Abstract
To initiate membrane fusion and virus entry, herpes simplex virus (HSV) gD binds to a cellular receptor such as herpesvirus entry mediator (HVEM). HVEM is a tumor necrosis factor (TNF) receptor family member with four natural ligands that either stimulate (LIGHT and LTα) or inhibit (BTLA and CD160) T cell function. We hypothesized that the interaction of gD with HVEM affects the binding of natural ligands, thereby modulating the immune response during infection. Here, we investigated the effect that gD has on the interaction of HVEM with its natural ligands. First, HSV gD on virions or cells downregulates HVEM from the cell surface. Similarly, trans-interaction with BTLA or LIGHT also downregulates HVEM from the cell surface, suggesting that HSV may subvert a natural mechanism for regulating HVEM activity. Second, we showed that wild-type gD had the lowest affinity for HVEM compared with the four natural ligands. Moreover, gD directly competed for binding to HVEM with BTLA but not LTα or LIGHT, indicating the possibility that gD selectively controls HVEM signals. On the other hand, natural ligands influence the use of HVEM by HSV. For instance, soluble BTLA, LTα, and LIGHT inhibited the binding of wild-type gD to HVEM, and soluble BTLA and LTα blocked HSV infection of HVEM-expressing cells. Thus, gD is at the center of the interplay between HVEM and its ligands. It can interfere with HVEM function in two ways, by competing with the natural ligands and by downregulating HVEM from the cell surface.
Herpes simplex virus (HSV) causes a primary infection in epithelial cells before establishing lifelong latency in sensory neurons. Four glycoproteins, gB, gD, and gH/gL, are essential for HSV entry and membrane fusion, and a fifth, gC, enhances the efficiency of entry by binding to heparan sulfate proteoglycans. When gD binds to a cell surface receptor, the fusion machinery of gB and gH/gL is activated, ultimately leading to virus entry (27).
HSV type 1 (HSV-1) can engage three unrelated receptors: herpesvirus entry mediator (HVEM), nectin-1, and 3-OS-modified heparan sulfate (20, 42, 62). HVEM is a member of the tumor necrosis factor receptor (TNFR) family. It is expressed mainly on lymphoid cells such as primary T and B cells and monocytes, but it is also expressed to a lesser extent on fibroblasts and endothelial cells (29, 32, 34, 71, 72). HVEM is the principal receptor for HSV entry into activated T cells (42). All clinical isolates of HSV-1 and HSV-2 tested have the ability to use HVEM as a receptor despite the fact that it is not expressed on the main targets of HSV infection in vivo (32). Thus, the interaction of HSV gD with HVEM may be more important for modulating the immune response to virus infection than for playing a direct role in HSV spread into its human host.
During HSV entry, binding of gD to its receptor induces conformational changes that activate gB and gH/gL to fuse the viral envelope with a cellular membrane. In addition, binding of gD to nectin-1 induces receptor downregulation and virion endocytosis (66). Here we show that interaction of gD with HVEM also causes receptor downregulation and virus endocytosis.
The TNFR family regulates the adaptive immune response by directing cell survival, proliferation, and differentiation of lymphocytes (36). Members of this family, including HVEM, have a common structure that is comprised of an ectodomain with four cysteine-rich domains (CRD), a transmembrane region (TMR), and a cytoplasmic tail (CT). Unlike many members of the TNFR family, HVEM does not have a death domain in its cytoplasmic tail, but it does have binding sites for TNFR-associated factor (TRAF) adaptor proteins (28, 37, 68). HVEM binds to four ligands in addition to HSV gD: “lymphotoxin-like, exhibits inducible expression, and competes with HSV gD for HVEM, a receptor expressed by T lymphocytes” (LIGHT), lymphotoxin alpha (LTα), B and T lymphocyte attenuator (BTLA), and CD160 (4, 22, 38, 58). LIGHT and LTα are members of the TNF family. LIGHT is expressed on the surfaces of activated T cells, natural killer cells, and immature dendritic cells, while LTα lacks a transmembrane region and is secreted (23, 25, 38). LIGHT and LTα each form homotrimers which bind to CRD2/3 of HVEM and are postulated to cause trimerization of HVEM on the cell surface (3). Binding of LIGHT to HVEM initiates signals that activate the transcription factors NF-κB and AP-1 (37, 67). This provides costimulation to activated T cells and leads to their proliferation and production of cytokines (67). Binding of LTα is also thought to provide T cell stimulation; however, studies looking directly at the interaction of LTα with HVEM have not been reported (56, 64).
Unlike LIGHT and LTα, BTLA and CD160 are members of the immunoglobulin (Ig) superfamily. BTLA is expressed on mature lymphocytes, macrophages, and dendritic cells (75) and is thought to be a dimer on the cell surface (10). CD160 forms multimers on the cell surface and is expressed on peripheral blood mononuclear cells (PBMCs), natural killer cells, and T cells (1, 21). Altogether, the functional data collected on HVEM and its ligands indicate that a complex combinatorial expression of these ligands by immune cells provides a highly regulated bidirectional mechanism to modulate cell survival, activation, or attenuation of the immune response (for complete reviews, see references 18, 44, 45, and 74). At the center of this network of interactions, HVEM function is likely to be affected in the presence of HSV gD, thereby altering the immune response during HSV infection.
Like BTLA and CD160, HSV gD also contains an Ig domain (7). The crystal structures of both gD-HVEM and BTLA-HVEM show that BTLA binds to HVEM directly through its Ig domain, while gD binds to HVEM through an N-terminal loop that extends from the Ig core (7, 12). The binding sites for gD and BTLA overlap extensively; both bind primarily to CRD1 of HVEM and have 20 contact residues in common (7, 12). The tyrosine residue at position 23 (Y23) plays a key role in HVEM binding to both gD and BTLA (12, 15). In fact, a form of gD with high affinity for HVEM, gD(Δ290-299), competitively inhibits binding of BTLA to HVEM (22, 48). HVEM CRD1 is also important for binding of CD160, and BTLA blocks the binding of CD160 to HVEM (4).
Competition studies between gD, LIGHT, and BTLA have yielded conflicting results. Using all soluble components, Sarrias et al. showed that gD(Δ290-299) does not inhibit binding of LIGHT to HVEM (55). However, in cell-based studies, Cheung et al. and Mauri et al. both demonstrated that gD inhibits the binding of HVEM to cells expressing LIGHT (9, 38). Cheung et al. found that BTLA could also block HVEM binding to LIGHT-expressing cells (9). However, Gonzalez et al. found that neither LIGHT nor LTα could block HVEM binding to BTLA, and in fact, BTLA and LIGHT were able to simultaneously bind HVEM to form a ternary complex (22). Thus, although HVEM and its ligands have been extensively studied, a number of issues remain unresolved.
