Abstract
Highly pathogenic avian influenza viruses of subtype H7N1 that emerged during an outbreak in 1999 and 2000 in Italy differ from their low-pathogenicity precursor viruses by changes in several genes, including three mutations in the NS1 protein. Two of them involve amino acid exchanges located within or closely adjacent to the nuclear export signal of NS1. The third mutation resulted in a new stop codon and thereby a C-terminal truncation of the NS1 protein of the highly pathogenic viruses. To find out whether these mutations contribute to the phenotypic differences between the highly pathogenic and low pathogenic viruses, we generated recombinants of the highly pathogenic A/ostrich/Italy/984/00 strain that contained the nuclear export signal and/or the extended C terminus of NS1 of a low pathogenic virus (A/chicken/Italy/1082/99). Using these recombinants we could demonstrate that replication rate and spread of infection in chicken fibroblast cultures, as well as infectivity for chicken embryos is reduced, whereas the mean death time for chicken embryos is increased, when the highly pathogenic virus acquires the NS1 motifs of the low pathogenic virus. Analysis of beta interferon transcription in chicken fibroblasts infected with the recombinants revealed that the mutations observed in the nuclear export signal of the highly pathogenic viruses were responsible for the enhanced interferon antagonism of these viruses. Cell fractionation and immunofluorescence studies in chicken fibroblasts showed that the nuclear export signal of the highly pathogenic viruses is responsible for cytoplasmic accumulation of NS1, whereas the C-terminal truncation promotes transport into the nucleoli. Comparative analysis in human A549 cells indicated that intracellular distribution of NS1 is host specific. Taken together, these observations support the concept that compartmentalization of NS1 within the cell contributes to the pathogenicity of avian influenza viruses.
The majority of avian influenza A viruses displays low pathogenicity in poultry. In contrast, some viruses of subtypes H5 and H7 periodically cause devastating outbreaks in chickens and turkeys, with mortality rates of up to 100%. On very rare occasions, they have also been responsible for disease in humans. They are therefore not only a serious economic threat to the poultry industry, but they are also of human concern because of their pandemic potential. It is now generally believed that these highly pathogenic avian influenza (HPAI) viruses are derived from the low pathogenic avian influenza (LPAI) viruses by mutational changes (1).
Thus, at the onset of an H7N1 outbreak in 1999/2000 in Northern Italy, only LPAI viruses were observed, whereas later on highly pathogenic isolates prevailed (3). Comparative sequence analysis of highly and low pathogenic isolates obtained at the outbreak revealed mutations in almost every gene, except M2, NS2, and PB1-F2 (unpublished data). The most prominent difference was an insertion of the amino acid sequence SRVR into the hemagglutinin, resulting in a multibasic cleavage site, which is a prime requisite for the highly pathogenic phenotype of avian influenza viruses (13). Of particular interest, however, were progressive truncations at the C-terminal end of the nonstructural protein NS1 (9), also known to be a major virulence factor. NS1 is a multifunctional protein that interacts with both antiviral cellular mRNAs and host defense proteins (11, 17), leading to a host-undisturbed replication of the virus. Thus, it prevents type I interferon (IFN) production by inhibiting activation of IRF-3 (35) and NF-κB (38), and it inhibits activation of IFN-induced antiviral proteins such as PKR and 2′-5′ oligo(A) synthetase (27, 28). NS1 shuttles between the cytoplasm, nucleus, and nucleolus by using its two nuclear localization signals (NLS1 and NLS2) (15), nuclear export signal (NES), and a recently discovered nucleolar localization signal (NoLS). NLS1 is conserved among all influenza A viruses and consists of residues R35, R38, and K41 in the N-terminal RNA-binding domain, whereas the C-terminal NLS2 comprising residues K219, R220, R231, and R232 is absent in many strains. NES lies within residues 138 and 147 and requires leucine at position 144 and 146 (17, 22). NoLS, which overlaps with NLS2 at the C-terminal end of NS1, consists of basic residues at positions 219, 220, 224, 229, 231, and 232, and it is destroyed by non-basic amino acids in position 229 (25).
In the present study we have generated recombinants of an H7N1 HPAI virus expressing NS1 with the NES and/or the C terminus of a LPAI virus. We show that these modifications alter the localization of NS1, as well as the virulence of the virus. These results corroborate the role of NS1 as an important pathogenicity determinant.
MATERIALS AND METHODS
Cells and viruses.
Chicken embryo fibroblasts (CF) were produced by digesting pieces of 11-day-old chicken embryos in buffered saline containing 2.5% trypsin (Gibco/BRL, Darmstadt, Germany); afterward, the cells were cultured in Dulbecco modified Eagle medium (DMEM) with 10% fetal calf serum (FCS). 293T human embryonic kidney cells and A549 cells were grown in DMEM supplemented with 10% FCS, whereas Madin-Darby canine kidney (MDCK) cells were passaged in minimal essential medium (MEM) containing 10% FCS. Wild-type (wt) and mutant viruses of A/ostrich/Italy/984/00 (HPAI) were propagated in 11-day-old embryonated chicken eggs. The highly pathogenic viruses A/ostrich/Italy/984/00 (isolate 984) and A/turkey/Italy/4580/99 (isolate 4580) and the low pathogenic viruses A/chicken/Italy/1082/99 (isolate 1082), A/turkey/Italy/1265/99 (isolate 1265), A/turkey/Italy/2379/99 (isolate 2379), and A/turkey/Italy/2505/99 (isolate 2505), all of subtype H7N1, were used for sequence analysis of their NS segments.
