Abstract
The nature of influenza virus to randomly mutate and evolve into new types with diverse antigenic determinants is an important challenge in the control of influenza infection. Particularly, variations within the amino acid sequences of major neutralizing epitopes of influenza virus hemagglutinin (HA) hindered the development of universal vaccines against H5N1 lineages. Based on distribution analyses of the identified major neutralizing epitopes of hemagglutinin, we selected three vaccine strains that cover the entire variants in the neutralizing epitopes among the H5N1 lineages. HA proteins of selected vaccine strains were expressed on the baculovirus surface (BacHA), and the preclinical efficacy of the vaccine formulations was evaluated in a mouse model. The combination of three selected vaccine strains could effectively neutralize viruses from clades 1, 2.1, 2.2, 4, 7, and 8 of influenza H5N1 viruses. In contrast, a vaccine formulation containing only adjuvanted monovalent BacHA (mono-BacHA) or a single strain of inactivated whole viral vaccine was able to neutralize only clade 1 (homologous), clade 2.1, and clade 8.0 viruses. Also, the trivalent BacHA vaccine was able to protect 100% of the mice against challenge with three different clades (clade 1.0, clade 2.1, and clade 7.0) of H5N1 strains compared to mono-BacHA or inactivated whole viral vaccine. The present findings provide a rationale for the development of a universal vaccine against H5N1 lineages. Furthermore, baculoviruses displaying HA will serve as an ideal choice for a vaccine in prepandemic or pandemic situations and expedite vaccine technology without the requirement of high-level-biocontainment facilities or tedious protein purification processes.
The nature of influenza virus to randomly mutate and evolve into new types with diverse antigenic determinants is an important challenge in the control of influenza infection (20). This has been evidently recognized by the recent outbreaks of H5N1 avian influenza virus infection and the current pandemic situation with H1N1 swine-origin influenza A virus (S-OIV). In fact, it has been well documented in the literature that H5N1 had acquired the ability to infect human tissues due mainly to the occurrence of mutation events (1). Highly pathogenic avian influenza (HPAI) H5N1 viruses are antigenically distinguishable owing to differences in hemagglutinin (HA) sequences, the principal determinant of immunity to influenza virus, resulting in different lineages or clades of H5N1 (13, 33). The control of infection with current H5N1 vaccines does not appear to be effective against heterologous strains or phylogenetically variant clades of H5N1 in part due to variations in the HA sequences, particularly within the neutralizing epitope region. Since present vaccines are based solely on the induction of neutralizing antibodies against these epitopes, differences in these sequences may render current vaccines unqualified for the prevention of influenza globally (15, 28, 31). To overcome such limitations and to completely realize the potential of vaccines worldwide, the concept of universal vaccines based on conserved viral proteins has recently been proposed. The highly conserved ion channel protein (M2) and the nucleoprotein (NP) of influenza virus have been evaluated for the induction of cross-protective cellular immunity and viral clearance (2, 35). Antibodies generated against these conserved proteins may reduce viral spread and accelerate recovery from influenza (14). However, antibodies specific to these proteins are poorly immunogenic and were found previously to be infection permissive (5-7, 13). Thus, the development of a vaccine based on influenza virus hemagglutinin appears to be the only viable option to prevent infections by HPAI viruses such as H5N1 viruses. Nevertheless, amino acid variations within the major antigenic neutralizing epitope regions among H5 subtypes restrict the development of such universal vaccines against different H5N1 lineages.
The development of a universal vaccine based entirely on HA of influenza virus is still feasible, if the variation or conservation of neutralizing epitopes among the several HPAI H5N1 virus clades can be identified. An understanding of the distribution pattern of such neutralizing epitopes could help in the design of future vaccines by incorporating two or more ideal H5N1 strains in the vaccine composition. The neutralizing epitopes of the selected viral strains should cover the variations among most H5 subtypes in order to acquire broad-range protective immunity against most H5N1 subtypes. Previous attempts to identify amino acid substitutions within HA sequences of variants that escaped from neutralization by monoclonal antibodies (MAbs) revealed the neutralizing epitope sites of HA (9, 10). Along with previous findings, we report here the identification of other major neutralizing epitopes of H5N1 by mapping their amino acid sequences using neutralizing monoclonal antibodies (n-MAbs). Analysis of the distribution of all identified neutralizing epitopes among H5 subtypes revealed variations within the antigenic determinants of H5N1 subtypes from both human and avian sources. Based on these results, we have selected three vaccine strains comprising the major neutralizing epitopes of HA to cover the entire variants within H5N1 lineages. In order to test our hypothesis in vivo, HA proteins of selected vaccine strains were expressed on the baculovirus surface (BacHA), and the efficacy of the vaccine formulations was evaluated with a mouse model challenged with phylogenetically variant H5N1 strains.
MATERIALS AND METHODS
Viruses.
H5N1 human influenza viruses from clade 2.1, A/Indonesia/CDC669/2006 and A/Indonesia/TLL013/2006, and one avian strain, A/Indonesia/TLL014/2006, were obtained from the Ministry of Health (MOH), Republic of Indonesia. The avian H5N2 (clade 1, A/chicken/Singapore/98) subtype was obtained from the Agri-Food and Veterinary Authority (AVA) of Singapore. The H5N1 viruses from different phylogenetic clades or subclades were rescued by reverse genetics (33). Briefly, the hemagglutinin (HA) and neuraminidase (NA) genes of H5N1 viruses from clades 1 (A/Vietnam1203/04), 2.1 (A/Indonesia/CDC1031/07), 2.2 (A/turkey/Turkey1/05), 2.2 [A/Nigeria/6e/07 (H5N1)], 2.3 (A/Anhui/1/05), 4 (A/goose/Guiyang/337/06), 7 (A/chicken/Shanxi/2/06), and 8 (A/chicken/Henan/12/04) were synthesized (GenScript) based on the sequences in the NCBI influenza virus database. The synthetic HA and NA genes were cloned into a dual-promoter plasmid for influenza A virus reverse genetics (22). The dual-promoter plasmids were obtained from the Centers for Disease Control and Prevention, Atlanta, GA. Reassortant viruses were rescued by transfecting plasmids containing HA and NA along with the remaining six influenza virus genes derived from high-growth master strain A/Puerto Rico/8/34 (H1N1) into cocultured 293T and MDCK cells by using Lipofectamine 2000 (Invitrogen Corp.). At 72 h posttransfection the culture medium was inoculated into embryonated eggs or MDCK cells. The HA and NA genes of reassortant viruses from the second passage were sequenced to confirm the presence of the introduced HA and NA genes and the absence of mutations. Stock viruses were propagated in the allantoic cavity of 10-day-old embryonated eggs, and virus containing allantoic fluid was harvested and stored in aliquots at −80°C. Virus content was determined by a standard hemagglutination assay as described previously (32). All experiments with highly pathogenic viruses were conducted in a biosafety level 3 (BSL-3) containment facility in compliance with CDC/NIH and WHO recommendations (17).