Our goal here was to characterize and compare the interactions of gD relative to LTα, LIGHT, BTLA, and CD160 under similar conditions using soluble proteins. To determine whether the affinity of gD for HVEM affected its ability to interfere with the binding of natural ligands, we used both wild-type (wt) gD [gD(306t)], similar to the form of gD found on virions, and a mutant gD that has a higher affinity for the receptor [gD(285t)]. gD(285t) is similar to the gD(Δ290-299) used in previous studies (22, 48, 55). Both of these mutants have a higher affinity for HVEM because the C terminus of the ectodomain, which normally occludes the receptor binding site on gD, is removed [e.g., in gD(285t)] or destabilized [e.g., in gD(Δ290-299)], allowing a faster association and yielding a higher affinity (33). Thus, the data presented here represent the first instance where the affinities and competition of each of the HVEM ligands have been determined under similar conditions. In addition, we found that BTLA and LTα can each block HSV infection that is mediated by HVEM. Furthermore, gD, BTLA, and LIGHT each downregulate HVEM from the cell surface. This suggests that HSV gD may tap into a regulatory mechanism used by the ligands of HVEM to control its expression. Thus, gD has the ability to interfere with HVEM function both by downregulating it from the cell surface and by directly blocking the binding of its natural ligands.
MATERIALS AND METHODS
Viruses.
HSV-1 KOS and KOStk12 were grown and titers were determined on Vero cells, and the viruses were purified as described previously (24, 42). The KOStk12 and gD-null KOSgDβ viruses were obtained from P. G. Spear (17). Noncomplemented KOSgDβ was produced in Vero cells, and complemented viruses were produced and titers were determined on VD60 cells (17, 35). Partial purification of KOSgDβ was done as described by Stiles et al. (66). Equivalent amounts of capsid were present in supernatant for the virus produced in VD60 cells and that for the virus produced in Vero cells as determined by Western blotting with antibody (Ab) NC-1.
Abs.
Anti-HVEM monoclonal Ab (MAb) CW10 and polyclonal Ab (PAb) R140 were described previously (69, 76). For fluorescence-activated cell sorting (FACS), CW10 was directly coupled with phycoerythrin (PE) at Molecular Probes/Invitrogen. Anti-BTLA MAb MIH26 was purchased from eBiosciences and anti-LIGHT MAb 115520 from R&D Systems. Anti-CD160 MAbs 5D.8E10 and 5D.10A11 were the generous gift of Gordon Freeman (Dana Farber Research Institute). Secondary anti-IgG antibodies coupled with PE or Alexa 488 were purchased from Molecular Probes/Invitrogen, and those coupled to horseradish peroxidase (HRP) were purchased from KPL.
Cell lines. (i) Culture conditions.
Murine melanoma B78H1 cells were grown in Dulbecco's modified Eagle's medium (DMEM) with 5% fetal calf serum (FCS) and penicillin-streptomycin (P-S). The previously described transfected B78H1 cell lines B78H1-control-16 (abbreviated B78 here), B78H1-HVEM, B78H1-HVEM-Y23A, B78H1-gD(wt), and B78H1-gD(W294A) were grown in the same medium supplemented with 500 μg/ml G418 (15, 40, 66). 293T cells were maintained in DMEM with 10% FCS and P-S. CHOK1 cells were maintained in Ham's F-12 medium with 10% FCS and P-S. CHO-nectin-1 (clone R3A) and CHO-HVEM (clone A12) were maintained in the same medium with 250 μg/ml G418 (16, 42). CHO-CD160 cells (a gift from G. Freeman, Dana Farber Research Institute) were maintained in the same medium with 300 μg/ml hygromycin B (1). Sf9 (Spodoptera frugiperda) cells, used for producing recombinant baculoviruses and recombinant glycoproteins, were propagated in Sf900II medium (GIBCO) (78). Human primary T cells (CD3+ CD14− CD11b− CD16−) were obtained from the Human Immunology Core of the University of Pennsylvania. Fresh resting cells were used immediately for FACS or cocultures without activation.
(ii) B78-gD(D30A) cell line.
B78H1 cells were transfected with plasmid pDL449 to express gD(D30A) (14). Selection of the cell line and determination of gD expression were done as described by Stiles et al. (66). The clonal cell line used in this study is B78H1-gD(D30A) no. 26.
(iii) Cell lines expressing BTLA and LIGHT.
The full-length open reading frame of human BTLA with flanking 5′ KpnI and 3′ XhoI sites for cloning were synthesized at Blue Heron Biotechnology. cDNA obtained from Blue Heron was digested with KpnI and XhoI and ligated into pcDNA3.1/hygro to make plasmid pKS846 for BTLA. The open reading frame of LIGHT was subcloned using BamHI and XhoI from plasmid pLIGHT (a gift from M. Lazaro and H. Ertl, Wistar Institute) into pcDNA3.1/hygro to generate plasmid pKS892. 293T cells were transfected with plasmids pKS846 (BTLA), pKS892 (LIGHT), and pcDNA3.1/hygro (for control) using Lipofectamine according to the manufacturer's instructions. Clonal cell lines were selected by limiting dilution in the presence of 250 μg/ml hygromycin B and further maintained in DMEM with 10% FCS, antibiotics, and 200 μg/ml hygromycin B. Clones were selected by FACS for the level of surface expression of BTLA using MAb MIH26 (eBiosciences) or of LIGHT using MAb α-LIGHT 115520 (R&D Systems). The clonal cell lines used in this study are 293T-BTLA no. 3, 293T-LIGHT no. 2, and 293T-control no. 1.
Baculovirus recombinants. (i) Construction of plasmids. (a) BTLA.
A 375-bp fragment of BTLA corresponding to amino acids 32 to 157 was PCR amplified from plasmid pKS846. Amino acid 32 is the first amino acid after the predicted signal peptide, and amino acid 157 is the last one before the predicted transmembrane region. Primers used were forward primer bac-hBTLA-F (5′-GCGGATCCAGAATCATGTGATGTACAGCTTTAT), which adds an upstream BamHI site, and reverse primer bac-hBTLA-R (5′-CGGCTGCAGTTAATGGTGATGATGGTGATGACTATACAGGAGCCAGGGTCT), which adds six histidine residues after amino acid 157 of BTLA, a stop codon, and a downstream PstI site. The PCR product was digested with BamHI and PstI and ligated into pVTBac (70) to create plasmid pKS779. This plasmid was recombined into baculovirus by cotransfecting with Baculogold DNA (BD Biosciences) (63). Baculovirus recombinants expressing BTLA were subjected to two rounds of plaque purification. The recombinant baculovirus was named bac-hBTLA(157t), and the recombinant protein was designated BTLA(157t) or BTLAt.
(b) LIGHT.
A 540-bp PCR fragment encoding amino acids 60 to 240 was PCR amplified from plasmid pLIGHT (a gift from M. Lazaro and H. Ertl, Wistar Institute) using forward primer bac-LIGHT-F (5′-GCGGATCCACATCACCATCACCATCACCAGCTGCACTGGCGTCTAGGAGAG), which adds a BamHI site and six histidine residues prior to amino acid 60 of LIGHT, and reverse primer bac-LIGHT-R (5′-GCGGAATTCTCACACCATGAAAGCCCCGAAGTAAGA), which adds a stop codon and a downstream EcoRI site. Since LIGHT is a type II membrane protein, amino acid 60 is the first amino acid after the transmembrane sequence in the ectodomain and amino acid 240 is the last amino acid of the protein. The PCR product was digested with BamHI and EcoRI and ligated into pVTBac to make plasmid pKS918. This plasmid was used to make baculovirus bac-LIGHT(t60). The recombinant protein was designated LIGHT(t60) or LIGHTt.
(c) CD160.
The sequence corresponding to amino acids 27 to 136 of CD160 with an additional six histidine residues after amino acid 136 was synthesized at Blue Heron Biotechnology. The sequence included an upstream BamHI site and a downstream EcoRI site for cloning. The DNA was digested with BamHI and EcoRI and ligated into pVTBac to generate plasmid pKS916. This plasmid was used to make baculovirus bac-CD160(136t). The recombinant protein was designated CD160(136t) or CD160t.
