Abstract
In the title compound, C31H27N3, the cyclohexene ring has an envelope configuration. In the crystal structure, there is an 34 Å3 void around the inversion center, but the low electron density (0.13 e Å−3) in the difference Fourier map suggests no solvent molecule occupying this void. No hydrogen bonding is found in the crystal structure.
Related literature
For background to organic compounds with light emitting properties, see: Tang et al. (1998 ▶); Li et al. (2003 ▶); Hye et al. (2004 ▶). For the synthesis, see: Lemke (1974 ▶); Tao & Miyata (2001 ▶). For related crystal structures, see: Kia et al. (2009 ▶); Ju et al. (2006 ▶).
Experimental
Crystal data
C31H27N3
M r = 441.56
Monoclinic,

a = 13.239 (5) Å
b = 16.757 (8) Å
c = 11.886 (3) Å
β = 104.073 (5)°
V = 2557.7 (17) Å3
Z = 4
Mo Kα radiation
μ = 0.07 mm−1
T = 293 K
0.48 × 0.19 × 0.16 mm
Data collection
Bruker SMART area-detector diffractometer
Absorption correction: none
20681 measured reflections
5879 independent reflections
3554 reflections with I > 2σ(I)
R int = 0.027
Refinement
R[F 2 > 2σ(F 2)] = 0.046
wR(F 2) = 0.167
S = 0.92
5879 reflections
309 parameters
H-atom parameters constrained
Δρmax = 0.13 e Å−3
Δρmin = −0.18 e Å−3
Data collection: SMART (Bruker, 2002 ▶); cell refinement: SAINT (Bruker, 2002 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809014378/xu2505sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809014378/xu2505Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (grant Nos. 50590403, 50402018 and 50603011).
supplementary crystallographic information
Comment
The organic compounds with donor-π-acceptor (D-π-A) structure have special light-emitting properties, and show potential application in organic light-emitting diodes (Tang et al., 1998; Hye et al., 2004). However, these molecules easily aggregate, which usually reduces their fluorescence intensity (Li et al., 2003). Therefore, it is important to study their intermolecular interaction in the solid state. Recently we synthesized the title compound and studied its crystal structure.
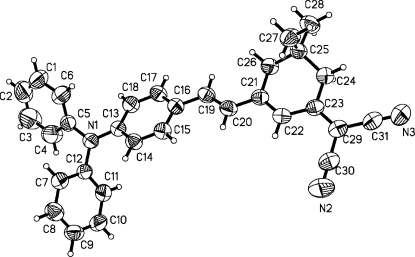
The molecular structure is shown in Fig. 1. Three benzenes of triphenyl amine group show a three-bladed propeller configuration due to repulsion force. The bond lengths of C13—N1 are shorter than the single C—N distance (1.47–1.50 Å) and longer than double C=N bond distance (1.34–1.38 Å), which is due to the conjugation of p-π in triphenyl amine group. Because of long conjugation length, all atoms are roughly coplanar (Kia et al., 2009). However, the cyclohexene group shows an envelope configuration due to its ring tension, which atoms are partly out of the plane (Ju et al., 2006). The triphenyl amine and cyclohexene groups could hold back farther gather of these molecules in the solid state, which is due to their non-coplanar conjugation. No hydrogen bonding is found in the crystal structure (Fig. 2).
Experimental
Hexahydropyridine (1 ml) and acetic acid (2 ml) were respectively added dropwise to a stirred benzene (100 ml) solution with 4-diphenylamino-benzaldehyde (1.1 g, 4 mmol) and 2-(3,5,5-Trimethylcyclohex-2-enylidene)-malononitrile (0.92 g, 5 mmol). The mixture was stirred at room temperature for 1 h, then separated water at refluxing temperature for another 5 h. Cooled to room temperature, the title compound was gotten (Lemke, 1974; Tao & Miyata, 2001). Single crystal suitable for X-ray diffraction analysis were obtained by slow evaporation of its ethanol saturated solution at room temperature.
Refinement
All H atoms were positioned geometrically, and allowed to ride on their parent atom with C—H = 0.93 (aromatic), 0.96 (methyl) and 0.97 Å (methylene). Uiso(H) = 1.5Ueq(C) for methyl group and 1.2Ueq(C) for others.
Figures
Fig. 1.