Production of n-MAbs.
A panel of five different virus-neutralizing MAbs (2D9, 3H11, 4C2, 4F8, and 6B8) to the HAs of H5 strains was used. The MAbs were produced as described previously (36). Briefly, BALB/c mice were immunized twice 2 weeks apart by the subcutaneous injection of purified formalin-inactivated A/Indonesia/CDC669/2006 (H5N1), A/Indonesia/TLL014/2006 (H5N1), or A/chicken/Singapore/98 (H5N2) virus mixed with Montanide ISA563 adjuvant (Seppic, France). Mice received an additional intravenous injection of the same viral antigen 3 days before the fusion of splenocytes with SP2/0 cells. Hybridoma culture supernatants were screened by immunofluorescence assays. Hybridomas that produced specific MAbs were cloned by limiting dilution, expanded, and further subcultured. The hybridoma culture supernatant was clarified and stored at −20°C. The MAbs were then tested for hemagglutination inhibition (HI) as described below. MAbs 2D9, 4F8, and 3H11 were obtained with mice immunized with A/Indonesia/CDC669/2006 (H5N1) virus. MAbs 4C2 and 6B8 were obtained with mice immunized with the A/Indonesia/TLL014/2006 (H5N1) and A/chicken/Singapore/98 (H5N2) viruses, respectively.
Mapping of H5 HA-neutralizing epitopes by using n-MAbs.
The neutralizing epitopes of H5 were mapped by the characterization of escape mutants (10, 23) with five different neutralization monoclonal antibodies (6B8, 4C2, 2D9, 4F8, and 3H11). Briefly, H5N1 virus was incubated with an excess amount of n-MAb for 1 h, and the mixture was inoculated into 10-day-old embryonated chicken eggs. The eggs were incubated at 37°C for 48 h. Virus was harvested and used for cloning in limiting dilution in embryonated chicken eggs, and the escape mutants were plaque purified. RNA was extracted from the allantoic fluid. The hemagglutinin gene was reverse transcriptase PCR (RT-PCR) amplified and cloned into a TA cloning vector (Promega), and several clones were sequenced. The sequences of individual clones were analyzed by comparisons with the sequences of the parent viruses listed in Table 1.
TABLE 1.
Neutralizing epitopes of H5 HA using n-MAbs by escape mutations
| Parental virusrb | MAb | Nucleotide | Nucleotide change | Amino acida | Amino acid change |
|---|---|---|---|---|---|
| A/Indonesia/CDC669/2006 | 3H11 | 416 | G to A | 139 | Gly to Glu |
| 4C2 | 566 | G to A | 189 | Arg to Lys | |
| 2D9 | 669 | T to A | 223 | Ser to Arg | |
| 565 | A to T | 189 | Arg to Trp | ||
| 4F8 | 464 | G to T | 155 | Ser to Ile | |
| A/Viet Nam/1203/2004 | 6B8 | 566 | A to C | 189 | Lys to Thr |
| 566 | A to T | 189 | Lys to Met | ||
| 567 | G to T | 189 | Lys to Asn | ||
| A/Indonesia/CDC594/2006 | 6B8 | 463 | A to G | 155 | Asn to Asp |
Amino acid numbering excludes the signal peptide of HA.
Viruses used for the selection of escape mutants.
Epitope distribution analysis.
Neutralizing epitopes of H5 HA identified previously by Kaverin et al. (10) and Stevens et al. (30) along with major epitopes identified in this study were taken into consideration for sequence analysis. To analyze the distribution of the neutralizing epitopes among the H5N1 viruses, full-length HA sequences were compared with avian and human H5N1 viruses from the NCBI influenza database. The protein polymorphism of H5N1 was analyzed with the Influenza Research Database (http://Fludb.org/), and up to 317 human strains and 2,028 avian strains were aligned in this study (13 March 2010, accession date). To assess variations in the epitopes, a positional-frequency table (Table 2) was produced from multiple sequence alignments for each H5N1 virus.
TABLE 2.
Epitope frequency in H5N1strains
| aa position | Frequency of aa (%) |
|
|---|---|---|
| Human H5N1 | Avian H5N1 | |
| 138 | Gln, 58.9; Leu, 41.1 | Gln, 80.4; Leu, 13.6 |
| 140 | Lys, 22.5; Ser, 28.5; Thr, 6 | Lys, 34.8; Ser, 6; Thr, 16.6 |
| 155 | Asn, 34.4; Ser, 63.4 | Asn, 50.6; Ser, 43.2 |
| 189 | Arg, 64.3; Lys, 34.7 | Arg, 43.3; Lys, 55.1 |
| 159 | Thr, 99.7 | Thr, 98.4 |
| 194 | Pro, 100 | Pro, 98 |
| 218 | Lys, 99.7 | Lys, 99.2 |
Selection of vaccine strains.
Three different H5N1 strains were selected to cover the variations within the neutralizing epitopes of HAs among most H5N1 lineages. Furthermore, the reactivity of the selected vaccine strains was confirmed by hemagglutination inhibition assays and microneutralization assays using appropriate neutralizing monoclonal antibodies.
Generation of recombinant baculovirus.
For the generation of recombinant baculovirus vectors, as described previously (21), white spot syndrome virus (WSSV) immediate-early 1 (ie1) promoter-controlled HA expression cassettes were inserted into shuttle vector pFastBac1 and integrated into the baculovirus genome within DH10BAC according to the protocol of the Bac-To-Bac system (Invitrogen). Briefly, the full-length HA gene was amplified from three different influenza virus strains with a standard PCR method (94°C for 20 s, 55°C for 30 s, and 72°C for 2 min for 30 cycles). H5 HA sequences were amplified with primers H5FSal (5′-ACGCGTCGACATGGAGAAAATAGTGCTTCT-3′) and H5RNot (5′-ATAAGGCGGCCGCTTAAATGCAAATTCTGCATTG-3′) from the A/Indonesia/CDC669/2006 (H5N1) (GenBank accession number ABI36428), A/Vietnam/1203/2004 (H5N1) (accession number ABW90135), and A/Anhui/1/2005 (H5N1) (accession number ABD28180) viruses. The WSSV ie1 promoter was inserted into the vector with SnabI and SalI sites, and HA genes were inserted into vector pFastBac1 with the NotI-SalI site. After the transformation of transfer vectors into DH10BAC, recombinant bacmids were transfected into Sf9 cells, and the budded baculovirus particles released into the medium were harvested at 4 days posttransfection.
The viral titers were determined by plaque assays and hemagglutination assays. A large-scale amplification of BacHA was carried out, and the virus particles were purified by two rounds of sucrose gradient centrifugation according to standard protocols (18). Recombinant baculovirus and influenza H5N1 viruses were inactivated with binary ethylenimine (BEI) as described previously by Rueda et al. (27) and King (11), respectively. The complete loss of infectivity of the inactivated BacHA and H5N1 viruses was determined by infection of Sf9 cell and MDCK cell monolayers, respectively. The cells were monitored daily under a microscope for cytopathic effects.