Cloning and mutagenesis of plasmids.
All eight gene segments of A/ostrich/Italy/984/00(984) were amplified by reverse transcription-PCR (RT-PCR) from isolated RNA and ligated into plasmid polISapI, except for the HA gene that was cloned into plasmid pHH21. Both the reconstitution of the C terminus and the changes (I136V and N139D) in the NES of the NS1 of the 984 virus were performed by using a QuikChange mutagenesis kit (Stratagene, Waldbronn, Germany), according to the manufacturer's protocol. Ile and Asn were exchanged for Val and Asp by using the oligonucleotides 5′-GCTAATTTCTCAGTCCTATTTGATCAACTAGAAAC-3′ and 5′-GTTTCTAGTTGATCAAATAGGACTGAGAAATTAGC-3′ as primers. The C terminus was reconstituted by using the primers 5′-CGCTACATGGCGAGATGAGTTGAGTCAGAAGTTTG-3′ and 5′-CAAACTTCTGACTCAACTCATCTCGCCATGTAGCG-3′. The presence of the introduced mutations and the absence of additional unwanted mutations were verified by sequencing of the whole cDNA.
Rescue of recombinant viruses.
293T cells were transfected with a mixture of eight plasmids encoding the eight gene segments of strain A/ostrich/Italy/984/00, together with expression plasmids of WSN PB1, PB2, PA, and NP (1 μg each) and Lipofectamine 2000 (Invitrogen, Darmstadt, Germany), as recommended by the manufacturer. Medium was removed 6 h later and replaced by DMEM containing 0.2% bovine serum albumin (BSA). After another 42 h, the supernatant was plated onto MDCK cells. Supernatant was used for double plaque purification of the rescued viruses before 11-day-old chicken eggs were inoculated for propagation of stock virus. The identity of propagated mutant viruses was ascertained by sequencing the NS1 gene after RT-PCR.
Replication of mutant viruses in cell culture.
For growth curves, CF monolayers were inoculated with wt and mutant viruses at a multiplicity of infection (MOI) of 0.0005 PFU per cell in phosphate-buffered saline (PBS) containing 0.2% BSA for 1 h. Unbound virus was washed away, and serum-free medium containing 0.2% BSA was added. For plaque assays, confluent MDCK cell monolayers in six-well plates were inoculated with 10-fold dilutions (in PBS-0.2% BSA) of mutant viruses for 30 min. Cells were then washed with PBS-0.2% BSA and overlaid with MEM containing 0.9% agar (Oxoid, Ltd., Hampshire, United Kingdom), 1% glutamine, and 0.3% BSA. Plaques were stained 3 days postinfection (p.i.) with 0.1% crystal violet in 10% formaldehyde. The spread of infection in CF monolayers was monitored by covering infected cells with Avicel overlay (1.25% Avicel [FMC, Barcelona, Spain], MEM, 1% glutamine, and 0.3% BSA) for 24 h (24). Afterward, the cells were washed twice with PBS-0.2% BSA, fixed with 4% paraformaldehyde, and stained by using rabbit anti-FPV (H7N1) serum, horseradish peroxidase (HRP)-conjugated donkey anti-rabbit antibody, and TrueBlue peroxidase substrate (KPL, Gaithersburg, MD). After further washing with H2O, the plates were scanned, and the sizes of two times 50 plaque areas of infected cells was measured by ImageJ software (National Institutes of Health).
Dissociation of nuclear and cytoplasmic fractions.
CF were grown in 60-mm dishes to 80% confluence and infected with virus at an MOI of 2 for 30 min. Cells were washed with PBS and incubated with 4 ml of DMEM containing 0.2% BSA for 7.5 h. Nuclear and cytoplasmic fractions were separated by using a nuclear extraction kit from Imgenex (San Diego, CA) according to the manufacturer's protocol. The amount of NS1 and chicken β-actin protein in each fraction was determined by SDS-PAGE and immunoblotting with a goat polyclonal antibody against the N terminus of NS1 (Santa Cruz Biotechnology, Santa Cruz, CA). β-Actin was detected by a mouse monoclonal antibody (Abcam, Cambridge, United Kingdom). The secondary antibodies IRDye800 (conjugated donkey anti-goat) and IRDye700 (conjugated goat anti-mouse) (Rockland, Gilbertsville, PA) were used for detection and quantification on a LI-COR Odyssey infrared imaging system (Bad Homburg, Germany). The results are shown as the average of three independent experiments.
RT-PCR for IFN-β.
