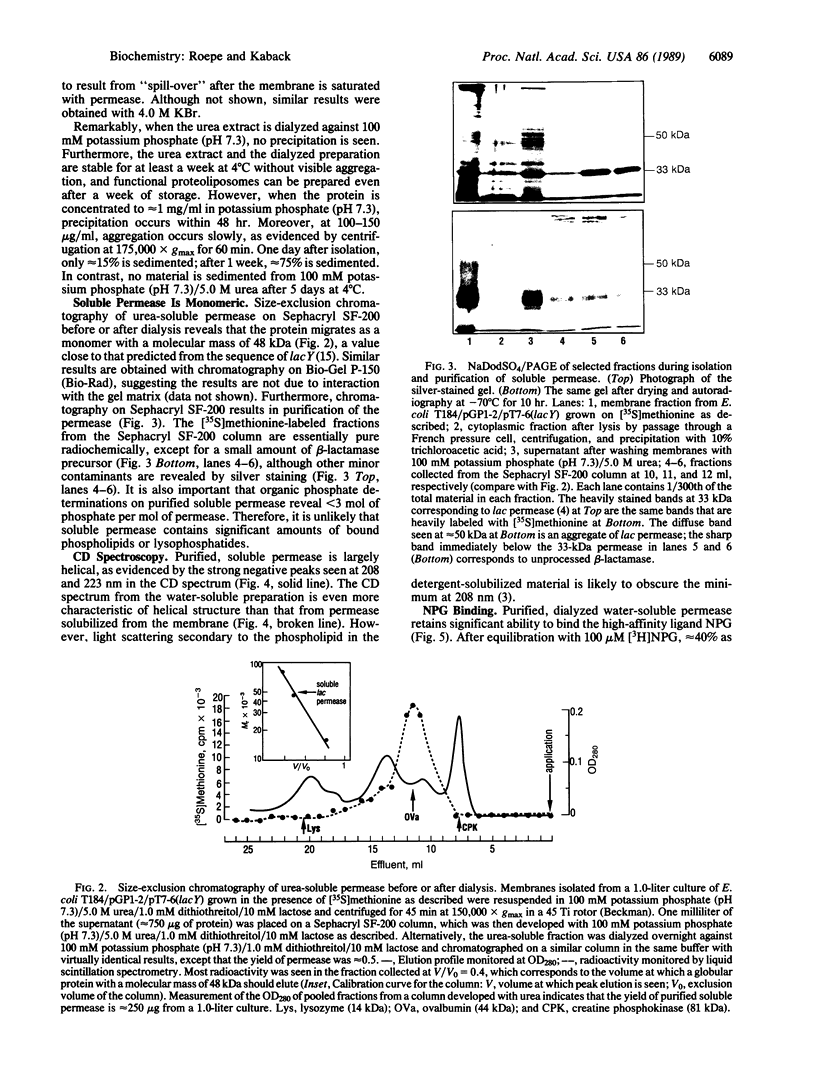
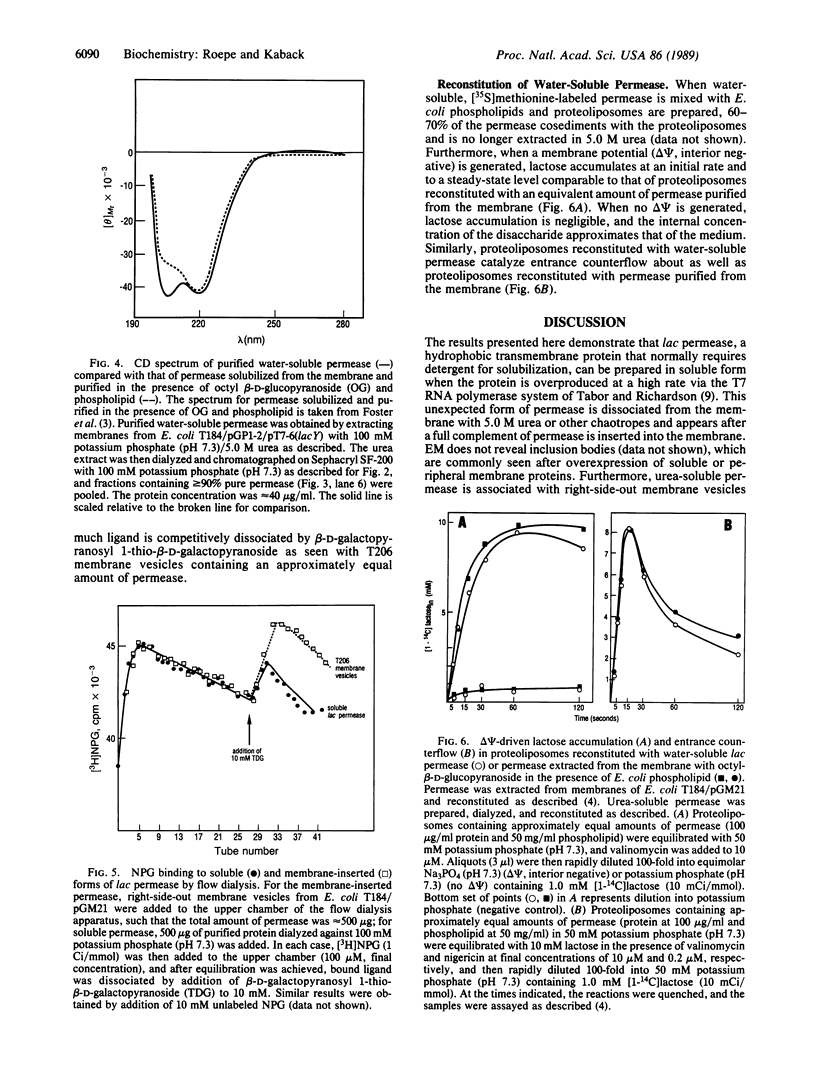
Lac permease, a polytopic membrane protein from Escherichia coli, has been purified in soluble form by overexpressing the lacY gene by means of the T7 RNA polymerase system. Soluble permease is dissociated from membranes with urea or other chaotropes and appears after the membrane is saturated with newly synthesized permease. Remarkably, this form of the permease appears to remain soluble in phosphate buffer at neutral pH after removal of urea, although it aggregates in a time- and concentration-dependent manner. Importantly, soluble permease behaves as a monomer during size-exclusion chromatography with or without urea, contains less than 3 mol of organic phosphate per mol of protein, and is largely helical. Soluble permease binds p-nitrophenyl alpha-D-galactopyranoside approximately 40% as well as permease in the native environment of the membrane and can be reconstituted into phospholipid vesicles that catalyze lactose counterflow or active transport in response to a membrane potential (interior negative). The results suggest that lac permease can assume a nondenatured conformation in aqueous solution.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldwin S. A., Henderson P. J. Homologies between sugar transporters from eukaryotes and prokaryotes. Annu Rev Physiol. 1989;51:459–471. doi: 10.1146/annurev.ph.51.030189.002331. [DOI] [PubMed] [Google Scholar]
- Büchel D. E., Gronenborn B., Müller-Hill B. Sequence of the lactose permease gene. Nature. 1980 Feb 7;283(5747):541–545. doi: 10.1038/283541a0. [DOI] [PubMed] [Google Scholar]