Vaccine trial.
Specific-pathogen-free female BALB/c mice (6 weeks old) were obtained from the Laboratory Animals Centre, National University of Singapore, and maintained at the Animal Holding Unit of the Temasek Life Sciences Laboratory. Twenty-four mice per each experimental group (n = 24 mice/group) were vaccinated subcutaneously two times at a regular interval of 28 days with 100 μl (HA titer, 128) of baculovirus displaying the HA of A/Vietnam/1203/2004 (H5N1) (mono-BacHA) or as a mixture of each baculovirus displaying the HAs from A/Indonesia/CDC669/2006 (H5N1), A/Vietnam/1203/2004 (H5N1), and A/Anhui/1/2005 (H5N1) (tri-BacHA) with or without the adjuvant Montanide ISA563 (water-in-oil emulsion; Seppic, France). Also, inactivated RG-H5N1 virus [A/VietNam/1203/2004 (H5N1)] (HA titer, 128) was used as a reference vaccine control. The serum was collected from 10 mice per experimental group on days 14, 28, and 42. Levels of hemagglutination inhibition activity and serum cross-clade neutralizing antibody were measured. The efficacy of the vaccine was assessed by host challenge against HPAI H5N1 influenza virus strains from different clades. All animal experiments were carried out in accordance with guidelines for animal experiments from the National Institute of Infectious Diseases (NIID), and experimental protocols were reviewed and approved by the Institutional Animal Care and Use Committee of the Temasek Life Sciences Laboratory, National University of Singapore, Singapore.
Hemagglutination inhibition assay.
Hemagglutination inhibition assays were performed as described previously (32). Receptor-destroying enzyme (RDE)-treated sera were serially diluted (2-fold) in V-bottom 96-well plates. Approximately 4 HA units of viral antigen was incubated with the serum for 30 min at room temperature, followed by the addition of 1% chicken red blood cells (RBCs) and incubation at room temperature for 40 min.
Microneutralization assay.
The microneutralization test was performed according to a previously described protocol (26). Briefly, MDCK cells were seeded into 96-well culture plates and cultured at 37°C to form a monolayer. Serial 2-fold dilutions of heat-inactivated (56°C for 30 min) serum samples were mixed separately with 100 50% tissue culture infective doses (TCID50) of H5N1 virus from different clades and incubated at room temperature for 1 h, and the mixtures were added to a monolayer of MDCK cells in triplicate wells. The neutralizing titers of mouse antiserum that completely prevented any cytopathic effect at reciprocal dilutions were calculated.
Diseases challenge test against H5N1 virus infection.
Three weeks after the final vaccination, mice were transferred into an animal BSL-3 containment facility. Six mice per group were challenged intranasally with 5 50% mouse lethal doses (MLD50) of homologous clade 1.0 (A/Vietnam/1203/2004), heterologous clade 2.1 (A/Indonesia/TLL13/06), and clade 7.0 (A/chicken/Shanxi/2/06) HPAI H5N1 virus strains. The MLD50 of influenza virus required for intranasal challenge experiments was predetermined. Mice were observed daily to monitor body weight and mortality. Monitoring continued until all animals died or until day 14 after challenge. For histopathology, mice were necropsied, and the lungs were stored in 10% (wt/vol) neutral buffered formalin, embedded in paraffin, and sectioned. Sections were stained with hematoxylin and eosin (H&E) prior to light microscopy examination and were evaluated for lung pathology. All challenge experiments were conducted at an animal biosafety level 3 containment facility.
Statistical analysis.
The data are expressed as arithmetic means ± standard deviations (SD) or standard errors (SE). An unpaired two-tailed Student's t test was performed to determine the level of significance in the difference between the means of two groups. One-way analysis of variance (ANOVA) was also used to test for differences between groups, and the Tukey honestly significant difference (HSD) post hoc test was used to determine which groups were significantly different from the rest. All statistical analyses were done with SigmaStat 2.0 (Jandel Corporation) software. The level of significance was expressed as a P value of <0.05.
RESULTS
Identification and characterization of neutralizing epitopes of H5 HA using n-MAbs.
A panel of five different neutralizing MAbs (6B8, 4C2, 2D9, 4F8, and 3H11) against influenza virus hemagglutinin (HA) was produced previously in our laboratory. All n-MAbs recognized the conformational epitopes of H5 and possessed the ability to neutralize influenza virus infection in vitro. Also, these n-MAbs were confirmed to have hemagglutination inhibition activity (data not shown). Amino acids involved in forming the epitopes of n-MAbs were analyzed by using virus escape mutants (Table 1). Sequencing of the complete HA gene isolated from multiple escape variants of n-MAb 6B8 carried single point mutations at amino acid (aa) position 189 (Lys to Asn) or 155 (Asn to Asp), n-MAb 4F8 carried a single point mutation at amino acid position 155 (Asn to Asp), and n-MAb 2D9 carried a single point mutation at amino acid position 189 (Arg to Trp) or 223 (Ser to Arg). A similar analysis of n-MAb 4C2 revealed the involvement of amino acid 155 (Ser to Ile) or 189 (Arg to Lys) in forming the epitope, while n-MAb 3H11 carried a single point mutation at amino acid position 138 (“Leu”), 139 (“Gly”), or 140 (“Ser”). All amino acid positions indicated here exclude the signal peptide of HA (Table 1).
Epitope distribution analysis.
Full-length HA sequences of avian and human H5N1 viruses were obtained from the Influenza Virus Database, maintained by the National Center for Biotechnology Information. HA sequences of human and avian H5N1 isolates were compared with those amino acids related to the major neutralizing epitopes. In addition, other major neutralizing epitopes (positions 140, 159, 194, and 218) previously identified by other groups were also taken into consideration for subsequent analyses. The results revealed that most of the major antigenic epitope regions contained significant variations among the human as well as avian H5N1 lineages (Table 2). For example, analysis of the highly variant 140′s loop (HA1 136 to 143) at position 140 (aa 140) indicated the predominant presence of lysine (22.5%) and serine (28.5%) amino acids at this position in all H5N1 human isolates, with only a minor presence of threonine at position 140 (6%) (Table 2). Furthermore, aa 155 of the loop (HA1 155 to 164) at position 150 has only two variants (63.4% of human H5N1 isolates have a “Ser” amino acid, and the remaining 34.4% have an “Asn” amino acid at this position). Also, the amino acid at position 189 on influenza virus hemagglutinin, located in the receptor binding site, contains an arginine amino acid in 64.26% of all H5N1 human strains, while the remaining 34.65% have a lysine amino acid at this position (Table 2).
Selection of vaccine strains.