CF were grown in 60-mm dishes to 80% confluence and infected with virus diluted in PBS with an MOI of 1.0 or PBS alone for 30 min. Cells were washed in PBS and incubated with 4 ml of DMEM containing 0.2% BSA for 7.5 h. As a positive control for IFN-β induction cells were transfected with 10 μg of poly(I:C)/ml using Lipofectamine 2000 according to the manufacturer's protocol. Total RNA from infected and/or transfected CF was isolated by using the RNeasy minikit (Qiagen Hilden, Germany) according to the protocol of the manufacturer for purification of total RNA from animal cells. RT-PCR was performed for 30 cycles using the Qiagen OneStep RT-PCR kit according to the manufacturer's protocol with the primers for chicken IFN-β mRNA (GenBank accession no. NM_ 001024836, primers from positions 194 to 213 and 398 to 379) and chicken β-actin mRNA (GenBank accession no. CHKBACTN, primers from positions 866 to 886 and 1278 to 1259). RT-PCR products were separated in 1.5% agarose gel electrophoresis and visualized under UV light after ethidium bromide staining. The intensities of visualized bands were measured by using ImageJ software (National Institutes of Health).
Immunofluorescence analysis of mutant viruses in cell culture.
CF and A549 cells at a confluence of 60% were infected with virus at an MOI of 1 as described above, fixed at 7.5 h p.i. with chilled methanol-acetone (1:1), and permeabilized with 0.1% Triton X-100 in PBS. After blocking with PBS containing 1% BSA, the cells were incubated with the primary antibody against NS1 (antibody as mentioned above) diluted in PBS containing 1% BSA and DAPI (4′,6′-diamidino-2-phenylindole; 10 μg/ml) for 1.5 h. The cells were washed three times with PBS and incubated with monoclonal donkey anti-goat antibody conjugated with Rhodamin (Dianova, Hamburg, Germany) for 1.5 h. The cells were washed again three times with PBS and covered by a glass slide with Mowiol. For quantification of cells displaying differences in NS1 localization, 100 cells were randomly examined by inspection in at least two independent experiments per recombinant virus on an Axiovert 200 ApoTome system from Zeiss (Goettingen, Germany).
EID50.
11-day-old embryonated eggs were infected with serial dilutions of the mutant viruses with decreasing 10-fold dilutions (in PBS). For determination of the 50% egg infectious dose (EID50), the allantoic fluid was collected 3 days p.i. and tested for viral infection by hemagglutination assay. The EID50 is the dose of PFU that is sufficient for infecting 50% of the chicken embryos.
Virus lethality for chicken embryos.
The allantoic cavity of 11-day-old embryonated eggs was inoculated with 10 PFU of the recombinant viruses. Ten eggs were infected with each virus and the mean death time (MDT) has been determined.
RESULTS
The NS1 protein of HPAI isolates has mutated at two distinct sites.
We have first compared by sequence analysis the NS genes of four LPAI (1082, 1265, 2379, and 2505) and two HPAI (984 and 4580) isolates obtained at different time points of the 1999 and 2000 outbreak. The results showed that the NS1 proteins of LPAI and HPAI viruses differed by only three mutations. Two of them involved amino acid exchanges at positions 136 and 139 located within or closely adjacent to the NES. The third mutation resulted in a new stop codon and the deletion of six amino acids at the C terminus of the NS1 protein of the HPAI viruses (Table 1 ). This finding was in line with the previous observation that C-terminal truncation of NS1 is a specific feature of the HPAI isolates (9). Since each of these mutations was located in functionally critical regions of NS1, it was of interest to find out whether they contributed to the phenotypic differences between LPAI and HPAI viruses.
TABLE 1.
Comparison of NS1 amino acid sequences of highly pathogenic (HP) and low pathogenic (LP) avian influenza viruses isolated in the 1999 and 2000 outbreak in Northern Italy
a *, Mutant viruses generated on the basis of the A/ostrich/Italy/984/00(984) reverse genetics system, which differ solely in their NS1 displaying either the HP or the LP sequence of NES and NoLS.
b Consensus sequence for NES (22). The NES sequence spans amino acids 138 to 147. Leucines at positions 144 and 146 (highlighted in dark gray) are thought to be required for cytoplasmic localization. Mutations detected between HPAI and LPAI viruses are highlighted in black.
c Consensus sequence for NoLS (25). The NoLS sequence is highlighted in dark and light gray at positions 219, 220, 224, 229, 231, and 232 of the NS1 protein. The nonbasic amino acid at position 229 (light gray) inhibits the nucleolar localization of NS1.
NS1 mutations modulate virus growth, virulence, and IFN response.
Using reverse genetics, we have therefore generated recombinants of HPAI virus 984 containing the NES and/or the C terminus of the LPAI strains. Mutant 984 H/H corresponds to the 984 wt virus showing the HPAI NES sequence and the truncated C terminus. Mutant 984 L/L has both the NES and the extended C terminus of the LPAI strains. Mutant 984 L/H has the LPAI NES sequence and the truncated C terminus of the HPAI virus, whereas mutant 984 H/L displays the HPAI NES in combination with the extended C terminus of the LPAI viruses.
Replication of these viruses was analyzed in primary CF cultures. Comparison of the growth curves shown in Fig. 1 A indicates that the high replication rate of the 984 H/H virus is moderately reduced when it contains either the LPAI NES (984 L/H) or the LPAI C terminus (984 H/L) and that there is a strong reduction when it displays the LPAI signature in both positions. We have also determined the size of the plaques formed by the mutants in CF monolayers 24 h p.i. In each case, 100 plaques were measured. Compared to the 984 H/H virus, which formed the largest plaques (100%), the size was reduced to 42 and 43% with 984 H/L and 984 L/H, respectively. The smallest plaques (24% of the average plaque size of the 984 H/H virus) were observed with the 984 L/L mutant (Fig. 1B). Taken together, these findings indicate that, upon conversion of the HPAI form of NS1 to the LPAI form, replication rates and spread of infection in tissue culture are reduced.