The molecular structure of the molecular structure of (I) showing the atom labels. Displacement ellipsoids are shown at the 50%.
Fig. 2.
The packing of (I), viewed down the b axis. H atoms not involved in hydrogen bonding have been omitted.
Crystal data
| C31H27N3 | F(000) = 936 |
| Mr = 441.56 | Dx = 1.147 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71069 Å |
| Hall symbol: -P 2ybc | Cell parameters from 962 reflections |
| a = 13.239 (5) Å | θ = 2.4–20.0° |
| b = 16.757 (8) Å | µ = 0.07 mm−1 |
| c = 11.886 (3) Å | T = 293 K |
| β = 104.073 (5)° | Block, red |
| V = 2557.7 (17) Å3 | 0.48 × 0.19 × 0.16 mm |
| Z = 4 |
Data collection
| Bruker SMART area-detector diffractometer | 3554 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.027 |
| graphite | θmax = 27.5°, θmin = 1.6° |
| φ and ω scans | h = −17→17 |
| 20681 measured reflections | k = −21→20 |
| 5879 independent reflections | l = −15→15 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.046 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.167 | H-atom parameters constrained |
| S = 0.92 | w = 1/[σ2(Fo2) + (0.1P)2 + 0.187P] where P = (Fo2 + 2Fc2)/3 |
| 5879 reflections | (Δ/σ)max < 0.001 |
| 309 parameters | Δρmax = 0.13 e Å−3 |
| 0 restraints | Δρmin = −0.18 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma (F2) is used only for calculating R-factors (gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.81570 (11) | 0.18274 (8) | 0.93407 (11) | 0.0627 (4) | |
| N2 | 0.43530 (17) | 0.55197 (11) | 0.20566 (18) | 0.1061 (6) | |
| N3 | 0.21605 (14) | 0.45416 (10) | −0.09927 (15) | 0.0845 (5) | |
| C1 | 0.7581 (2) | 0.01018 (13) | 1.1028 (2) | 0.0974 (7) | |
| H1 | 0.7009 | −0.0186 | 1.1131 | 0.117* | |
| C2 | 0.8549 (3) | −0.01085 (14) | 1.1632 (2) | 0.1187 (10) | |
| H2 | 0.8641 | −0.0534 | 1.2148 | 0.142* | |
| C3 | 0.9377 (2) | 0.03051 (15) | 1.1477 (2) | 0.1226 (10) | |
| H3 | 1.0043 | 0.0163 | 1.1891 | 0.147* | |
| C4 | 0.92488 (17) | 0.09351 (13) | 1.07133 (19) | 0.0943 (7) | |
| H4 | 0.9830 | 0.1213 | 1.0615 | 0.113* | |
| C5 | 0.82738 (14) | 0.11594 (10) | 1.00942 (13) | 0.0585 (4) | |
| C6 | 0.74278 (16) | 0.07411 (11) | 1.02576 (16) | 0.0741 (5) | |
| H6 | 0.6758 | 0.0884 | 0.9856 | 0.089* | |
| C7 | 0.86935 (13) | 0.28927 (10) | 1.07559 (14) | 0.0600 (4) | |
| H7 | 0.8285 | 0.2669 | 1.1209 | 0.072* | |
| C8 | 0.92347 (14) | 0.35847 (11) | 1.11063 (15) | 0.0692 (5) | |
| H8 | 0.9191 | 0.3828 | 1.1796 | 0.083* | |