Based on an analysis of amino acid sequences within the neutralizing epitopes, a panel of H5N1 viruses (317 human strains and 2,028 avian strains) was analyzed for the frequency of amino acid variations within the major neutralizing epitopes of HA. We have selected three different H5N1 strains [A/Indonesia/CDC669/2006 (H5N1) (clade 2.1), A/Vietnam/1203/2004 (H5N1) (clade 1.0), and A/Anhui/1/2005 (H5N1) (clade 2.3)] such that the combination of these strains would cover all major amino acid variations of neutralizing epitopes of H5 HA (Table 3). For example, the A/Vietnam/1203/2004 and A/Indonesia/CDC669/2006 strains contain Ser at position 155, while the A/Anhui/1/2005 strain has “Asn” at the same position. Furthermore, the A/Vietnam/1203/2004 and A/Anhui/1/2005 strains contain Lys at position 189, while the A/Indonesia/CDC669/2006 strain has an “Arg” amino acid at the same position (Table 3).
TABLE 3.
Immunogenic epitopes in the three strains
| Strain | Clade | Immunogenic epitope at position: |
Host | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 138 | 140 | 155 | 189 | 194 | 159 | 218 | |||
| A/Vietnam/1203/04 (H5N1) | 1 | Gln | Lys | Ser | Lys | Pro | Thr | Lys | Human |
| A/Indonesia/CDC669/06 (H5N1) | 2.1 | Leu | Ser | Ser | Arg | Pro | Thr | Lys | Human |
| A/Anhui/1/05 (H5N1) | 2.3 | Gln | Thr | Asn | Lys | Pro | Thr | Lys | Human |
Differential recognition of selected vaccine strains by n-MAbs.
The variations within the selected vaccine strains [A/Vietnam/1203/04 (clade 1.0), A/Indonesia/CDC669/06, and A/Anhui/1/05 (H5N1) (clade 2.3)] were confirmed based on the results of virus neutralization and HI with different n-MAbs. The exposure of vaccine strains to n-MAbs resulted in a differential reactivity pattern with n-MAbs. As shown in Tables 4 and 5, n-MAb 6B8 shows preferential binding to the A/Vietnam/1203/04 and A/Anhui/1/05 strains, while the neutralization of A/Indonesia/CDC669/06 by the same n-MAb is absent, as indicated by hemagglutination inhibition and microneutralization assays. However, n-MAb 4C2 neutralized only the A/Indonesia/CDC669/06 strain. A similar difference in the pattern of recognition was observed with other n-MAbs. These findings also indicated the existence of strong antigenic variance between the A/Indonesia/CDC669/06 and A/Vietnam/1203/04 strains (Tables 4 and 5). In addition, the inclusion of A/Anhui/1/05 (H5N1) in vaccine compositions covers the variation of the amino acid at position 155 (Asn) of vaccine strains, which comprises 34% of human H5N1 isolates.
TABLE 4.
Hemagglutination inhibition titers of n-MAbs against three H5N1 strains
| n-MAba | Titer |
||
|---|---|---|---|
| A/Vietnam/1203/04 | A/Indonesia/CDC669/06 | A/Anhui/1/05 | |
| 6B8 | 512 | <8 | 256 |
| 4C2 | <8 | 512 | <8 |
| 4F8 | 64 | 512 | 32 |
| 3H11 | <8 | 512 | <8 |
| 2D9 | 128 | 512 | 128 |
n-MAb concentration, 1 mg/ml.
TABLE 5.
Virus microneutralization titers of n-MAbs against three H5N1 strains
| n-MAba | Virus titer (concn of n-MAb to neutralize 100 TCID50 of virus [μg/ml]b) |
||
|---|---|---|---|
| A/Vietnam/1203/04 | A/Indonesia/CDC669/06 | A/Anhui/1/05 | |
| 6B8 | 320 (2.75) | <10 (175) | 160 (6.25) |
| 4C2 | <10 (200) | 320 (2.52) | <10 (235.5) |
| 4F8 | 40 (32.5) | 320 (3.25) | 20 (75.5) |
| 3H11 | <10 (250) | 320 (2.75) | <10 (305) |
| 2D9 | 80 (17.5) | 320 (3.25) | 160 (7.12) |
Concentration of each n-MAb at 1 mg/ml.
One hundred TCID50 of each virus strain used for microneutralization assays.
Serum HI assay.
Hemagglutination inhibition titers measure the efficacy of the antibody response to inhibit HA function. The HI titer results showed that mice immunized with tri-BacHA adjuvanted with the Montanide ISA vaccine significantly enhanced (P < 0.001) the serum HI titer compared to the unadjuvanted tri-BacHA on days 28 and 42 (Fig. 1 A). Moreover, the HI titer of mice vaccinated with adjuvanted tri-BacHA was comparable to the HI titer of mice either vaccinated with adjuvanted RG-H5N1 virus or adjuvanted with mono-BacHA (Fig. 1A). The serum HI titers against different clades (clades 2.2, 4.0, and 7.0) of H5N1 strains on day 42 also revealed that mice immunized with adjuvanted tri-BacHA significantly enhanced (P < 0.001 and P < 0.02) serum HI titers against clade 2.2, clade 4.0, and clade 7.0 compared to mice either vaccinated with adjuvanted RG-H5N1 virus or adjuvanted with mono-BacHA (Fig. 1B).
FIG. 1.
Serum hemagglutination inhibition assay. Groups of mice were subcutaneously immunized two times on days 0 and 28 with tri-BacHA, mono-BacHA (BacHA-A/Vietnam/1203/2004), or inactivated whole viral vaccine (RG-A/Vietnam/1203/2004). (A) The serum HI titer against clade 1.0 homologous H5N1 (RG-A/Vietnam/1203/2004) virus was measured on days 14, 28, and 42. The results are expressed as log2 HI titers, and each point represents the arithmetic mean value (n = 10) ± SD. (***, P < 0.001 for point a compared with an unadjuvanted counterpart). (B) The serum HI titer on day 42 against heterologous H5N1 strains from clade 2.2 (A/Nigeria/6e/07(H5N1), clade 4.0 (A/goose/Guiyang/337/06), and clade 7.0 (A/chicken/Shanxi/2/06) was measured. Sera (1:10 dilutions) from vaccinated mice were further serially diluted (2-fold), and HI assays were performed. Each point represents the arithmetic mean value (n = 10) ± SE. (*, P < 0.05; **, P < 0.02; ***, P < 0.001).
Serum cross-clade neutralizing antibody titers.
The serum neutralizing antibody titer against 100 TCID50 of different clades of H5N1 strains on day 42 showed that vaccination with adjuvanted tri-BacHA significantly neutralized viruses from different clades (clade 1.0, clade 2.1, clade 2.2, clade 4.0, clade 7.0, and clade 8.0) compared with mice vaccinated with unadjuvanted tri-BacHA (Fig. 2).
FIG. 2.