FIG. 1.
Growth characteristics of viruses differing in the NES and C terminus of NS1. Recombinants of strain 984 have been analyzed that contain HPAI NES and the HPAI C terminus (H/H), HPAI NES and the LPAI C terminus (H/L), LPAI NES and the HPAI C terminus (L/H), and LPAI NES and the LPAI C terminus (L/L). (A) Virus replication in CF cultures. Cells were inoculated at an MOI of 0.0005 PFU per cell. Growth curves show the averages of two independent experiments. (B) Spread of infection in CF monolayers. After infection, cells were covered with an Avicel overlay for 24 h to avoid virus spread by diffusion. Infected cells were stained with rabbit anti-FPV (H7N1) serum, HRP-conjugated donkey anti-rabbit serum, and TrueBlue peroxidase substrate. Plaque sizes of 100 were measured by using ImageJ software in two independent experiments. Pictures display a section of infected and stained cells. The sizes of yellow-edged plaques were measured for quantitative analysis. (C) Infectivity of the mutant viruses was measured by determining the EID50 using 11-day-old embryonated chicken eggs. Eggs were infected with dilutions of 0.01 to 100 PFU for determination of the EID50.
We have also determined the EID50 for each mutant. There is a distinct EID50 increase from 0.5 PFU with 984 H/H over 1.2 PFU (984 H/L) and 1.4 PFU (984 L/H) to 6.8 PFU (984 L/L). The results demonstrate that introduction of the LPAI motifs into NS1 reduces the infectivity of the virus for chicken embryos.
To find out whether the mutations affected the virulence of the recombinant viruses we analyzed their lethality for chicken embryos. The MDT values were 50 ± 5, 46 ± 5, 44 ± 3, and 36 ± 6 h p.i. for 984 L/L, 984 H/L, 984 L/H, and 984 H/H, respectively. There is a small, though significant difference between mutants 984 L/L and 984 H/H, indicating that acquisition of NS1 of the LPAI viruses reduces the pathogenicity of the HPAI 984 virus.
It was then of interest to analyze the IFN response to the mutants. Induction of IFN-β transcription was measured by RT-PCR using IFN-β mRNA and β-actin mRNA specific primers. The amount of induced IFN-β mRNA was strongly reduced after infection with 984 H/H and 984 H/L. In comparison, IFN-β mRNA levels were significantly higher when cells were infected with 984 L/L or 984 L/H (Fig. 2). Thus, strong IFN-β antagonism correlates with the presence of the HPAI NES signal in NS1.
FIG. 2.
IFN-β induction by virus infection. CF cultures were infected with 984 H/H, 984 H/L, 984 L/H, and 984 L/L at an MOI of 1.0 or transfected with poly(I:C) as a positive and PBS as a negative control. At 7.5 h p.i., isolated cellular RNA samples were analyzed by RT-PCR for the amount of IFN-β and β-actin mRNA. The intensity of the bands was measured by using ImageJ software. The IFN-β mRNA levels were normalized by determining the amount of β-actin mRNA.
HPAI specific mutations enhance the cytoplasmic and nucleolar localization of NS1.
To analyze the intracellular localization of NS1, we first looked at the distribution between nucleus and cytoplasm. CF were infected at a MOI of 2.0 and separated into a nuclear and a cytoplasmic fraction at 7.5 h p.i. The amount of NS1 protein was determined by Western blot analysis. As shown in Fig. 3, 78.7 and 84% of NS1 are found in the cytoplasm after infection with 984 H/H and 984 H/L, respectively, whereas with 984 L/L and 984 L/H only ca. 50% are in the cytoplasm. These observations suggest that the HPAI NES promotes accumulation of NS1 in the cytoplasm.
FIG. 3.
NS1 in nuclear and cytoplasmic fractions. (A) Confluent CF cultures were infected with each virus at an MOI of 2.0 PFU per cell. At 7.5 h p.i., cells were harvested and fractionated. Amounts of NS1 in nuclear (NC) and cytoplasmic (CY) fractions were analyzed by Western blot analysis. β-Actin was used as a control. (B) Quantification of bands was performed using Odyssey infrared imaging and ImageJ software and is displayed as the ratio of NS1 present in nucleus and cytoplasm. The averaged results of three independent experiments are shown.