| C9 | 0.98393 (15) | 0.39192 (11) | 1.04424 (17) | 0.0719 (5) | |
| H9 | 1.0205 | 0.4388 | 1.0682 | 0.086* | |
| C10 | 0.99010 (14) | 0.35617 (12) | 0.94293 (16) | 0.0742 (5) | |
| H10 | 1.0304 | 0.3791 | 0.8975 | 0.089* | |
| C11 | 0.93693 (14) | 0.28622 (11) | 0.90749 (15) | 0.0670 (5) | |
| H11 | 0.9425 | 0.2616 | 0.8392 | 0.080* | |
| C12 | 0.87536 (12) | 0.25291 (9) | 0.97371 (13) | 0.0531 (4) | |
| C13 | 0.73979 (12) | 0.18623 (9) | 0.82857 (12) | 0.0521 (4) | |
| C14 | 0.69523 (13) | 0.25939 (9) | 0.78731 (13) | 0.0543 (4) | |
| H14 | 0.7134 | 0.3051 | 0.8321 | 0.065* | |
| C15 | 0.62532 (12) | 0.26470 (9) | 0.68204 (14) | 0.0532 (4) | |
| H15 | 0.5975 | 0.3143 | 0.6563 | 0.064* | |
| C16 | 0.59430 (11) | 0.19754 (9) | 0.61181 (13) | 0.0504 (4) | |
| C17 | 0.63661 (12) | 0.12448 (10) | 0.65608 (13) | 0.0571 (4) | |
| H17 | 0.6158 | 0.0784 | 0.6131 | 0.069* | |
| C18 | 0.70818 (13) | 0.11848 (10) | 0.76140 (14) | 0.0578 (4) | |
| H18 | 0.7355 | 0.0689 | 0.7878 | 0.069* | |
| C19 | 0.52268 (12) | 0.20214 (10) | 0.49796 (13) | 0.0533 (4) | |
| H19 | 0.4978 | 0.1541 | 0.4626 | 0.064* | |
| C20 | 0.48912 (12) | 0.26930 (10) | 0.43923 (14) | 0.0552 (4) | |
| H20 | 0.5141 | 0.3171 | 0.4753 | 0.066* | |
| C21 | 0.41880 (11) | 0.27527 (9) | 0.32670 (13) | 0.0501 (4) | |
| C22 | 0.40255 (12) | 0.34749 (9) | 0.27357 (13) | 0.0549 (4) | |
| H22 | 0.4357 | 0.3918 | 0.3131 | 0.066* | |
| C23 | 0.33747 (12) | 0.35870 (9) | 0.16069 (13) | 0.0512 (4) | |
| C24 | 0.28127 (13) | 0.28808 (9) | 0.09855 (13) | 0.0561 (4) | |
| H24A | 0.3210 | 0.2671 | 0.0465 | 0.067* | |
| H24B | 0.2144 | 0.3056 | 0.0515 | 0.067* | |
| C25 | 0.26297 (12) | 0.22075 (9) | 0.17857 (13) | 0.0520 (4) | |
| C26 | 0.36565 (12) | 0.20259 (9) | 0.26656 (14) | 0.0545 (4) | |
| H26A | 0.3521 | 0.1660 | 0.3243 | 0.065* | |
| H26B | 0.4123 | 0.1761 | 0.2269 | 0.065* | |
| C27 | 0.17984 (13) | 0.24505 (11) | 0.24035 (16) | 0.0686 (5) | |
| H27A | 0.1163 | 0.2569 | 0.1840 | 0.103* | |
| H27B | 0.2026 | 0.2915 | 0.2869 | 0.103* | |
| H27C | 0.1681 | 0.2021 | 0.2891 | 0.103* | |
| C28 | 0.22779 (15) | 0.14665 (11) | 0.10523 (16) | 0.0787 (6) | |
| H28A | 0.2813 | 0.1307 | 0.0679 | 0.118* | |
| H28B | 0.1651 | 0.1584 | 0.0475 | 0.118* | |
| H28C | 0.2148 | 0.1042 | 0.1541 | 0.118* | |
| C29 | 0.32897 (13) | 0.43195 (9) | 0.10715 (14) | 0.0590 (4) | |
| C30 | 0.38730 (16) | 0.49914 (11) | 0.16151 (17) | 0.0730 (5) | |
| C31 | 0.26565 (15) | 0.44413 (10) | −0.00723 (17) | 0.0649 (5) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0691 (9) | 0.0547 (8) | 0.0565 (8) | −0.0110 (7) | −0.0001 (7) | 0.0077 (6) |