- Costello M. J., Escaig J., Matsushita K., Viitanen P. V., Menick D. R., Kaback H. R. Purified lac permease and cytochrome o oxidase are functional as monomers. J Biol Chem. 1987 Dec 15;262(35):17072–17082. [PubMed] [Google Scholar]
- Dornmair K. An improved purification of lactose permease. FEBS Lett. 1988 Aug 1;235(1-2):35–39. doi: 10.1016/0014-5793(88)81229-8. [DOI] [PubMed] [Google Scholar]
- Engelman D. M., Steitz T. A. The spontaneous insertion of proteins into and across membranes: the helical hairpin hypothesis. Cell. 1981 Feb;23(2):411–422. doi: 10.1016/0092-8674(81)90136-7. [DOI] [PubMed] [Google Scholar]
- Foster D. L., Boublik M., Kaback H. R. Structure of the lac carrier protein of Escherichia coli. J Biol Chem. 1983 Jan 10;258(1):31–34. [PubMed] [Google Scholar]
- Kaback H. R. Molecular biology of active transport: from membrane to molecule to mechanism. Harvey Lect. 1987;83:77–105. [PubMed] [Google Scholar]
- Kaback H. R. Site-directed mutagenesis and ion-gradient driven active transport: on the path of the proton. Annu Rev Physiol. 1988;50:243–256. doi: 10.1146/annurev.ph.50.030188.001331. [DOI] [PubMed] [Google Scholar]
- Li J., Tooth P. Size and shape of the Escherichia coli lactose permease measured in filamentous arrays. Biochemistry. 1987 Jul 28;26(15):4816–4823. doi: 10.1021/bi00389a032. [DOI] [PubMed] [Google Scholar]
- Parker M. W., Pattus F., Tucker A. D., Tsernoglou D. Structure of the membrane-pore-forming fragment of colicin A. Nature. 1989 Jan 5;337(6202):93–96. doi: 10.1038/337093a0. [DOI] [PubMed] [Google Scholar]
- Pattus F., Heitz F., Martinez C., Provencher S. W., Lazdunski C. Secondary structure of the pore-forming colicin A and its C-terminal fragment. Experimental fact and structure prediction. Eur J Biochem. 1985 Nov 4;152(3):681–689. doi: 10.1111/j.1432-1033.1985.tb09248.x. [DOI] [PubMed] [Google Scholar]