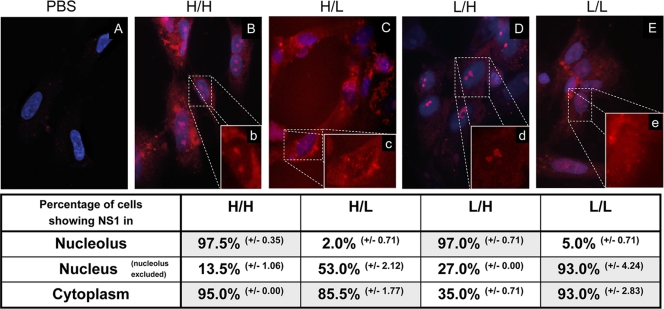
Finally, it was of interest to find out if NS1 was transported into the nucleolus. CF were infected with MOI of 1.0, and immunofluorescence studies were performed 7.5 h p.i. Nuclei were stained with DAPI, and NS1 was visualized by using a goat polyclonal anti-NS1 antibody and a rhodamine-conjugated secondary anti-goat antibody. In 984 H/H-infected cultures, the majority of cells showed cytoplasmic localization of NS1. Interestingly, in most cells NS1 was also present in the nucleolus (Fig. 4 B) at this time point. Taken together with the results from the cell fractionation experiment (Fig. 3), these data show that in 984 H/H-infected cells NS1 is predominantly localized in the cytoplasm, but that a smaller fraction specifically accumulates in the nucleolus. After infection with 984 L/H there is also abundant nucleolar localization of NS1, whereas only one-third of the cells display NS1 in the cytoplasm (Fig. 4D). Again, in conjunction with the cell fractionation data, these results show that in 984 L/H-infected cells NS1 is equally distributed between nucleoli and cytoplasm. Thus, the C-terminal truncation appears to be responsible for the transport of NS1 into nucleoli. In contrast, when NS1 has a nontruncated C terminus, as is the case with 984 H/L and 984 L/L., it is completely absent from nucleoli (Fig. 4C and E) and appears only in the cytoplasm or in both cytoplasm and nuclei (Fig. 3). This NS1 distribution pattern was only visible at early times (5 h and 7.5 h p.i.), whereas at later stages of infection (10 h p.i.) all NS1 variants remigrated from the cytoplasm into the nucleus (data not shown). Furthermore, NS1 of 984 H/H and 984 L/H was no longer detectable in the nucleoli of the infected cells, as was the case with 984 L/H and 984 L/L.
FIG. 4.
Nucleolar localization of NS1. CF cells were inoculated with PBS (A), viruses H/H (B, b), L/H (C, c), H/L (D, d) and L/L (E, e) at an MOI of 1.0 PFU per cell. Cells were fixed at 7.5 h postinoculation, and NS1 was detected by indirect immunofluorescence with goat anti-NS1 polyclonal antibody and rhodamine-labeled anti-goat antibody. Nuclei were stained with DAPI. Magnification, ×630. Insets have an additional ×2 digital magnification. For each virus, the percentage of cells expressing NS1 protein in the nucleus, nucleolus, or cytoplasm was calculated by analyzing 2 × 100 infected CF in two independent experiments.
Interestingly, NS localization in CF strikingly differed from the patterns observed in human A549 cells (Fig. 5). Thus, in A549 cells NS1 of 984 H/H is observed only in the nucleus, whereas in chicken cells it is localized in the cytoplasm and the nucleolus. Likewise, NS1 of 984 L/H and 984 L/L was also present exclusively in the nucleus of A549 cells, as opposed to the nucleolar and/or cytoplasmic targeting in chicken cells. Only NS1 of 984 H/L showed a similar compartmentation in both cells, with accumulation in the cytoplasm and little or nothing present in nucleus and nucleolus. These results indicate that the localization signals of NS1 are differently interpreted by the cellular protein transport machineries of different cells.
FIG. 5.
Comparative analysis of NS1 localization in chicken fibroblasts and A549 cells. CF and human A549 cells were infected with viruses H/H, H/L, L/H, L/L, or PBS at an MOI of 1.0 PFU per cell. (A) Cells were fixed at 7.5 h postinoculation, and NS1 was detected by indirect immunofluorescence using goat anti-NS1 polyclonal antibody and rhodamine-labeled anti-goat antibody. Nuclei were stained with DAPI. (B) For each virus, the percentage of cells expressing NS1 protein in the nucleus, nucleolus, or cytoplasm was calculated by analyzing 2 × 100 infected chicken fibroblast or human A549 cells in two independent experiments.
DISCUSSION
We have analyzed here mutations in the NS1 protein that were observed when highly pathogenic H7N1 viruses emerged from their low pathogenic precursors. The mutations are located in two important targeting signals of NS1, the NES and the NoLS, and have therefore distinct effects on the cellular compartmentalization of the protein. Our data show that the NES of the HPAI viruses, as present in mutants H/H and H/L, enhances cytoplasmic localization of NS1 (Fig. 3) and reduces IFN-β transcription (Fig. 2) compared to the NES of the LPAI viruses present in mutants L/H and L/L. These observations suggest that enhanced IFN antagonism as a result of NES-mediated accumulation of NS1 in the cytoplasm increases pathogenicity. On the other hand, mutants H/L and L/H have similar infectivity and replication rates ranging between the extremes of mutants H/H and L/L (Fig. 1). Thus, it appears that NoLS-dependent functions of NS1 also determine the replication and presumably the pathogenicity of these viruses. When we analyzed the MDT for chicken embryos we observed that mutant 984 L/L, indeed, had a lower virulence than the 984 H/H virus. The difference was significant but not dramatic. This was not unexpected since mutant 984 L/L still has retained the multibasic hemagglutinin cleavage site as a major determinant of high pathogenicity. Mutant 984 L/L is therefore still a HPAI virus, yet with reduced pathogenicity. Taken together, these observations provide further support to the concept that NS1 is an important determinant of pathogenicity.