| N2 | 0.1275 (16) | 0.0607 (11) | 0.1231 (15) | −0.0240 (11) | 0.0169 (12) | −0.0041 (10) |
| N3 | 0.1009 (13) | 0.0683 (11) | 0.0790 (11) | −0.0048 (9) | 0.0118 (9) | 0.0167 (8) |
| C1 | 0.141 (2) | 0.0771 (15) | 0.0818 (15) | −0.0301 (15) | 0.0420 (15) | 0.0037 (11) |
| C2 | 0.190 (3) | 0.0671 (15) | 0.0774 (15) | −0.0170 (18) | −0.0100 (18) | 0.0142 (11) |
| C3 | 0.133 (2) | 0.0778 (16) | 0.122 (2) | −0.0014 (16) | −0.0367 (17) | 0.0290 (15) |
| C4 | 0.0822 (14) | 0.0771 (14) | 0.1062 (16) | −0.0069 (11) | −0.0110 (12) | 0.0232 (12) |
| C5 | 0.0716 (11) | 0.0487 (9) | 0.0526 (9) | −0.0029 (8) | 0.0101 (8) | 0.0012 (7) |
| C6 | 0.0856 (13) | 0.0656 (12) | 0.0772 (12) | −0.0033 (10) | 0.0315 (10) | 0.0035 (9) |
| C7 | 0.0588 (10) | 0.0628 (11) | 0.0595 (9) | −0.0036 (8) | 0.0162 (8) | 0.0009 (8) |
| C8 | 0.0745 (12) | 0.0604 (11) | 0.0696 (11) | 0.0016 (9) | 0.0112 (9) | −0.0073 (8) |
| C9 | 0.0724 (12) | 0.0521 (10) | 0.0809 (12) | −0.0102 (9) | −0.0011 (9) | 0.0066 (9) |
| C10 | 0.0684 (12) | 0.0802 (13) | 0.0709 (12) | −0.0198 (10) | 0.0111 (9) | 0.0164 (10) |
| C11 | 0.0672 (11) | 0.0787 (12) | 0.0552 (9) | −0.0115 (9) | 0.0149 (8) | 0.0008 (8) |
| C12 | 0.0512 (9) | 0.0529 (9) | 0.0517 (8) | −0.0048 (7) | 0.0059 (7) | 0.0057 (7) |
| C13 | 0.0522 (9) | 0.0521 (9) | 0.0507 (8) | −0.0033 (7) | 0.0099 (7) | 0.0008 (7) |
| C14 | 0.0576 (9) | 0.0484 (9) | 0.0559 (9) | −0.0010 (7) | 0.0117 (7) | −0.0049 (7) |
| C15 | 0.0517 (9) | 0.0481 (9) | 0.0597 (9) | 0.0047 (7) | 0.0132 (7) | 0.0000 (7) |
| C16 | 0.0429 (8) | 0.0542 (9) | 0.0544 (9) | 0.0022 (7) | 0.0126 (6) | −0.0010 (7) |
| C17 | 0.0558 (9) | 0.0525 (10) | 0.0606 (9) | −0.0009 (8) | 0.0092 (7) | −0.0085 (7) |
| C18 | 0.0595 (10) | 0.0469 (9) | 0.0635 (10) | 0.0054 (7) | 0.0080 (8) | 0.0017 (7) |
| C19 | 0.0437 (8) | 0.0570 (10) | 0.0589 (9) | −0.0002 (7) | 0.0118 (7) | −0.0057 (7) |
| C20 | 0.0462 (9) | 0.0565 (10) | 0.0612 (9) | −0.0058 (7) | 0.0098 (7) | −0.0016 (7) |
| C21 | 0.0413 (8) | 0.0512 (9) | 0.0588 (9) | −0.0006 (7) | 0.0141 (7) | −0.0009 (7) |
| C22 | 0.0528 (9) | 0.0501 (9) | 0.0619 (9) | −0.0073 (7) | 0.0139 (7) | −0.0040 (7) |
| C23 | 0.0491 (8) | 0.0484 (9) | 0.0592 (9) | −0.0017 (7) | 0.0193 (7) | −0.0003 (7) |
| C24 | 0.0558 (9) | 0.0552 (10) | 0.0573 (9) | −0.0034 (7) | 0.0135 (7) | −0.0001 (7) |
| C25 | 0.0471 (8) | 0.0459 (8) | 0.0597 (9) | −0.0019 (7) | 0.0068 (7) | 0.0021 (7) |