(d) LTα.
The sequence corresponding to amino acids 35 to 205 of lymphotoxin α with six histidine residues after amino acid 205 was synthesized at Blue Heron Biotechnology. Since LTα is normally a secreted protein, no truncation was necessary. Additionally, the sequence included an upstream BamHI site and a downstream EcoRI site. The DNA was digested with BamHI and EcoRI and ligated into pVTBac to create plasmid pKS917. This plasmid was used to make baculovirus bac-LTα. The recombinant protein was designated LTα(r).
(ii) Purification of recombinant proteins. (a) BTLAt and LTα(r).
Sf9 cells grown in a 1-liter suspension culture were infected with recombinant baculovirus (multiplicity of infection [MOI] = 4) for 48 h. The clarified supernatant was concentrated, exchanged into phosphate-buffered saline (PBS) by tangential flow filtration (10-kDa-molecular-mass cutoff membrane; Millipore), and incubated overnight with 2.5 ml of nickel-nitriloacetic acid resin (Qiagen) at 4°C. The resin was washed first with PBS and then with stepwise-increasing concentrations of imidazole (0.01 to 0.25 M) in 0.02 M phosphate buffer (pH 7.5) containing 0.5 M NaCl. The 0.25 M imidazole fraction, containing the purest recombinant protein, was dialyzed against PBS and concentrated (10-kDa-molecular-mass cutoff centrifugal membrane; Millipore). The yields of purified BTLAt and LTα(r) were 2 mg/liter of supernatant.
(b) LIGHTt and CD160t.
The purification of LIGHTt and CD160t was done using the protocol described above with the following modifications. These proteins were purified for 4 h at 4°C using only 0.5 ml of nickel-nitriloacetic acid resin in the presence of 5 mM imidazole. The 0.1 M and 0.25 M fractions, which contained most of the pure proteins, were combined before dialysis against PBS and concentrated (5-kDa-molecular-mass cutoff centrifugal membrane). These modifications improved the purities of LIGHTt and CD160t but did not improve their yields, which remained low. The yields of purified LIGHTt and CD160t were 2 and 20 μg/liter of supernatant, respectively.
(c) HVEMt-Y23A.
The expression plasmid for HVEM(200t)-Y23A was derived using QuikChange mutagenesis of plasmid pCW275 (77) with forward primer pmr-HveA-Y23A-F (5′-CAAGTGCAGTCCAGGTGCTCGTGTGAAAGGAGGC) and reverse primer pmr-HveA-Y23A-R (5′-GCCTCCTTCACACGAGCACCTGGACTGCACTTG). The resulting plasmid was named pKS919. Recombinant baculovirus bac-HVEM(200t)-Y23A was made using the methods described above for BTLA. The recombinant protein was designated HVEM(200t)-Y23A or HVEMt-Y23A and was purified using the protocol previously described for HVEM(200t) (76). The yield of purified HVEMt-Y23A was 1.5 mg/liter of supernatant.
Gel filtration analysis of molecular weight.
Dextran blue was used to determine the void volume of a Superdex 200 column (GE Healthcare Life Sciences), which was calibrated with high- and low-molecular-weight standards using PBS as the running buffer. Two hundred microliters of concentrated LIGHTt, LTα(r), BTLAt, and CD160t was loaded independently. The molecular weights of the eluted proteins were calculated from a standard plot according to the molecular weight standards.
FACS analysis of cell cocultures.
Target cells expressing HVEM were detached with trypsin and then counted and divided in aliquots for labeling with Qdots using the Qtracker 655 labeling kit (Quantum Dot Corp/Invitrogen, Hayward CA) for 1 h at 37°C in DMEM supplemented with 5% FCS according to the manufacturer's instructions. Qdot labeling was done to identify target cells during FACS analysis (66). Target cells were washed three times with culture medium and mixed with twice as many unlabeled effector cells. A total of 0.75 × 106 cells were plated in each well of a 12-well plate and cultivated overnight in DMEM supplemented with 5% FCS and antibiotics. Target and effector cells were also cultivated separately as controls. For FACS analysis, cells were detached with EDTA (Versene) and resuspended in cold PBS containing 3% FCS (PBS-FCS). Labeling with anti-HVEM MAb CW10-PE was performed on 0.5 × 106 cells (50 μl) for 30 min on ice. Cells were washed with cold PBS-FCS and fixed with 3% paraformaldehyde (PFA) in PBS with 3% FCS. In order to gate target cells during FACS analysis, Qtracker655 was detected by excitation at 630 nm and reading at 660 ± 15 nm. Qtracker655-positive target cells were positively selected for measurement of PE fluorescence for HVEM detection.
FACS analysis of downregulation during virus infection.
A total of 0.75 × 106 B78-HVEM cells were seeded in 12-well plates in DMEM with 5% FCS and antibiotics and allowed to attach 4 h at 37°C. The medium was removed, and virus was added in 1 ml chilled medium and allowed to bind for 45 min at 4°C. The cells were then shifted to 37°C for 30 min. Cells were detached with EDTA and resuspended in cold PBS-FCS. B78-HVEM cells were stained with anti-HVEM MAb CW10-PE (20 μg/ml) in 50 μl PBS-FCS. Cells were then washed and fixed in 3% PFA in PBS with 3% FCS.
Determination of binding of HVEMt to CHO-CD160 cells by cell-based enzyme-linked immunosorbent assay (CELISA).
HVEMt was diluted in Ham's F-12 medium with 10% FCS, added to 4 × 104 CHO-CD160 or CHO cells in 96-well plates, and incubated for 1 h at 4°C. Cells were washed three times with PBS and fixed with 100 μl 3% paraformaldehyde in PBS at room temperature (RT) for 60 min. Cells were washed twice, and anti-HVEM PAb R140 (3 μg/ml) was added in Ham's F-12 medium with 10% FCS and left for 1 h at RT. Cells were washed as before and incubated with anti-rabbit IgG coupled to HRP (2 μg/ml) for 1 h. Cells were washed with 20 mM citrate buffer (pH 4.5) before the addition of substrate [2,29-azinobis(3-ethylbenzthiazolinesulfonic acid) (ABTS)] (Moss Inc.). Absorbance at 405 nm was read on a microtiter plate reader (Bio-Tek).
SPR experiments. (i) Kinetics and affinity measurements.
Surface plasmon resonance (SPR) experiments were carried out on a Biacore 3000 optical biosensor (Biacore Life Sciences) at 25°C. The running buffer for the experiments was HBS-EP (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% polysorbate 20). HVEMt was coupled to a CM5 sensor chip through primary amine groups. The flow cells were activated with 1-ethyl-3-(3-dimethylamino-propyl)carbodiimide hydrochloride and N-hydroxysuccinimide (EDC-NHS). HVEMt (0.05 mg/ml) in 10 mM sodium acetate (NaAc) (pH 4.5) was injected until an amount corresponding to approximately 2,000 response units (RU) was coupled to flow cell 2 (Fc2). The surface was quenched by injection of 1 M ethanolamine-HCl (pH 8.5). The Fc1 control surface was activated and quenched without the addition of protein.