The strong nucleolar accumulation of the 984 virus NS1 in chicken cells evolved by a truncation of 6 amino acids at its C terminus. This observation throws new light on the structure of NoLS. Table 2 shows a comparison of the C-terminal amino acid sequences and the nucleolar localization of NS1 observed with several viruses including the ones analyzed here. Our data support the concept that NoLS is destroyed by a nonbasic amino acid (E) in position 229 (25). In addition, they indicate that not only the replacement of E by K but also the removal of E by a deletion can restore the nucleolar localization of NS1. Thus, the sequence K219-R220-X-X-X-R224 appears to be sufficient for a functional NoLS, at least in the system analyzed here. Volmer et al. (36) reported very recently that NS1 accumulated in the nucleoli of avian cells in the absence of a C-terminal NoLS, suggesting that other signals may be responsible for nucleolar localization in such cells. Although there is no direct evidence supporting this concept from the data presented here, they do not exclude the possibility that, besides the C-terminal NoLS, additional nucleolar targeting signals exist.
TABLE 2.
Variation of amino acid sequences at the NS1 C termini of different influenza A viruses
a Consensus sequence for NoLS (25). The NoLS sequence is highlighted in dark gray and light gray at positions 219, 220, 224, 229, 231, and 232 of the NS1 protein. The nonbasic amino acid at position 229 (highlighted in light gray) inhibits the nucleolar localization of NS1.
b *, Data from Melen et al. (25).
C-terminal deletions of the NS1 protein have been observed in many influenza A virus subtypes, such as H1N1, H3N2 (20), H3N8 (2), and H5N1 (14). They have often been found to be responsible for reduced IFN antagonism and therefore for virus attenuation (4, 10, 23, 34). NS1 of the 2009 pandemic H1N1 influenza A virus also has a C-terminal truncation, but elongation has no effect on pathogenicity (18). In contrast, we show here that C-terminal truncation of NS1 contributes to enhanced pathogenicity without significantly suppressing IFN transcription. Thus, there are striking variations in the consequences of such deletions that may reflect the multiple functions associated with the C terminus of NS1 (8).
When localized in the nucleolus, influenza NS1 protein was shown to inhibit rRNA synthesis (21) and to interact with nucleolin, a multifunctional nucleolar protein (30). As described for other viruses (19), this might favor viral transcription and translation or alter the cell cycle for the viral needs. The nucleolar localization of dengue, kunjin, and Japanese encephalitis virus core proteins apparently plays a critical role in virus replication and pathogenesis in mammalian cells (29, 37, 39). Apart from the nucleolar accumulation, there are more functions related with the C terminus of influenza NS1, such as the proposed PABII-binding site located between amino acid positions 223 and 237 (5). This binding site is destroyed by the truncation found in the H7N1 HPAI NS1. The interaction between NS1 and PABII is thought to cause a reduction in cellular mRNA export from the nucleus, leading to shutoff cellular protein translation and, in addition, to hold Cap structures available in the nucleus for the viral polymerase (7). Such a mechanism, however, does not provide an explanation for the observations made here. A missing interaction between the NS1 and PABII would probably lead to decreased virulence; the opposite of what we can detect in infected chicken embryos.
For the last four amino acids at the NS1 C terminus, another important function was identified. These amino acids can act as a ligand that binds to PDZ domains of proteins playing important roles in many cellular signaling pathways. NS1 of avian influenza viruses is able to bind to many PDZ domain-containing human proteins, whereas NS1 of human strains cannot (31). In addition, mice that were infected with A/WSN viruses containing NS1 proteins with avian PDZ ligands showed increased pathogenicity (20). PDZ domain-containing proteins were shown to regulate cell junctions, cellular signal transduction, and protein sorting (33). Although little is known about PDZ domains in avian cells, cellular signaling pathways might be disturbed when NS1 binds to avian proteins containing PDZ domains. The inability to bind might therefore be of benefit for the virus.
The NES of NS1 has the consensus sequence X138-X-X-X-X-X-L144-X-L146-X147 (22). Although mutations V136I and D139N do not change this motif, it is reasonable to assume that they modulate the NES, since they shift NS1 localization from the nucleus to the cytoplasm. Increased amounts of NS1 in the cytoplasm might reduce IFN-β transcription by various mechanisms involving cytoplasmic factors. Thus, NS1 can directly block the function of two cytoplasmic antiviral proteins, 2′-5′ oligo(A) synthetase (OAS) (28) and the double-stranded RNA-dependent serine/threonine protein kinase R (PKR). Both OAS and PKR are key regulators of viral transcription/translation processes but play additional roles in other innate defense mechanisms, such as IFN-β induction and apoptosis (12, 40). Another very important mechanism is the interaction of NS1 with RIG-I in the cytoplasm. It was demonstrated that RIG-I senses single-stranded RNA viral genomes bearing 5′-phosphates and activates the induction of IFN-β (17, 26, 32). This recognition can be blocked by NS1 through binding directly or via an additional factor to RIG-I (16, 32). Furthermore, the enhanced amount of NS1 in the cytoplasm might lead to decreased IFN-β transcription by inhibiting the activation of the cytoplasmic proteins IRF-3 and NF-κB (35, 37). Finally, cytoplasmic accumulation of NS1 might enhance translation of viral proteins and thus contribute to the elevated growth rates observed (6).