Cross-clade serum microneutralization in mice. Groups of mice were subcutaneously immunized two times on days 0 and 28 with tri-BacHA, mono-BacHA (BacHA-A/Vietnam/1203/2004), or inactivated whole H5N1 viral vaccine (RG-A/Vietnam/1203/2004). (A) Microneutralization titers of vaccinated mouse sera against H5N1 viruses from clade 1.0 (A/Vietnam/1203/2004), clade 2.1 (A/Indonesia/CDC1031/2007), and clade 2.2 [A/turkey/Turkey1/05 (H5N1)]. (B) Microneutralization titers of vaccinated mouse sera against H5N1 viruses from clade 4.0 (A/goose/Guiyang/337/06), clade 7.0 (A/chicken/Shanxi/2/06), and clade 8.0 (A/chicken/Henan/12/2004) were used for the assay. The sera from day 42 (14 days after the final immunization) were used for the assay. Each point represents the arithmetic mean value (n = 10) ± SE. (*, P < 0.05; **, P < 0.02; ***, P < 0.001).
Moreover, mice immunized with adjuvanted tri-BacHA significantly (P < 0.02) enhanced the neutralization titer against clade 2.2, clade 4, and clade 7 viruses compared to mice vaccinated with adjuvanted RG-H5N1 vaccine. Also, adjuvanted tri-BacHA significantly (P < 0.05) enhanced the neutralization titer against clade 2.2 and clade 7 viruses compared to mice vaccinated with adjuvanted mono-BacHA vaccine (Fig. 2A and B). Furthermore, a vaccine composition containing mono-BacHA or RG-H5N1, both adjuvanted with Montanide ISA563, was able to neutralize clade 1 (homologous), clade 2.1, and clade 8.0 viruses but did not result in the efficient neutralization of viruses of other clades (clade 2.2, clade 4.0, and clade 7.0 H5N1 strains).
Challenge studies after vaccination.
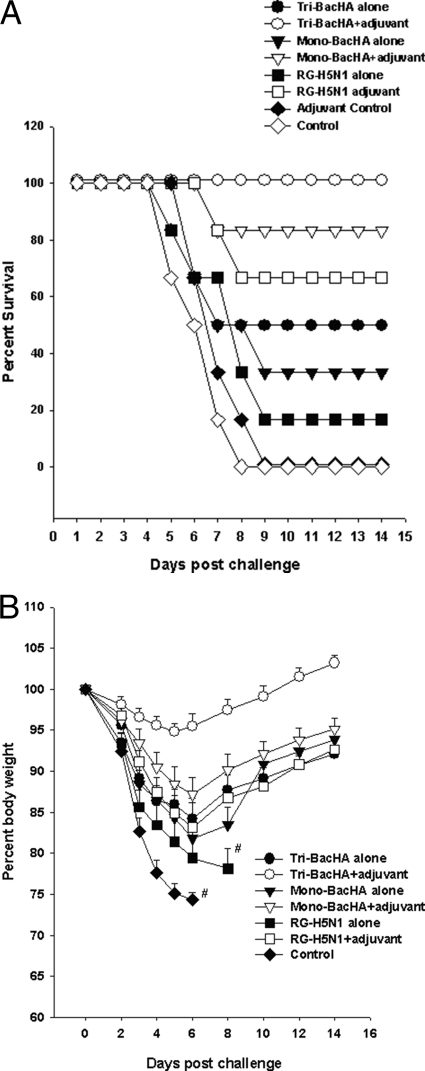
Three weeks after the final immunization, all groups of mice were challenged intranasally with 5 MLD50 of HPAI H5N1 virus strains from clade 1.0, clade 2.1, or clade 7.0. Groups of mice immunized with adjuvanted tri-BacHA or mono-BacHA or RG-H5N1 obtained 100% protection against clade 1.0 and clade 2.1 viruses (Fig. 3). Moreover, adjuvanted tri-BacHA provided 100% protection against clade 7.0 H5N1 infection. However, mice immunized with adjuvanted RG-H5N1 vaccine and mono-BacHA vaccine provided only 66.6% and 83.3% protections against clade 7.0 H5N1 infection, respectively (Fig. 4 A).
FIG. 3.
Protection of mice against lethal challenge with clade 1.0 or clade 2.1 H5N1 virus. Groups of mice were subcutaneously immunized two times on days 0 and 28 with tri-BacHA, mono-BacHA (BacHA-A/Vietnam/1203/2004), or inactivated whole H5N1 viral vaccine (RG-A/Vietnam/1203/2004). Three weeks after the final vaccination, mice were intranasally infected with 5 MLD50 of clade 1.0 (A/Vietnam/1203/2004) (A) and clade 2.1 (A/Indonesia/TLL013/06) (B) HPAI H5N1 virus strains. Mice were monitored for survival throughout a 14-day observation period. The results are expressed in terms of percent survival.
FIG. 4.
Protection of mice against lethal challenge with clade 7.0 H5N1 virus. Groups of mice were subcutaneously immunized two times on days 0 and 28 with tri-BacHA, mono-BacHA (BacHA-A/Vietnam/1203/2004), or inactivated whole H5N1 viral vaccine (RG-A/Vietnam/1203/2004). Three weeks after the final vaccination, mice were intranasally infected with 5 MLD50 of a clade 7.0 (A/chicken/Shanxi/2/06) HPAI H5N1 virus strain. (A) Mice were monitored for survival against clade 7.0 H5N1 viral challenge throughout a 14-day observation period. The results are expressed in terms of percent survival. (B) Mice were monitored for weight loss after clade 7.0 H5N1 viral challenge throughout a 14-day observation period. The results are expressed in terms of percent body weight (at the beginning of the trial).
The progression of infection was indicated by various trends of a decrease in body weight for the different groups. In groups of mice challenged with an H5N1 clade 7.0 (A/chicken/Shanxi/2/06) strain, no significant decreases in body weight were observed for mice vaccinated with adjuvanted tri-BacHA after the challenge (Fig. 4B). However, mice vaccinated with mono-BacHA with adjuvant showed up to a 12% loss of body weight, although the body weight was gradually regained after 6 days postchallenge. The groups that were vaccinated with adjuvanted RG-H5N1 vaccine showed a higher loss of body weight, up to 17% on day 6, but they then slowly regained their body weights by day 14 after challenge (Fig. 4B). Histopathology studies were performed for the mice vaccinated and challenged with clade 1.0 (homologous) and clade 7.0 (heterologous) H5N1 viruses. On day 6 postinfection, lungs of unvaccinated mice had pulmonary lesions consisting of moderate to severe necrotizing bronchitis and moderate to severe histiocytic alveolitis with associated pulmonary edema (Fig. 5 I and J). The uninfected mice lacked lesions in the lungs (Fig. 5A and B). Mice vaccinated with tri-BacHA had only minimal bronchitis against clade 1 and clade 7 H5N1 viral challenges (Fig. 5C and D). Moreover, mice vaccinated with mono-BacHA or inactivated whole H5N1 had minimal bronchitis against clade 1 (homologous) virus challenge (Fig. 5E and G), however, which showed moderate to severe bronchitis against clade 7.0 virus challenge (Fig. 5F and H).