To measure affinities, 2-fold serial dilutions of each ligand were injected across Fc1 and Fc2 at 50 μl/min. Ligand-HVEM association was allowed to occur for 2 min, with the wash delay set for an additional 2 min to allow for a smooth dissociation curve. The chip surface was regenerated by injecting brief pulses of 0.2 M Na2CO3 (pH 10) until the response signal returned to baseline. SPR data were analyzed with BIAevaluation software version 4.1, which employs global curve-fitting analysis. Sensorgrams were corrected for nonspecific binding by subtracting the control sensorgram (Fc1) from the HVEMt surface sensorgram (Fc2). Model curve fitting was done with a 1:1 Langmuir binding model with a drifting baseline. This is the simplest model for the interaction between receptor and ligand; it follows the equation A + B ⇆ AB. The rate of association (kon) is measured from the forward reaction, while the rate of dissociation (koff) is measured from the reverse reaction. The binding affinity (KD) is koff/kon. In each case, the chi-square (χ2) value for goodness of fit was less than 1 and the residuals were within ±3 RU, indicating that the data were a good fit to the binding model.
BTLAt binding to HVEMt could not be analyzed using the global curve-fitting analysis because the rates of association and dissociation were too high. Therefore, various concentrations of BTLAt were injected to equilibrium for measurement of affinity (KD) by Scatchard analysis. Equilibrium binding was reached within 2 min, and thus the amount of binding (RU) at 2 min was used for Scatchard analysis. For each concentration, RU bound versus RU bound/concentration was plotted and fitted to a linear model. The negative inverse of the slope of the line is equal to the KD.
(ii) Competition between two ligands.
Each HVEM ligand (acting as ligand 1) was covalently coupled to the surface of a CM5 sensor chip through primary amines as described above. The coinject function was used to inject 300 to 400 RU of HVEMt immediately followed by the test ligand (acting as ligand 2). To regenerate the sensor chip surface, brief pulses of 0.2 M Na2CO3 (pH 9) were injected as necessary until the response signal returned to baseline. This process was repeated for each of the ligands. All ligands were able to block themselves using this method.
(iii) Binding of HVEMt-Y23A.
Each ligand was coupled to a CM5 sensor chip using primary amines as described above. HVEMt or HVEMt-Y23A was flowed across the chip at 5 μl/min for 4 min. The amount of binding at the end of the injection was recorded. The sensor chip surface was regenerated with brief pulses of 0.2 M Na2CO3 (pH 10) until the signal returned to baseline.
(iv) Competition by anti-HVEM MAbs.
MAb CW3, CW7, CW10, CW12, or CW13 was coupled to the surface of a CM5 sensor as described above. Coinjection of HVEMt followed by the test ligand was performed as described above for competition between two ligands. The sensor chip was regenerated with 0.2 M Na2CO3 (pH 11) until the response returned to baseline.
Blocking of virus entry with HVEM ligands.
CHO-HVEM, CHO-nectin-1, or B78-HVEM cells were preincubated with gD(306t), gD(285t), BTLAt, or LTα(r) diluted in culture medium (50 μl/well) for 1 h at 4°C. An equal volume of culture medium containing purified KOStk12 virus (MOI = 5) was added, and cells were placed at 37°C for 6 h. β-Galactosidase activity was used to monitor HSV entry. The results were plotted as percentages of values for controls where no ligand was added.
RESULTS
gD downregulates HVEM from the cell surface.
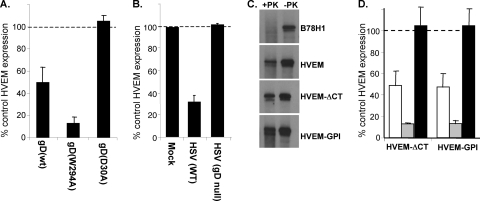
Our previous studies with the HSV receptor nectin-1 showed that trans-interaction with gD expressed on the cell surface and on virions downregulated this receptor from the cell surface (65, 66). Downregulation of nectin-1 consisted of internalization into the endosome/lysosome pathway followed by degradation (65, 66). Since gD-mediated internalization of nectin-1 also directs virus to the endocytic pathway during entry, we investigated whether this was also true for HVEM. To test for HVEM downregulation, we first used B78H1 cells expressing gD in a coculture assay with B78-HVEM target cells (66). As effectors, we used cell lines expressing either wild-type gD [gD(wt)], gD(W294A) [which is a mutant form of gD that binds to HVEM with 100-fold-higher affinity than gD(wt)], or gD(D30A) (which is a mutant gD that cannot bind to HVEM but still binds nectin-1) (13, 33, 66). B78 cells expressing no gD were used as a control. After coculture, cells were detached and stained with the anti-HVEM monoclonal antibody (MAb) CW10. When target B78-HVEM cells were cocultured with B78 cells (no gD), we detected a high level of HVEM, and this value was set at 100%. We detected a similar level of HVEM expression in cocultures with B78-gD(D30A) cells, indicating that HVEM-gD interaction is required for downregulation (Fig. 1A). However, when B78-HVEM cells were cocultured with either B78-gD(wt) or B78-gD(W294A) cells, considerably less HVEM was detected on the cell surface, being reduced to 50% or less than 20%, respectively (Fig. 1A). These results were confirmed by immunofluorescence microscopy of HVEM in cocultures (data not shown). Therefore, HVEM, like the structurally unrelated nectin-1, is downregulated from the cell surface by binding of gD expressed on an adjacent cell. Interestingly, gD(W294A) downregulated HVEM to a greater extent than gD(wt), suggesting that the affinity of the binding interaction affects the efficiency of receptor downregulation. This affinity effect was also observed for nectin-1 downregulation (66).
FIG. 1.
Downregulation of HVEM from the cell surface induced by gD. (A) Downregulation of HVEM expression in cocultures with gD-expressing cells. Target B78-HVEM cells were labeled with Qtracker655 and mixed with effector B78 cells or cells expressing gD. Cells were stained with MAb CW10-PE to detect HVEM by FACS. Target cells were positively selected based on Qtracker655 fluorescence for measurement of HVEM expression. Bar graphs represent PE fluorescence of Qtracker655-positive HVEM-expressing cells as a percentage of the PE fluorescence in cocultures with control B78 cells. The averages from three experiments ± standard errors (SE) are shown. (B) gD on HSV virions downregulates HVEM. HSV KOS (MOI = 50) or KOSgDβ (equivalent of MOI = 50) was added to B78-HVEM cells and left for 45 min at 4°C before shifting to 37°C for 30 min. Cells were then stained with CW10-PE to detect HVEM and analyzed by FACS. The level of HVEM detected on the cell surface when no virus was added was set to 100%. (C) Protease protection assay on HVEM cells. The indicated cell lines were incubated with virus at 4°C before being shifted to 37°C for 15 min. Cells were then treated with proteinase K (+PK) or mock digested (−PK). Cells were lysed, and gB was immunoprecipitated with MAb DL16 and detected by Western blotting (PAb R68). (D) Downregulation of HVEM-ΔCT and HVEM-GPI detected by FACS. Target cells were labeled with Qtracker655 and mixed with effector cells expressing no gD, gD(wt) (white bars), gD(W294A) (gray bars), or gD(D30A) (black bars). Bar graphs represent PE fluorescence of Qtracker655-positive HVEM-expressing cells as a percentage of the PE fluorescence in cocultures with control B78 cells (no gD). The averages from three experiments ± SE are shown.