We also present evidence that intracellular distribution of NS1 is host cell specific. None of the 984 NS1 mutants was able to accumulate in the nucleolus of human A549 cells. In the case of 984 L/L and H/L, this had to be expected since these viruses have an NS1 with the NoLS destroying nonbasic residue E at position 229 (25). However, truncation of the C terminus of NS1 did not restore the NoLS activity of mutants 984 H/H and L/H in human cells as it did in chicken cells, suggesting that the NoLS sequences might differ depending on the host cell.
In conclusion, we have identified mutations in the NS1 protein of a highly pathogenic avian influenza virus that affect the intracellular location of this protein in a host-dependent manner. We show that a strong NES and a restored NoLS resulting from C-terminal truncation lead to the accumulation of NS1 in the cytoplasm and enhanced recruitment of the protein into the nucleolus of chicken cells. We also demonstrate that the altered compartmentation of NS1 is associated with an increase of virulence for the avian host.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft (SFB 593, TP B1 and SPP1190) and by the AVIFLU program of the European Commission.
Footnotes
Published ahead of print on 15 September 2010.
REFERENCES
- 1.Alexander, D., and I. Brown. 2009. History of highly pathogenic avian influenza. Rev. Sci. Tech. 28:19-38. [DOI] [PubMed] [Google Scholar]
- 2.Bryant, N., A. Rash, C. Russell, J. Ross, A. Cooke, S. Bowman, S. MacRae, N. Lewis, R. Paillot, R. Zanoni, H. Meier, L. Griffiths, J. Daly, A. Tiwari, T. Chambers, J. Newton, and D. Elton. 2009. Antigenic and genetic variations in European and North American equine influenza virus strains (H3N8) isolated from 2006 to 2007. Vet. Microbiol. 138:41-52. [DOI] [PubMed] [Google Scholar]
- 3.Capua, I., G. Cattoli, C. Terregino, and S. Marangon. 2008. Avian influenza in Italy, 1997-2006, p. 59-70. In Avian influenza, vol. 27. Karger, Basel, Switzerland. [Google Scholar]
- 4.Cauthen, A., D. Swayne, M. Sekellick, P. Marcus, and D. Suarez. 2007. Amelioration of influenza virus pathogenesis in chickens attributed to the enhanced interferon-inducing capacity of a virus with a truncated NS1 gene. J. Virol. 81:1838-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, Z., Y. Li, and R. Krug. 1999. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 18:2273-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de la Luna, S., P. Fortes, A. Beloso, and J. Ortín. 1995. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J. Virol. 69:2427-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dias, A., D. Bouvier, T. Crépin, A. McCarthy, D. Hart, F. Baudin, S. Cusack, and R. Ruigrok. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458:914-918. [DOI] [PubMed] [Google Scholar]
- 8.Dundon, W., and I. Capua. 2009. A closer look at the NS1 of influenza virus. Viruses 1:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dundon, W., A. Milani, G. Cattoli, and I. Capua. 2006. Progressive truncation of the non-structural 1 gene of H7N1 avian influenza viruses following extensive circulation in poultry. Virus Res. 119:171-176. [DOI] [PubMed] [Google Scholar]
- 10.Falcón, A., R. Marión, T. Zürcher, P. Gómez, A. Portela, A. Nieto, and J. Ortín. 2004. Defective RNA replication and late gene expression in temperature-sensitive influenza viruses expressing deleted forms of the NS1 protein. J. Virol. 78:3880-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García, M., J. Gil, I. Ventoso, S. Guerra, E. Domingo, C. Rivas, and M. Esteban. 2006. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 70:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology 279:375-384. [DOI] [PubMed] [Google Scholar]
- 13.Garten, W., and H. Klenk. 2008. Cleavage activation of the influenza virus hemagglutinin and its role in pathogenesis, p. 156-167. In Avian influenza, vol. 27. Karger, Basel, Switzerland. [Google Scholar]
- 14.Govorkova, E., J. Rehg, S. Krauss, H. Yen, Y. Guan, M. Peiris, T. Nguyen, T. Hanh, P. Puthavathana, H. Long, C. Buranathai, W. Lim, R. Webster, and E. Hoffmann. 2005. Lethality to ferrets of H5N1 influenza viruses isolated from humans and poultry in 2004. J. Virol. 79:2191-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenspan, D., P. Palese, and M. Krystal. 1988. Two nuclear location signals in the influenza virus NS1 nonstructural protein. J. Virol. 62:3020-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo, Z., L. Chen, H. Zeng, J. Gomez, J. Plowden, T. Fujita, J. Katz, R. Donis, and S. Sambhara. 2007. NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am. J. Respir. Cell Mol. Biol. 36:263-269. [DOI] [PubMed] [Google Scholar]
- 17.Hale, B., R. Randall, J. Ortín, and D. Jackson. 2008. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89:2359-2376. [DOI] [PubMed] [Google Scholar]