FIG. 5.
Histopathology of lung tissue in vaccinated mice. Shown are photomicrographs of hematoxylin- and eosin-stained lung sections of mice 6 days after challenge with clade 1.0 and 7.0 H5N1 strains. (A and B) Normal morphology seen in uninfected mice. (C and D) Mice vaccinated with tri-BacHA and challenged with clade 1 (C) and clade 7 (D) H5N1 viruses. (E and F) Mice vaccinated with mono-BacHA and challenged with clade 1.0 (E) and clade 7.0 (F) H5N1 strains. (G and H) Mice vaccinated with inactivated whole H5N1 vaccine and challenged with clade 1 (G) and clade 7 (H) H5N1 viruses. (I and J) Unvaccinated mice challenged with clade 1.0 (I) and 7.0 (J) H5N1 strains.
DISCUSSION
Influenza virus evades the immune system by randomly changing antigenic determinants (25), such as the major neutralizing epitopes, present in the globular head region of hemagglutinin. Upon infection, the host response is characterized mainly by the induction of antibodies against such neutralizing epitopes, which blocks the attachment of viral hemagglutinin to the target cell receptor (16, 29). However, considerable amino acid variations within these neutralizing epitopes of HA lead to the emergence of antigenically distinct influenza H5N1 viruses. In fact, it was reported previously that seasonal influenza viruses were able to efficiently escape from vaccine-induced immunity in the human population through antigenic drift (19). Moreover, the mutation of a few key amino acids in HA1 variable regions is sufficient to allow viral escape from vaccine-induced antibody responses (34). Although current H5N1 vaccine candidates continue to provide good antigenic coverage of most isolates within corresponding clades, it was recently recognized that some viruses within clades 1, 2.2, and 2.3 themselves show evidence of antigenic heterogeneity (8). Since H5N1 viruses have already split into numerous sublineages or clades, it becomes imperative to select vaccine strains that represent the variations among H5 subtypes, particularly in the region of neutralizing epitopes. Such a vaccine strain selection will lead to a broad range of protection against most H5N1 lineages.
Our universal vaccine development strategy involved three steps: the mapping of neutralizing epitopes of H5N1 virus hemagglutinin by using n-MAbs, an analysis of the distribution of neutralizing epitopes among all H5N1 lineages, and the selection of ideal vaccine strains to cover the variations within the neutralizing epitopes of entire H5N1 viruses. In this present study, a panel of five n-MAbs (6B8, 4C2, 2D9, 4F8, and 3H11) was used to map the neutralizing epitopes of H5N1 virus. The mapping of neutralizing epitopes with n-MAbs revealed that amino acids at position 138, 155, 189, or 223 were involved in the formation of major neutralizing epitopes of H5N1 virus hemagglutinin. These amino acids are located in the receptor binding site. Position 155 corresponds to an amino acid in antigenic site B of H3 (9). The amino acid at position 189 is within an α-helix, and that at position 223 is in the loop structure (30). n-MAb 3H11 carried a single point mutation at amino acid position 139 (Gly), and any independent mutations in position 138 (Leu) or 140 (Ser) of the parent virus cause the neutralization evasion from 3H11. These findings indicate that 3H11 targets an epitope that includes the sequence “LeuGlySer” (positions 138 to 140), which is located in antigenic site A of H5 (9). In addition, other amino acids at positions 159, identified previously by Kaverin et al. (10), and at positions 194 and 218, identified previously by Stevens et al. (30), were also taken into consideration for subsequent analyses. A comparison of major neutralizing epitope sequences of H5N1 viruses with an influenza virus research database revealed the variations within the epitope region of all human and avian H5N1 virus hemagglutinins (Table 2).
Based on our epitope distribution analysis, we have selected three different strains, A/Indonesia/CDC669/2006 (H5N1) (clade 2.1), A/Vietnam/1203/2004 (H5N1) (clade 1.0), and A/Anhui/1/2005 (H5N1) (clade 2.3), to collectively represent the variations among all H5N1 subtypes. The selected vaccine strains were confirmed for the reactivity pattern with different neutralizing MAbs by assays of virus neutralization and HI titers. As shown in Tables 4 and 5, n-MAbs 4C2 and 4F8 recognize only A/Indonesia/CDC669/06 and did not react with the A/Vietnam/1203/04 and A/Anhui/1/05 strains. This pattern of reactivity could be due possibly to the change in the amino acid at position 189, as the A/Indonesia/CDC669/06 strain has an Arg at position 189, while Lys is present in the A/Vietnam/1203/04 and A/Anhui/1/05 strains at the same position. On the other hand, n-MAb 6B8 reacts with both the A/Vietnam/1203/04 and A/Anhui/1/05 strains, possibly due to the presence of the common residue Lys at position 189. Similarly, the amino acid at position 155 has also been found to have a significant impact on the antibody recognition of H5N1 strains. Moreover, as shown in Table 3, the loop at position 150 has two variants, with 63.4% of human H5N1 isolates having a Ser amino acid and the remaining 34.4% having an Asn amino acid at this position. Also, the amino acid at position 189, located in the receptor binding site of HA, is an arginine amino acid in 64.26% of all H5N1 human strains, while the remaining 34.65% have a lysine amino acid at this position. Hence, it is reasonable to speculate that the selected vaccine strains should represent the variations within the major antigenic epitopes of almost 99% of all H5N1 lineages, including both human and avian viruses.
HA proteins of selected strains were individually expressed on the baculovirus surface, and the vaccine formulation was evaluated in a mouse model. A recombinant baculovirus with the immediate-early 1 (ie1) promoter of WSSV was constructed to facilitate the high-level expression of influenza virus H5 hemagglutinin in both insect and mammalian cells. The nature of ie1 as an immediate-early promoter supports protein expression at an early phase of the baculoviral life cycle, resulting in an enhanced display of functional hemagglutinin on the baculovirus envelope (24). As oligomerization is required for the efficient transport of the HA proteins to the host cell membrane (4), a prerequisite for the baculovirus to acquire the protein, it is presumed that HAs displayed on the baculovirus surface should have been presented in their oligomeric forms. Hence, this model will help mimic the native structure of the protein, thus imitating the wild-type influenza virus. HA displayed on the baculovirus surface has retained its native structure, as evidenced by the hemagglutination activity and authentic cleavage of HA0 into HA1 and HA2 (data not shown). Although baculovirus-expressed influenza virus HAs are generally not cleaved in insect cells, HAs of highly pathogenic avian influenza viruses (such as H5 and H7) with multiple basic amino acids at the cleavage site have been shown to be cleaved into HA1 and HA2 subunits in the absence of trypsin or trypsin-like proteases (12). The partial cleavage of HA0 in the current study may be due to the presence of subtilisin-like proprotein convertases (PCs) in insect cells (3), whose substrate specificities and inhibitor profiles are identical to those of mammalian PCs.