Downregulation of endogenous HVEM was assessed using primary human resting T cells in coculture experiments. After overnight cocultures, the level of surface HVEM was reduced by about 30% in the presence of B78-gD(W294A) cells, while cells expressing wt gD had no detectable effect. Thus, a high affinity for HVEM is more important to mediate downregulation in primary T cells, which express smaller amounts of HVEM than B78-HVEM cells. Furthermore, since T cells are nonadherent in culture, they may have less contact with adherent effector cells, which may also limit the overall downregulation in this assay. This confirms that endogenous HVEM is downregulated by gD in primary T cells in an affinity-dependent manner.
HSV enters cells either by fusion at the plasma membrane or by endocytosis (41, 47, 80). Since the interaction of virion gD with nectin-1 induces receptor downregulation and endocytosis of HSV into B78H1 cells (65), we determined whether gD on virions also causes HVEM downregulation. B78-HVEM cells were exposed to HSV KOS (MOI = 50) and incubated at 4°C for 45 min for virus attachment and then at 37°C for 30 min to allow virus entry. Cells were detached with EDTA and analyzed by FACS for HVEM surface expression. Infected cells expressed only 30% as much HVEM as mock-infected cells (Fig. 1B). To be certain that HVEM downregulation was caused by virion gD, we inoculated cells with the gD-null virus KOSgDβ at the equivalent MOI (66). In this case, the level of surface HVEM expression was the same as that of mock-infected control cells (Fig. 1B). Thus, downregulation of HVEM from the cell surface is caused by virion gD and rapidly follows exposure to virus.
We next used a protease protection assay to determine whether downregulation of HVEM was accompanied by virus endocytosis. In this assay, glycoproteins on virions that enter by endocytosis are protected from degradation by protease treatment of intact infected cells (41). However, when virions enter cells by direct fusion, glycoproteins are left on the cell surface and are degraded by protease treatment. B78 or B78-HVEM cells were incubated with virus at 4°C prior to shifting the temperature to 37°C for 15 min to allow endocytosis. The cells were then incubated with proteinase K or mock digested. Cell lysates were prepared, and gB was immunoprecipitated and analyzed by Western blotting (Fig. 1C). In the mock-digested samples, equal amounts of gB were detected on both B78 and B78-HVEM cells. However, gB was not protected from proteinase K digestion of B78 cells because they lack a gD receptor. In contrast, when B78-HVEM cells were infected, gB was protected from proteinase K, indicating that HSV enters these cells by endocytosis. Therefore, the rapid gD-mediated downregulation of HVEM correlates with endocytic entry of HSV into B78-HVEM cells.
The cytoplasmic tail of HVEM has binding sites for the TRAF family of adaptor proteins, which are involved in activation of NF-κB by HVEM (28, 37). Therefore, we wanted to determine if the cytoplasmic tail (CT) or transmembrane region (TMR) of HVEM plays a role in HVEM downregulation and HSV endocytosis. B78 cells were engineered to express HVEM with a truncated cytoplasmic tail (HVEM-ΔCT) or a glycosylphosphatidylinositol (GPI)-anchored HVEM (HVEM-GPI). HVEM-ΔCT and HVEM-GPI function as well as full-length HVEM in mediating HSV entry into B78H1 cells (data not shown). We used the protease protection assay to examine the pathway of HSV entry into B78-HVEM-ΔCT and B78-HVEM-GPI cells. We found no difference between full-length HVEM and either HVEM-ΔCT or HVEM-GPI in this assay (Fig. 1C). Moreover, these modified forms of HVEM were downregulated by gD (Fig. 1D). These results indicate that the cytoplasmic tail and the transmembrane region of HVEM are dispensable both for HSV endocytosis and for gD-mediated downregulation of HVEM. This suggests that other cellular proteins may be involved in endocytosis of HVEM by connecting the receptor to the endocytic machinery in the cytoplasm. The extent of endocytosis triggered by cell-bound gD or virion gD is unclear. Whether it is limited or generalized, our data indicate that the endocytosis is initially triggered by gD binding to HVEM.
BTLA and LIGHT, but not CD160, downregulate HVEM from the cell surface.
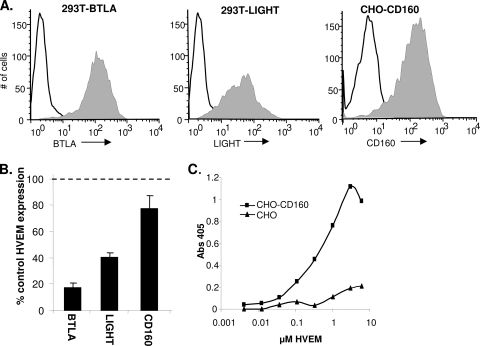
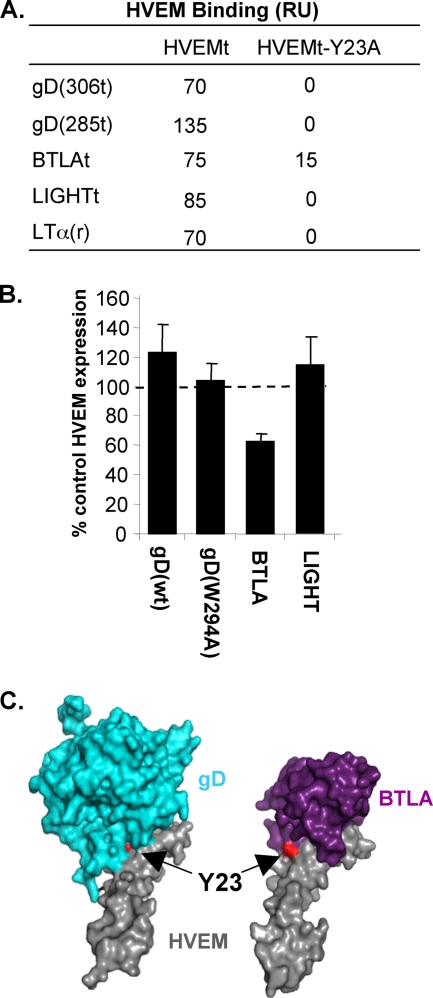
It is possible that gD activates a natural response that is normally generated by binding of HVEM to one or several of its cellular ligands. HVEM has three other ligands that are expressed on the cell surface: LIGHT, BTLA, and CD160. We used the coculture assay to test whether any of the natural ligands might also downregulate HVEM. To do this, we used human 293T cells to create effector cell lines that stably express full-length BTLA or LIGHT. We also obtained a CHO cell line that expresses human CD160 (1). Cell lines were screened by FACS for cell surface expression of HVEM ligands using MAbs MIH26 (BTLA), αLIGHT 115520 (LIGHT), and 5D.10A11 (CD160) (4, 49). All of the cell lines expressed high levels of ligand on their surfaces (Fig. 2A), while no ligands were detected on parental cells. Target B78-HVEM cells were cocultured overnight with 293T-LIGHT, 293T-BTLA, or CHO-CD160 effector cells. Surface expression of HVEM on target cells was analyzed by FACS. We detected a high level of HVEM when B78-HVEM cells were cocultured with either 293T or CHO cells, and this level was set to 100%. In contrast, when B78-HVEM cells were cocultured with 293T-BTLA cells, HVEM expression was only 20% of the control level (Fig. 2B). This was comparable to what was observed in cocultures with effector cells expressing gD(W294A) (Fig. 1A). In cocultures with 293T-LIGHT cells, HVEM expression on the surface of target cells was 40% of control levels (Fig. 2B). In contrast, there was only a slight reduction in HVEM expression when the target cells were cocultured with CHO-CD160 effector cells (Fig. 2B).