- 18.Hale, B., J. Steel, B. Manicassamy, R. Medina, J. Ye, D. Hickman, A. Lowen, D. Perez, and A. Garcia-Sastre. 2010. Mutations in the NS1 C-terminal tail do not enhance replication or virulence of the 2009 pandemic H1N1 influenza A virus. J. Gen. Virol. 91:1737-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiscox, J. 2002. The nucleolus: a gateway to viral infection? Arch. Virol. 147:1077-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson, D., M. Hossain, D. Hickman, D. Perez, and R. Lamb. 2008. A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc. Natl. Acad. Sci. U. S. A. 105:4381-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krug, R., W. Yuan, D. Noah, and A. Latham. 2003. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology 309:181-189. [DOI] [PubMed] [Google Scholar]
- 22.Li, Y., Y. Yamakita, and R. Krug. 1998. Regulation of a nuclear export signal by an adjacent inhibitory sequence: the effector domain of the influenza virus NS1 protein. Proc. Natl. Acad. Sci. U. S. A. 95:4864-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipatov, A., S. Andreansky, R. Webby, D. Hulse, J. Rehg, S. Krauss, D. Perez, P. Doherty, R. Webster, and M. Sangster. 2005. Pathogenesis of Hong Kong H5N1 influenza virus NS gene reassortants in mice: the role of cytokines and B- and T-cell responses. J. Gen. Virol. 86:1121-1130. [DOI] [PubMed] [Google Scholar]
- 24.Matrosovich, M., T. Matrosovich, W. Garten, and H. Klenk. 2006. New low-viscosity overlay medium for viral plaque assays. Virol. J. 3:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melén, K., L. Kinnunen, R. Fagerlund, N. Ikonen, K. Twu, R. Krug, and I. Julkunen. 2007. Nuclear and nucleolar targeting of influenza A virus NS1 protein: striking differences between different virus subtypes. J. Virol. 81:5995-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mibayashi, M., L. Martínez-Sobrido, Y. Loo, W. Cárdenas, M. J. Gale, and A. García-Sastre. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 81:514-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Min, J., and R. Krug. 2006. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: inhibiting the 2′-5′ oligo(A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. U. S. A. 103:7100-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Min, J., S. Li, G. Sen, and R. Krug. 2007. A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology 363:236-243. [DOI] [PubMed] [Google Scholar]
- 29.Mori, Y., T. Okabayashi, T. Yamashita, Z. Zhao, T. Wakita, K. Yasui, F. Hasebe, M. Tadano, E. Konishi, K. Moriishi, and Y. Matsuura. 2005. Nuclear localization of Japanese encephalitis virus core protein enhances viral replication. J. Virol. 79:3448-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murayama, R., Y. Harada, T. Shibata, K. Kuroda, S. Hayakawa, K. Shimizu, and T. Tanaka. 2007. Influenza A virus nonstructural protein 1 (NS1) interacts with cellular multifunctional protein nucleolin during infection. Biochem. Biophys. Res. Commun. 362:880-885. [DOI] [PubMed] [Google Scholar]
- 31.Obenauer, J., J. Denson, P. Mehta, X. Su, S. Mukatira, D. Finkelstein, X. Xu, J. Wang, J. Ma, Y. Fan, K. Rakestraw, R. Webster, E. Hoffmann, S. Krauss, J. Zheng, Z. Zhang, and C. Naeve. 2006. Large-scale sequence analysis of avian influenza isolates. Science 311:1576-1580. [DOI] [PubMed] [Google Scholar]
- 32.Pichlmair, A., O. Schulz, C. Tan, T. Näslund, P. Liljeström, F. Weber, and C. Reis e Sousa. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997-1001. [DOI] [PubMed] [Google Scholar]
- 33.Sheng, M., and C. Sala. 2001. PDZ domains and the organization of supramolecular complexes. Annu. Rev. Neurosci. 24:1-29. [DOI] [PubMed] [Google Scholar]
- 34.Solórzano, A., R. Webby, K. Lager, B. Janke, A. García-Sastre, and J. Richt. 2005. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J. Virol. 79:7535-7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Talon, J., C. Horvath, R. Polley, C. Basler, T. Muster, P. Palese, and A. García-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Volmer, R., B. Mazel-Sanchez, C. Volmer, S. Soubies, and J. Guérin. 2010. Nucleolar localization of influenza A NS1: striking differences between mammalian and avian cells. Virol. J. 7:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, S., W. Syu, K. Huang, H. Lei, C. Yao, C. King, and S. Hu. 2002. Intracellular localization and determination of a nuclear localization signal of the core protein of dengue virus. J. Gen. Virol. 83:3093-3102. [DOI] [PubMed] [Google Scholar]
- 38.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. Beg, and A. García-Sastre. 2000. Influenza A virus NS1 protein prevents activation of NF-κB and induction of alpha/beta interferon. J. Virol. 74:11566-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Westaway, E., A. Khromykh, M. Kenney, J. Mackenzie, and M. Jones. 1997. Proteins C and NS4B of the flavivirus Kunjin translocate independently into the nucleus. Virology 234:31-41. [DOI] [PubMed] [Google Scholar]
- 40.Zhirnov, O., T. Konakova, T. Wolff, and H. Klenk. 2002. NS1 protein of influenza A virus downregulates apoptosis. J. Virol. 76:1617-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]