- Reeves J. P., Hong J. S., Kaback H. R. Reconstitution of D-lactate-dependent transport in membrane vesicles from a D-lactate dehydrogenase mutant of Escherichia coli. Proc Natl Acad Sci U S A. 1973 Jul;70(7):1917–1921. doi: 10.1073/pnas.70.7.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roepe P. D., Zbar R. I., Sarkar H. K., Kaback H. R. A five-residue sequence near the carboxyl terminus of the polytopic membrane protein lac permease is required for stability within the membrane. Proc Natl Acad Sci U S A. 1989 Jun;86(11):3992–3996. doi: 10.1073/pnas.86.11.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnick G., Schildiner S., Kaback H. R. Equilibrium between two forms of the lac carrier protein in energized and nonenergized membrane vesicles from Escherichia coli. Biochemistry. 1976 Nov 16;15(23):5126–5131. doi: 10.1021/bi00668a028. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teather R. M., Bramhall J., Riede I., Wright J. K., Fürst M., Aichele G., Wilhelm U., Overath P. Lactose carrier protein of Escherichia coli. Structure and expression of plasmids carrying the Y gene of the lac operon. Eur J Biochem. 1980;108(1):223–231. doi: 10.1111/j.1432-1033.1980.tb04715.x. [DOI] [PubMed] [Google Scholar]
- Viitanen P., Newman M. J., Foster D. L., Wilson T. H., Kaback H. R. Purification, reconstitution, and characterization of the lac permease of Escherichia coli. Methods Enzymol. 1986;125:429–452. doi: 10.1016/s0076-6879(86)25034-x. [DOI] [PubMed] [Google Scholar]
- Wright J. K., Overath P. Purification of the lactose:H+ carrier of Escherichia coli and characterization of galactoside binding and transport. Eur J Biochem. 1984 Feb 1;138(3):497–508. doi: 10.1111/j.1432-1033.1984.tb07944.x. [DOI] [PubMed] [Google Scholar]