FIG. 2.
Downregulation of HVEM by BTLA, LIGHT, or CD160. (A) Ligand expression on the indicated effector cell lines detected by FACS using MAb MIH26 (BTLA), LIGHT 115520 (LIGHT), or 5D.10A11 (CD160) and goat anti-mouse secondary Ab conjugated to Alexa 488. The white histogram indicates unstained cells. (B) Downregulation of HVEM in coculture experiments. B78-HVEM target cells were labeled with Qtracker655 and mixed with effector 293T-BTLA, 293T-LIGHT, CHO-CD160, or control (293T or CHO) cells. Cells were stained with MAb CW10-PE to detect HVEM. Bar graphs represent PE fluorescence of Qtracker655-positive HVEM-expressing cells as a percentage of the PE fluorescence in cocultures with control cells from the averages from three experiments ± SE. (C) Detection of HVEM binding to CHO-CD160 cells by CELISA. HVEMt was incubated with CHO or CHO-CD160 cells for 1 h at 4°C. HVEM binding was detected with PAb R140 and goat anti-rabbit Ig conjugated to HRP.
We used a cell-based ELISA to confirm that CD160 on the surface of CHO cells could bind to HVEM. HVEMt binding to either CHO or CHO-CD160 cells was detected with the anti-HVEM PAb R140. HVEMt bound to CHO-CD160 cells in a dose-dependent manner but did not bind to parental CHO cells. Thus, the low level of HVEM downregulation noted by FACS was not due to a failure of CD160 to interact with HVEM. We conclude that downregulation of HVEM is a common robust response to the binding of gD, BTLA, or LIGHT but not to the binding of CD160. We suggest that HVEM downregulation could be a natural means for regulation of HVEM expression that is subverted by HSV gD.
Comparison of binding affinities of HVEM for gD and its natural ligands.
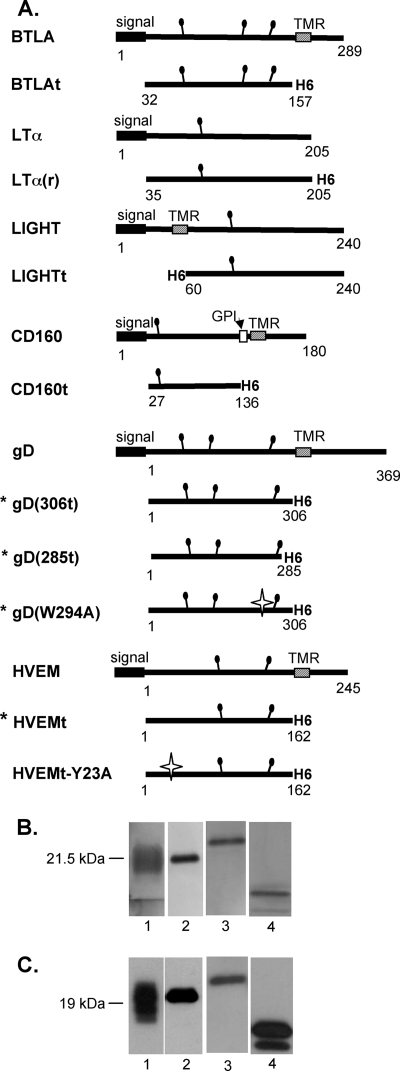
Because gD induced a response similar to that of two of the natural ligands of HVEM, we compared how gD binds to HVEM relative to these ligands. To do this, we constructed recombinant baculoviruses that produced soluble forms of human LIGHT, LTα, BTLA, or CD160. For LTα, BTLA, and CD160, the expressed protein consisted of its ectodomain with a C-terminal six-histidine tag (Fig. 3A). Since LIGHT is a type II membrane protein, the six-histidine tag was placed on the N terminus of the ectodomain. All proteins were purified by nickel affinity chromatography and analyzed by SDS-PAGE. Purity was assessed by silver staining, and antigenicity was evaluated by Western blotting (Fig. 3B and C). The multiple bands for BTLA and CD160 resulted from various amounts of N-linked glycosylation (data not shown). Additionally, each protein was recognized by MAbs to conformational epitopes by ELISA, suggesting that the conformation of the proteins is similar to that of the native protein (data not shown). Each protein monomer was the expected size as determined by Western blotting (Fig. 3C), and their oligomeric states in solution were estimated by gel filtration chromatography (Table 1). LTα(r) and LIGHTt were each determined to be a trimer in solution, as previously reported for LTα and LIGHT produced in bacteria and mammalian cells, respectively (2, 3, 52). In contrast, CD160t and BTLAt both appeared to be monomeric in solution (Table 1). Therefore, in calculating binding by surface plasmon resonance (SPR), we considered LTα(r) and LIGHTt to bind to immobilized HVEMt as trimers and CD160t and BTLAt to bind to HVEMt as monomers. Thus, we have produced and characterized soluble forms for all of the ligands of HVEM in the same expression system. This allowed us to directly compare HVEM cellular and viral ligands under the same experimental conditions.
FIG. 3.
HVEM ligands produced in baculovirus. (A) Schematics of full-length and truncated constructs of each ligand. Constructs indicated with an asterisk were previously published (33, 53, 63, 77). The white stars indicate point mutations. (B and C) Soluble proteins purified from the supernatant of Sf9 cells infected with recombinant baculovirus were analyzed by silver staining (B) and Western blotting with anti-histidine tag MAb (C). Lanes 1, BTLAt; lanes 2, LTα(r); lanes 3, LIGHTt; lanes 4, CD160t.
TABLE 1.
Mass analysis data
| Protein | Mass (kDa) determined by: |
Oligomeric state | |
|---|---|---|---|
| Formula (no. of glycosylation sites)a | Gel filtrationb | ||
| LIGHTt | 20.8 (1) | Major peak, 70.9 | Trimer |
| Minor peak, 24.5 | Monomer | ||
| LTα(r) | 20.2 (1) | 69.0 | Trimer |
| CD160t | 13.2 (1) | 14.4 | Monomer |
| BTLAt | 14.5 (3) | 22.4 | Monomer |
Based on amino acid composition; the mass of N-CHO not included.
Estimated based on the elution volume of the maximum of the peak compared to protein standards. The broadness of peaks varied based on the number of N-linked carbohydrates.