| C26 | 0.0476 (8) | 0.0490 (9) | 0.0646 (9) | 0.0025 (7) | 0.0089 (7) | −0.0003 (7) |
| C27 | 0.0488 (9) | 0.0758 (12) | 0.0828 (12) | −0.0002 (8) | 0.0192 (8) | 0.0162 (9) |
| C28 | 0.0809 (13) | 0.0588 (11) | 0.0841 (13) | −0.0143 (9) | −0.0039 (10) | −0.0047 (9) |
| C29 | 0.0636 (10) | 0.0501 (9) | 0.0652 (10) | −0.0030 (8) | 0.0189 (8) | 0.0023 (7) |
| C30 | 0.0869 (14) | 0.0490 (10) | 0.0813 (12) | −0.0072 (10) | 0.0173 (10) | 0.0056 (9) |
| C31 | 0.0738 (12) | 0.0497 (10) | 0.0730 (12) | −0.0035 (9) | 0.0215 (9) | 0.0079 (8) |
Geometric parameters (Å, °)
| N1—C13 | 1.405 (2) | C15—H15 | 0.9300 |
| N1—C5 | 1.418 (2) | C16—C17 | 1.395 (2) |
| N1—C12 | 1.431 (2) | C16—C19 | 1.453 (2) |
| N2—C30 | 1.141 (2) | C17—C18 | 1.378 (2) |
| N3—C31 | 1.143 (2) | C17—H17 | 0.9300 |
| C1—C2 | 1.354 (4) | C18—H18 | 0.9300 |
| C1—C6 | 1.392 (3) | C19—C20 | 1.342 (2) |
| C1—H1 | 0.9300 | C19—H19 | 0.9300 |
| C2—C3 | 1.347 (4) | C20—C21 | 1.435 (2) |
| C2—H2 | 0.9300 | C20—H20 | 0.9300 |
| C3—C4 | 1.375 (3) | C21—C22 | 1.358 (2) |
| C3—H3 | 0.9300 | C21—C26 | 1.498 (2) |
| C4—C5 | 1.374 (3) | C22—C23 | 1.420 (2) |
| C4—H4 | 0.9300 | C22—H22 | 0.9300 |
| C5—C6 | 1.374 (2) | C23—C29 | 1.375 (2) |
| C6—H6 | 0.9300 | C23—C24 | 1.494 (2) |
| C7—C8 | 1.373 (2) | C24—C25 | 1.533 (2) |
| C7—C12 | 1.375 (2) | C24—H24A | 0.9700 |
| C7—H7 | 0.9300 | C24—H24B | 0.9700 |
| C8—C9 | 1.373 (3) | C25—C27 | 1.520 (2) |
| C8—H8 | 0.9300 | C25—C28 | 1.524 (2) |
| C9—C10 | 1.365 (3) | C25—C26 | 1.531 (2) |
| C9—H9 | 0.9300 | C26—H26A | 0.9700 |
| C10—C11 | 1.379 (2) | C26—H26B | 0.9700 |
| C10—H10 | 0.9300 | C27—H27A | 0.9600 |
| C11—C12 | 1.381 (2) | C27—H27B | 0.9600 |
| C11—H11 | 0.9300 | C27—H27C | 0.9600 |
| C13—C18 | 1.392 (2) | C28—H28A | 0.9600 |
| C13—C14 | 1.397 (2) | C28—H28B | 0.9600 |
| C14—C15 | 1.366 (2) | C28—H28C | 0.9600 |
| C14—H14 | 0.9300 | C29—C31 | 1.428 (3) |
| C15—C16 | 1.402 (2) | C29—C30 | 1.428 (3) |
| C13—N1—C5 | 122.75 (13) | C17—C18—C13 | 120.40 (15) |
| C13—N1—C12 | 118.53 (12) | C17—C18—H18 | 119.8 |
| C5—N1—C12 | 118.29 (13) | C13—C18—H18 | 119.8 |
| C2—C1—C6 | 121.1 (2) | C20—C19—C16 | 125.98 (15) |
| C2—C1—H1 | 119.4 | C20—C19—H19 | 117.0 |
| C6—C1—H1 | 119.4 | C16—C19—H19 | 117.0 |
| C3—C2—C1 | 119.4 (2) | C19—C20—C21 | 126.94 (15) |
| C3—C2—H2 | 120.3 | C19—C20—H20 | 116.5 |
| C1—C2—H2 | 120.3 | C21—C20—H20 | 116.5 |
| C2—C3—C4 | 120.8 (2) | C22—C21—C20 | 119.28 (14) |
| C2—C3—H3 | 119.6 | C22—C21—C26 | 119.98 (14) |