The subcutaneous immunization of adjuvanted tri-BacHA significantly enhanced the serum HI titer compared to that of its unadjuvanted counterpart. Moreover, the HI titer (against A/Vietnam/1203/04) of mice vaccinated with adjuvanted tri-BacHA was comparable to those of mice vaccinated with either adjuvanted whole RG-H5N1 virus or adjuvanted mono-BacHA. In addition, adjuvanted tri-BacHA induced higher neutralization antibody titers, which efficiently neutralized 100 TCID50 of heterologous H5N1 strains from various clades (clade 1.0, clade 2.1, clade 2.2, clade 4.0, clade 7.0, and clade 8.0) compared to unadjuvanted tri-BacHA. A vaccine formulation containing only adjuvanted mono-BacHA or inactivated RG-H5N1 vaccine was able to neutralize clade 1 (homologous), clade 2.1, and clade 8.0 viruses but did not efficiently neutralize H5N1 viruses from other clades (clade 2.2, clade 4.0, and clade 7.0). The strong cross-clade immunity of tri-BacHA adjuvanted with the Montanide ISA563 vaccine formulation could be due to the coverage of variations within the neutralizing epitopes of H5N1 lineages. The protective efficacy of the vaccine was evaluated by challenging the vaccinated mice with H5N1 strains from clade 1, clade 2.1, and clade 7. A 100% survival rate was obtained with the group vaccinated with adjuvanted tri-BacHA or mono-BacHA or inactivated whole viral vaccine against clade 1.0 and clade 2.1 viruses. In addition, adjuvanted tri-BacHA provided 100% protection against clade 7.0 H5N1 virus without any infection symptoms. However, adjuvanted inactivated whole viral vaccine and mono-BacHA provided only 66.6% and 83.3% protection against clade 7.0 H5N1 infection, respectively. Also, the progression of infection was indicated by various trends of a decrease in body weight for the different groups. Mice vaccinated with adjuvanted mono-BacHA or adjuvanted whole viral vaccine showed a higher loss of body weight of up to 17% on day 6 against clade 7.0 viruses. This indicates the inability of monovalent vaccines to confer protection against diverse H5N1 subtypes, which might be due to the variation within the antigenic determinants (such as neutralizing epitopes) of different virus subtypes.
In summary, the subcutaneous immunization of mice with baculovirus displaying hemagglutinin from three selected vaccine strains induced systemic immune responses and exhibited cross-protection against H5N1 viral infection without any clinical symptoms. Also, the present findings revealed that the selection of vaccine strains based on the variations within the neutralizing epitopes among the subtypes will help prevent infection mediated by newly emerged H5N1 mutants. The vaccine formulation used in this study was produced rapidly without any biosafety concerns. Baculoviruses displaying HA will serve as an ideal choice for a vaccine in pandemic and prepandemic situations and expedite the vaccine technology without the requirement for high-level biocontainment facilities or tedious protein purification processes.
Acknowledgments
We are grateful for the financial support received from Temasek Life Science Laboratory, Singapore.
We thank the Ministry of Health (MOH), Indonesia, for technical support and collaboration. We thank Ruben Donis, Influenza Division, Centers for Disease Control and Prevention, Atlanta, GA, for providing the plasmids for reverse genetics. We also thank Nayana Prabhu and Hui Ting Ho for the generation of RG-H5N1 viruses.
We declare that no competing interests exist.
Footnotes
Published ahead of print on 15 September 2010.
REFERENCES
- 1.Ayora-Talavera, G., H. Shelton, M. A. Scull, J. Ren, I. M. Jones, R. J. Pickles, and W. S. Barclay. 2009. Mutations in H5N1 influenza virus hemagglutinin that confer binding to human tracheal airway epithelium. PLoS One 4:e7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, G. L., and K. Subbarao. 2009. Attacking the flu: neutralizing antibodies may lead to universal vaccine. Nat. Med. 15:1251-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cieplik, M., H. D. Klenk, and W. Garten. 1998. Identification and characterization of Spodoptera frugiperda furin: a thermostable subtilisin-like endopeptidase. Biol. Chem. 379:1433-1440. [DOI] [PubMed] [Google Scholar]
- 4.Copeland, C. S., R. W. Doms, E. M. Bolzau, R. G. Webster, and A. Helenius. 1986. Assembly of influenza hemagglutinin trimers and its role in intracellular transport. J. Cell Biol. 103:1179-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Filette, M., W. M. Jou, A. Birkett, K. Lyons, B. Schultz, A. Tonkyro, S. Resch, and W. Fiers. 2005. Universal influenza A vaccine: optimization of M2-based constructs. Virology 337:149-161. [DOI] [PubMed] [Google Scholar]
- 6.Fan, J., X. Liang, M. S. Horton, H. S. Perry, M. P. Citron, G. J. Heidecker, T. M. Fu, J. Joyce, C. T. Przysiecki, P. M. Keller, V. M. Garsky, R. Ionescu, Y. Rippeon, L. Shi, M. A. Chasain, J. H. Condra, M. E. Davies, J. Liao, E. A. Emini, and J. W. Shiver. 2004. Preclinical study of influenza virus A M2 peptide conjugate vaccines in mice, ferrets, and rhesus monkeys. Vaccine 22:2993-3003. [DOI] [PubMed] [Google Scholar]
- 7.Frace, A. M., A. I. Klimov, T. Rowe, R. A. Black, and J. M. Katz. 1999. Modified M2 proteins produce heterotypic immunity against influenza A virus. Vaccine 17:2237-2244. [DOI] [PubMed] [Google Scholar]
- 8.Govorkova, E. A., R. J. Webby, J. Humberd, J. P. Seiler, and R. G. Webster. 2006. Immunization with reverse-genetics-produced H5N1 influenza vaccine protects ferrets against homologous and heterologous challenge. J. Infect. Dis. 194:159-167. [DOI] [PubMed] [Google Scholar]
- 9.Kaverin, N. V., I. A. Rudneva, N. A. Ilyushina, N. L. Varich, A. S. Lipatov, Y. A. Smirnov, E. A. Govorkova, A. K. Gitelman, D. K. Lvov, and R. G. Webster. 2002. Structure of antigenic sites on the hemagglutinin molecule of H5 influenza virus and phenotypic variation of escape mutants. J. Gen. Virol. 83:2497-2505. [DOI] [PubMed] [Google Scholar]
- 10.Kaverin, N. V., I. A. Rudneva, E. A. Govorkova, T. A. Timofeeva, A. A. Shilov, K. S. Kochergin-Nikitsky, P. S. Krylov, and R. G. Webster. 2007. Epitope mapping of the hemagglutinin molecule of a highly pathogenic H5N1 influenza virus by using monoclonal antibodies. J. Virol. 81:12911-12917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King, D. J. 1991. Evaluation of different methods of inactivation of Newcastle disease virus and avian influenza virus in egg fluids and serum. Avian Dis. 35:505-514. [PubMed] [Google Scholar]
- 12.Kuroda, K., C. Hauser, R. Rott, H. D. Klenk, and W. Doerfler. 1986. Expression of the influenza virus haemagglutinin in insect cells by a baculovirus vector. EMBO J. 5:1359-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lam, T. T., C. C. Hon, O. G. Pybus, S. L. K. Pond, R. T. Wong, C. W. Yip, F. Zeng, and F. C. Leung. 2008. Evolutionary and transmission dynamics of reassortant H5N1 influenza virus in Indonesia. PLoS Pathog. 4:e1000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, W., H. Li, and Y-H. Chen. 2003. N-terminus of M2 protein could induce antibodies with inhibitory activity against influenza virus replication. FEMS Immunol. Med. Microbiol. 35:141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manzoli, L., F. Schioppa, A. Boccia, and P. Villari. 2007. The efficacy of influenza vaccine for healthy children: a meta-analysis evaluating potential sources of variation in efficacy estimates including study quality. Pediatr. Infect. Dis. J. 26:97-106. [DOI] [PubMed] [Google Scholar]
- 16.Marasco, W. A., and J. Sui. 2007. The growth and potential of human antiviral monoclonal antibody therapeutics. Nat. Biotechnol. 25:1421-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Institutes of Health and Centers for Disease Control and Prevention. 1999. Biosafety in microbiological and biomedical laboratories, 4th ed. U.S. Department of Health and Human Services, Washington, DC.