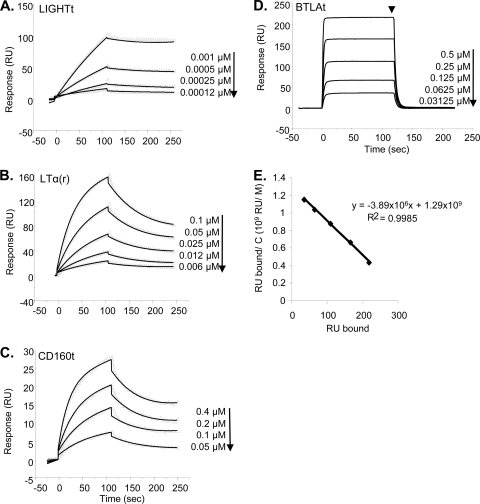
Previous SPR studies showed that gD(306t) and gD(285t) bind to immobilized HVEMt with affinities of 3.2 × 10−6 M and 3.7 × 10−8 M, respectively (53, 76, 79). The mutant gD(Δ290-299), produced in our laboratory and used in other binding studies (12, 22, 38), has an affinity (i.e., KD = 3.3 × 10−8 M [53]) and binding properties similar to those of gD(285t) used here. We coupled HVEMt to the surface of a CM5 sensor chip and examined the kinetics of binding of LTα(r), LIGHTt, CD160t, and BTLAt to HVEM. Each ligand was serially diluted in buffer and injected across the surface of the sensor chip. We first showed that the affinities of gD(306t) and gD(285t) for HVEM were 2.1 × 10−6 M and 3 × 10−8 M, respectively, values consistent with previous reports. LIGHT had the lowest KD for binding to HVEM, at 3 × 10−9 M (Fig. 4A; Table 2). This was consistent with the affinity of LIGHTt66 produced in mammalian cells (52). LTα(r) had a lower association rate (kon) and a higher dissociation rate (koff) than LIGHTt, resulting in a 20-fold-lower binding affinity for HVEM, with a KD of 5.7 × 10−8 M (Fig. 4B; Table 2). The affinity of CD160t for HVEM was 1.7 × 10−7 M, which is about 60-fold lower than that of LIGHTt and 3-fold lower than that of LTα(r) (Fig. 4C; Table 2). This is because CD160t has both a lower association rate and a higher dissociation rate than either LIGHTt or LTα(r). The kinetics of BTLAt binding to HVEMt were clearly different from those of the other ligands, as it both associated and dissociated very quickly. In fact, the on and off rates were too high to allow the determination of affinity by global fitting (Fig. 4D). Therefore, we used Scatchard analysis to determine the KD by injecting BTLAt at various concentrations until equilibrium was reached (Fig. 4D). This method was previously demonstrated to yield the same affinity value as the global fitting analysis; however, on and off rates cannot be calculated (53, 79). Our analysis yielded a KD of 2.6 × 10−7 M for binding of BTLAt to HVEMt (Fig. 4E; Table 2), which is in a range similar to that of the KD between CD160t and HVEMt. By determining the binding affinities and kinetics of each of the HVEM ligands under the same conditions, we were able to directly compare them to each other. The two TNF family ligands (LTα and LIGHT) had a higher affinity for HVEM than the ligands from the Ig superfamily (BTLA and CD160) (Table 2). The affinity of gD(285t) was similar to that of LTα but higher than that of either BTLA or CD160. Interestingly, the affinities of all of the natural ligands for HVEM were 10- to 1,000-fold higher than the affinity of wild-type gD(306t).
FIG. 4.
Sensorgrams of ligand binding to HVEM. (A to C) Repeated injections of LIGHTt (A), LTα(r) (B), or CD160t (C) across immobilized HVEMt at the indicated concentrations are represented by the triangles. The solid line is the best fit used for kinetics determination. (D) Equilibrium binding sensorgrams of the binding of BTLAt to immobilized HVEMt. The arrowhead indicates the time point used for Scatchard analysis. (E) Scatchard analysis for BTLAt binding. RU bound is plotted against RU bound/concentration of BTLAt flowed across the surface. The negative inverse of the slope is equal to the binding affinity (KD). Representative curves from two experiments are shown.
TABLE 2.
Kinetic and affinity values for ligand-HVEMt binding
Competition between ligands for binding to HVEM.
gD binds to a site on HVEM that overlaps the binding site of BTLA and is proposed to be on the opposite side of that of LIGHT or LTα. To define how the viral and cellular HVEM ligands affect binding of each other, we carried out a series of competition studies by SPR using soluble proteins to examine all the possible ligand combinations. Previous studies showed that the high-affinity gD(Δ290-299) competes with BTLA for binding to HVEM as determined by ELISA (22). BTLA competes with CD160 as determined by ELISA as well (4). However, there was no information about whether wild-type gD(306t) could compete with either of these proteins. Additionally, previous reports conflicted in regard to the ability of LIGHT or LTα to bind to HVEM at the same time as gD (38, 55).
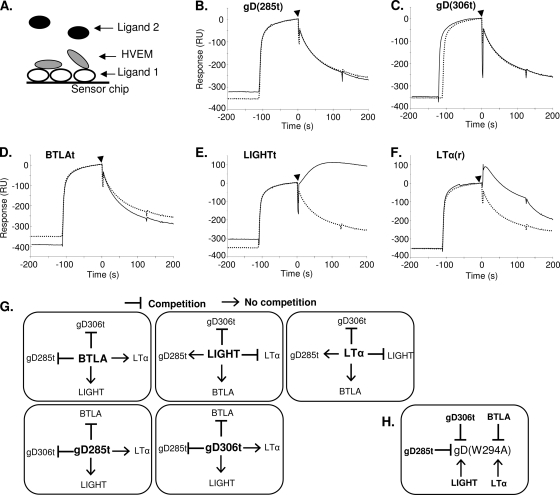
In this assay, the competing ligand (ligand 1) was directly coupled to a CM5 sensor chip (Fig. 5A). The capture of HVEMt by ligand 1 was followed immediately by injection of ligand 2. Thus, if ligand 2 is able to bind to HVEM that is already bound and presented by ligand 1, there is no competition between the ligands. However, if ligand 2 cannot bind in this situation, this means that ligand 1 competes with ligand 2 for binding to HVEM (Fig. 5A). Results of SPR competition experiments in which gD(285t) was coupled to the sensor chip as ligand 1 are shown in Fig. 5B to F. As expected, gD(285t) did not bind to HVEM(200t) that was already bound to gD(285t) on the chip (Fig. 5B). Thus, gD(285t) effectively competes with itself, which validates this method for looking at competition between the ligands of HVEM. In addition, gD(285t) competed with both gD(306t) and BTLAt for binding to HVEMt (Fig. 5C and D). This is in agreement with a previous study which showed that gD(Δ290-299) competes with BTLA for binding of HVEM as determined by ELISA (22). Interestingly, we found that both LIGHTt and LTα(r) bound to HVEMt presented by gD(285t), suggesting that gD(285t) does not compete with either LIGHTt or LTα(r) for binding to HVEM.
FIG. 5.
Competition between ligands for HVEM binding by SPR. (A) Schematic of method used for competition. Ligand 1 was coupled directly to the sensor chip. HVEMt was flowed across and allowed to bind, followed immediately by injection of ligand 2. (B to F) Series of experiments with gD(285t) (ligand 1) in competition with the indicated ligand 2. The reference sensorgram (dotted line) represents a parallel experiment with buffer injected in place of ligand 2 to monitor the loss of signal due to natural dissociation of HVEMt from ligand 1. Increased signal after injection of ligand 2 (arrowhead, continuous line) relative to the reference sensorgram indicates binding of ligand 2 and the absence of competition [e.g., for LIGHTt and LTα(r)]. (G) Summary of results from competition experiments with gD(285t), gD(306t), BTLAt, LIGHTt, and LTα(r). The ligand that is coupled to the sensor chip (ligand 1) is in the center in bold. The ability of ligand 1 to compete with ligand 2 is indicated with a T bar for competition or an arrow for no competition. (H) Summary of the effects of HVEM ligands on gD(W294A) binding. T bars indicate that ligand 1 (bold) prevents gD(W294) (ligand 2) from binding HVEM, and the arrow indicates an absence of competition.