| C4—C3—H3 | 119.6 | C20—C21—C26 | 120.72 (14) |
| C5—C4—C3 | 120.8 (2) | C21—C22—C23 | 123.27 (14) |
| C5—C4—H4 | 119.6 | C21—C22—H22 | 118.4 |
| C3—C4—H4 | 119.6 | C23—C22—H22 | 118.4 |
| C4—C5—C6 | 118.46 (17) | C29—C23—C22 | 121.22 (14) |
| C4—C5—N1 | 119.87 (17) | C29—C23—C24 | 120.23 (14) |
| C6—C5—N1 | 121.62 (16) | C22—C23—C24 | 118.50 (13) |
| C5—C6—C1 | 119.5 (2) | C23—C24—C25 | 114.28 (13) |
| C5—C6—H6 | 120.3 | C23—C24—H24A | 108.7 |
| C1—C6—H6 | 120.3 | C25—C24—H24A | 108.7 |
| C8—C7—C12 | 120.24 (16) | C23—C24—H24B | 108.7 |
| C8—C7—H7 | 119.9 | C25—C24—H24B | 108.7 |
| C12—C7—H7 | 119.9 | H24A—C24—H24B | 107.6 |
| C9—C8—C7 | 120.29 (17) | C27—C25—C28 | 109.77 (14) |
| C9—C8—H8 | 119.9 | C27—C25—C26 | 110.51 (14) |
| C7—C8—H8 | 119.9 | C28—C25—C26 | 109.03 (13) |
| C10—C9—C8 | 119.75 (17) | C27—C25—C24 | 110.18 (13) |
| C10—C9—H9 | 120.1 | C28—C25—C24 | 108.51 (13) |
| C8—C9—H9 | 120.1 | C26—C25—C24 | 108.80 (13) |
| C9—C10—C11 | 120.45 (17) | C21—C26—C25 | 113.60 (12) |
| C9—C10—H10 | 119.8 | C21—C26—H26A | 108.8 |
| C11—C10—H10 | 119.8 | C25—C26—H26A | 108.8 |
| C10—C11—C12 | 119.83 (17) | C21—C26—H26B | 108.8 |
| C10—C11—H11 | 120.1 | C25—C26—H26B | 108.8 |
| C12—C11—H11 | 120.1 | H26A—C26—H26B | 107.7 |
| C7—C12—C11 | 119.43 (15) | C25—C27—H27A | 109.5 |
| C7—C12—N1 | 120.57 (14) | C25—C27—H27B | 109.5 |
| C11—C12—N1 | 119.96 (14) | H27A—C27—H27B | 109.5 |
| C18—C13—C14 | 118.16 (14) | C25—C27—H27C | 109.5 |
| C18—C13—N1 | 121.87 (14) | H27A—C27—H27C | 109.5 |
| C14—C13—N1 | 119.96 (14) | H27B—C27—H27C | 109.5 |
| C15—C14—C13 | 120.86 (14) | C25—C28—H28A | 109.5 |
| C15—C14—H14 | 119.6 | C25—C28—H28B | 109.5 |
| C13—C14—H14 | 119.6 | H28A—C28—H28B | 109.5 |
| C14—C15—C16 | 121.88 (14) | C25—C28—H28C | 109.5 |
| C14—C15—H15 | 119.1 | H28A—C28—H28C | 109.5 |
| C16—C15—H15 | 119.1 | H28B—C28—H28C | 109.5 |
| C17—C16—C15 | 116.57 (14) | C23—C29—C31 | 122.11 (15) |
| C17—C16—C19 | 120.59 (14) | C23—C29—C30 | 121.35 (15) |
| C15—C16—C19 | 122.84 (14) | C31—C29—C30 | 116.48 (15) |
| C18—C17—C16 | 122.08 (15) | N2—C30—C29 | 178.8 (2) |
| C18—C17—H17 | 119.0 | N3—C31—C29 | 179.1 (2) |
| C16—C17—H17 | 119.0 | ||
| C6—C1—C2—C3 | −0.5 (4) | C14—C15—C16—C17 | 1.5 (2) |
| C1—C2—C3—C4 | −0.1 (4) | C14—C15—C16—C19 | −177.95 (14) |
| C2—C3—C4—C5 | 0.2 (4) | C15—C16—C17—C18 | −2.3 (2) |
| C3—C4—C5—C6 | 0.2 (3) | C19—C16—C17—C18 | 177.10 (14) |
| C3—C4—C5—N1 | 177.9 (2) | C16—C17—C18—C13 | 0.9 (2) |
| C13—N1—C5—C4 | 144.71 (18) | C14—C13—C18—C17 | 1.4 (2) |