- 18.O'Reilly, D. R., L. K. Miller, and V. A. Luckow. 1992. Baculovirus expression vectors: a laboratory manual. W. H. Freeman and Company, New York, NY.
- 19.Park, A. W., J. M. Daly, N. S. Lewis, D. J. Smith, J. L. Wood, and B. T. Grenfell. 2009. Quantifying the impact of immune escape on transmission dynamics of influenza. Science 326:726-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plotkin, J. B., J. Dushoff, and S. A. Levin. 2002. Hemagglutinin sequence clusters and the antigenic evolution of influenza A virus. Proc. Natl. Acad. Sci. U. S. A. 99:6263-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prabakaran, M., S. Velumani, F. He, A. K. Karuppannan, G. Y. Geng, L. K. Yin, and J. Kwang. 2008. Protective immunity against influenza H5N1 virus challenge in mice by intranasal co-administration of baculovirus surface-displayed HA and recombinant CTB as an adjuvant. Virology 380:412-420. [DOI] [PubMed] [Google Scholar]
- 22.Prabakaran, M., H. T. Ho, N. Prabhu, S. Velumani, M. Szyporta, F. He, K. P. Chan, L. M. Chen, Y. Matsuoka, R. O. Donis, and J. Kwang. 2009. Development of epitope-blocking ELISA for universal detection of antibodies to human H5N1 influenza viruses. PLoS One 4:e4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prabakaran, M., N. Prabhu, F. He, Q. Hongliang, H. T. Ho, J. Qiang, T. Meng, M. Goutama, and J. Kwang. 2009. Combination therapy using chimeric monoclonal antibodies protects mice from lethal H5N1 infection and prevents formation of escape mutants. PLoS One 4:e5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prabakaran, M., S. Madhan, N. Prabhu, J. Qiang, and J. Kwang. 2010. Gastrointestinal delivery of baculovirus displaying influenza virus hemagglutinin protects mice against heterologous H5N1 infection. J. Virol. 84:3201-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Recker, M., O. G. Pybus, S. Nee, and S. Gupta. 2007. The generation of influenza outbreaks by a network of host immune responses against a limited set of antigenic types. Proc. Natl. Acad. Sci. U. S. A. 104:7711-7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rowe, T., R. A. Abernathy, J. Hu-Primmer, W. W. Thompson, X. Lu, W. Lim, K. Fukuda, N. J. Cox, and J. M. Katz. 1999. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J. Clin. Microbiol. 37:937-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rueda, P., J. Fominaya, J. P. M. Langeveld, C. Bruschke, C. Vela, and J. I. Casal. 2000. Effect of different baculovirus inactivation procedures on the integrity and immunogenicity of porcine parvovirus-like particles. Vaccine 19:726-734. [DOI] [PubMed] [Google Scholar]
- 28.Russell, C. A., T. C. Jones, I. G. Barr, N. J. Cox, R. J. Garten, V. Gregory, I. D. Gust, A. W. Hampson, A. J. Hay, A. C. Hurt, J. C. de Jong, A. Kelso, A. I. Klimov, T. Kageyama, N. Komadina, A. S. Lapedes, Y. P. Lin, A. Mosterin, M. Obuchi, T. Odagiri, A. D. Osterhaus, G. F. Rimmelzwaan, M. W. Shaw, E. Skepner, K. Stohr, M. Tashiro, R. A. Fouchier, and D. J. Smith. 2008. Influenza vaccine strain selection and recent studies on the global migration of seasonal influenza viruses. Vaccine 12(Suppl. 4):D31-D34. [DOI] [PubMed] [Google Scholar]
- 29.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 30.Stevens, J., O. Blixt, T. M. Tumpey, J. K. Taubenberger, J. C. Paulson, and I. A. Wilson. 2006. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312:404-410. [DOI] [PubMed] [Google Scholar]
- 31.Wang, T. T., and P. Palese. 2009. Universal epitopes of influenza virus hemagglutinins? Nat. Struct. Mol. Biol. 16:233-234. [DOI] [PubMed] [Google Scholar]
- 32.Webster, R. G., Y. Kawaoka, J. Taylor, R. Weinberg, and E. Paoletti. 1991. Efficacy of nucleoprotein and hemagglutinin antigens expressed in fowlpox virus as vaccine for influenza in chickens. Vaccine 9:303-308. [DOI] [PubMed] [Google Scholar]
- 33.WHO. 2005. Evolution of H5N1 avian influenza viruses in Asia. Emerg. Infect. Dis. 11:1515-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson, I. A., and N. J. Cox. 1990. Structural basis of immune recognition of influenza virus hemagglutinin. Annu. Rev. Immunol. 8:737-771. [DOI] [PubMed] [Google Scholar]
- 35.Wu, F., J. H. Huang, X. Y. Yuan, W. S. Huang, and Y. H. Chen. 2007. Characterization of immunity induced by M2e of influenza virus. Vaccine 25:8868-8873. [DOI] [PubMed] [Google Scholar]
- 36.Yokoyama, W. M. 2001. Production of monoclonal antibody, p. 2.5.1-2.5.17. In J. E. Coligan, A. M. Kruisbeek, D. H. Margulies, E. M. Shevach, and W. Strober (ed.), Current protocols in immunology. John Wiley & Sons, Inc., Newcastle, United Kingdom.