| C12—N1—C5—C4 | −42.9 (2) | N1—C13—C18—C17 | −177.36 (14) |
| C13—N1—C5—C6 | −37.7 (2) | C17—C16—C19—C20 | −169.57 (15) |
| C12—N1—C5—C6 | 134.71 (17) | C15—C16—C19—C20 | 9.8 (2) |
| C4—C5—C6—C1 | −0.7 (3) | C16—C19—C20—C21 | 179.76 (14) |
| N1—C5—C6—C1 | −178.40 (17) | C19—C20—C21—C22 | −172.13 (15) |
| C2—C1—C6—C5 | 0.9 (3) | C19—C20—C21—C26 | 6.3 (2) |
| C12—C7—C8—C9 | 0.0 (3) | C20—C21—C22—C23 | 177.62 (13) |
| C7—C8—C9—C10 | 0.1 (3) | C26—C21—C22—C23 | −0.8 (2) |
| C8—C9—C10—C11 | −0.7 (3) | C21—C22—C23—C29 | −175.45 (14) |
| C9—C10—C11—C12 | 1.3 (3) | C21—C22—C23—C24 | 2.0 (2) |
| C8—C7—C12—C11 | 0.5 (3) | C29—C23—C24—C25 | −158.00 (14) |
| C8—C7—C12—N1 | −177.28 (15) | C22—C23—C24—C25 | 24.5 (2) |
| C10—C11—C12—C7 | −1.2 (3) | C23—C24—C25—C27 | 72.25 (18) |
| C10—C11—C12—N1 | 176.63 (15) | C23—C24—C25—C28 | −167.55 (13) |
| C13—N1—C12—C7 | 116.95 (17) | C23—C24—C25—C26 | −49.05 (18) |
| C5—N1—C12—C7 | −55.8 (2) | C22—C21—C26—C25 | −26.7 (2) |
| C13—N1—C12—C11 | −60.8 (2) | C20—C21—C26—C25 | 154.85 (14) |
| C5—N1—C12—C11 | 126.46 (17) | C27—C25—C26—C21 | −71.15 (17) |
| C5—N1—C13—C18 | −33.5 (2) | C28—C25—C26—C21 | 168.12 (13) |
| C12—N1—C13—C18 | 154.15 (15) | C24—C25—C26—C21 | 49.95 (18) |
| C5—N1—C13—C14 | 147.73 (15) | C22—C23—C29—C31 | 178.69 (14) |
| C12—N1—C13—C14 | −24.6 (2) | C24—C23—C29—C31 | 1.2 (2) |
| C18—C13—C14—C15 | −2.3 (2) | C22—C23—C29—C30 | 1.7 (2) |
| N1—C13—C14—C15 | 176.54 (14) | C24—C23—C29—C30 | −175.73 (15) |
| C13—C14—C15—C16 | 0.8 (2) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: XU2505).
References
- Bruker (2002). SAINT and SMART Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Hye, J. L., Jiwon, S., Jaehoon, H. & Soo, Y. P. (2004). Chem. Mater.16, 456–465.
- Ju, H. D., Wan, Y., Yu, W. T. & Tao, X. T. (2006). Thin Solid Films, 515, 2403–2409.
- Kia, R., Fun, H.-K. & Kargar, H. (2009). Acta Cryst. E65, o682–o683. [DOI] [PMC free article] [PubMed]
- Lemke, R. (1974). Synthesis, pp. 359–361.
- Li, J. Y., Liu, D., Hong, Z. R. & Tong, S. W. (2003). Chem. Mater.15, 1486–1490.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Tang, C. W., Vanslyke, S. A. & Chen, C. H. (1998). J. Appl. Phys.65, 3610–3616.
- Tao, X. T. & Miyata, S. (2001). Appl. Phys. Lett.78, 279–281.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536809014378/xu2505sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536809014378/xu2505Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report