Figure 5G summarizes competition experiments involving each of the ligand pairings except CD160, where binding was too low to be included. gD(306t) blocked the binding of BTLAt and gD(285t), suggesting that wild-type gD is just as effective at blocking BTLA as gD(285t). Reciprocally, BTLAt blocked the binding of both forms of gD. Furthermore, gD(285t), gD(306t), and BTLAt did not compete with LIGHTt or LTα(r). LIGHTt competed with LTα(r) but not with BTLAt, confirming the formation of a ternary complex, HVEM-BTLA-LIGHT (22). Interestingly, although LIGHTt and LTα(r) could not block binding of gD(285t), they did block gD(306t).
There are two differences between gD(306t) and gD(285t) that may account for this difference: (i) the affinity of gD(285t) for HVEM is 100-fold higher than that of gD(306t), and (ii) gD(306t) contains a longer C terminus of the ectodomain, which is known to change conformation upon binding to HVEM (33). To determine if the presence of the C terminus directly affects competition, we used a mutant form of gD(306t) carrying a W294A point mutation. gD(W294A) retains the ectodomain C terminus like gD(306t) but has the same affinity for HVEM as gD(285t) (33). As shown with gD(306t) and gD(285t), we found that BTLAt blocked the binding of gD(W294A) (Fig. 5H). However, neither LIGHTt nor LTα(r) blocked binding of gD(W294A) to HVEM. We conclude that the difference in affinity between the wild-type and high-affinity forms of gD caused the disparity in blocking by LIGHTt or LTα(r).
Mapping of ligand binding sites with mutants and antibodies.
Competition between ligands can be due to overlapping binding sites or to conformational changes of the receptor which prevent interaction of a ligand at a different site. We therefore characterized the binding sites of gD and cellular ligands to HVEM using receptor mutants and antireceptor MAbs. The crystal structures of gD-HVEM and BTLA-HVEM complexes showed that gD and BTLA bind to overlapping sites and suggested that the tyrosine at position 23 (Y23) of HVEM is critical for binding of either ligand (7, 12). Previous studies showed that mutation of Y23 to alanine (HVEM-Y23A) abolishes binding to gD and BTLA as determined by ELISA (12, 15). Here we examined the binding of the HVEM ligands to HVEM-Y23A by SPR. A soluble form of HVEMt-Y23A was produced from baculovirus and purified similarly to HVEMt. This mutant was recognized by conformation-dependent MAbs CW3 and CW10, indicating that it was folded properly (data not shown). Each ligand was directly coupled to a CM5 sensor chip, and HVEMt or HVEMt-Y23A was flowed across the surface. HVEMt bound to each of the ligands (Fig. 6A). Under similar conditions, HVEMt-Y23A did not bind to gD(306t), gD(285t), LIGHT, or LTα. Unexpectedly, HVEMt-Y23A did bind to BTLAt, although it bound less well than wild-type HVEMt (Fig. 6A).
FIG. 6.
Effect of the Y23A mutation on the interaction of HVEM with its ligands. (A) Binding determined by SPR. HVEMt or HVEMt-Y23A was flowed across gD(306t), gD(285t), LIGHTt, LTα(r), or BTLAt immobilized to the sensor chip surface. The total amount of binding (RU) was recorded at end of a 4-min association period. (B) Target cells expressing HVEM-Y23A were cocultured with control cells or cells expressing gD(wt), gD(W29A), BTLA, or LIGHT. Cells were stained with MAb CW10-PE, and HVEM-Y23A expression was detected by FACS as described for HVEM in Fig. 1A. (C) Surface representations of the gD-HVEM (left) and BTLA-HVEM (right) crystal structures. Residue Y23 is shown in red and is indicated with the arrow.
To determine if the absence or reduction of binding to HVEMt-Y23A had functional implications, we tested whether the Y23A mutation would affect downregulation of full-length HVEM by gD, BTLA, or LIGHT. We used B78-HVEM-Y23A cells (15) as targets in the coculture assay with effector cells that expressed either gD(wt), gD(W294A), BTLA, or LIGHT. After coculture with cells expressing gD or LIGHT, there was no downregulation of HVEM-Y23A from the cell surface (Fig. 6B). However, expression was reduced by 40% in cocultures with 293T-BTLA cells (Fig. 6B). These results correlate with binding data in which BTLA retained some ability to interact with HVEM-Y23A, whereas gD and LIGHT did not. A close examination of crystal structures reveals that although BTLA does interact with Y23 of HVEM, this residue is at the periphery of the interface. In contrast, Y23 is at the center of the binding site for gD (Fig. 6C).
We next examined the ability of anti-HVEM MAbs to prevent the binding of gD and natural ligands by SPR. Each MAb was directly coupled to the chip and used to capture HVEM before injection of the ligands. We first tested MAb CW3, which binds to HVEM CRD1 and blocks gD binding to HVEMt as determined by ELISA (76). In addition to blocking binding of gD(306t) and gD(285t), MAb CW3 also prevented binding of BTLAt, LIGHTt, and LTα(r) to HVEM (Table 3). Other MAbs with epitopes mapped to different regions (CW7, CW10, CW12, and CW13) had no effect on the binding of any of the ligands. Together, the HVEM-Y23A and MAb binding data suggest that the binding sites for each of the ligands are located closely enough together that their binding could be blocked with a single MAb or by a point mutation.
TABLE 3.
Binding of ligands to HVEM presented by MAbsa
| MAb | CRDb | Binding |
||||
|---|---|---|---|---|---|---|
| gD(306t) | gD(285t) | BTLAt | LIGHTt | LTα(r) | ||
| CW3 | 1 | − | − | − | − | − |
| CW7 | 3, 4 | + | + | + | + | + |
| CW10 | 3, 4 | + | + | + | + | + |
| CW12 | 3, 4 | + | + | + | + | + |
| CW13 | 3, 4 | + | + | + | + | + |
HVEMt was captured by the indicated MAbs on a CM5 biosensor chip. The indicated ligands were flowed across these surfaces, and binding was recorded. Each ligand bound similarly (>50 RU) to HVEM presented by CW7, -10, -12, and -13.
Cysteine-rich domains (CRD) are numbered as described by Montgomery et al. (42). MAb CW3 binds to CRD1, while the other epitopes were mapped to CRD3 and -4.
Blocking of HSV entry.
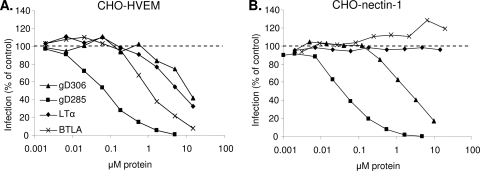
Since BTLA and LTα were able to compete with gD for binding to HVEM, we wanted to use a functional assay to determine if these ligands could affect HSV entry into HVEM-expressing cells. To determine if the natural ligands of HVEM blocked HSV entry similarly to soluble gD, we added various concentrations of gD(306t), gD(285t), BTLAt, or LTα(r) to either CHO-HVEM or CHO-nectin-1 cells at 4°C for 1 h. We then infected the cells with HSV KOStk12, which expresses the lacZ gene, and measured entry by β-galactosidase expression at 6 h postinfection. LIGHTt and CD160t were not tested, as the protein yield was too low to be used in this assay. As expected, gD(306t) and gD(285t) blocked entry into both CHO-HVEM and CHO-nectin-1 cells (16, 77). Because it has a higher affinity for HVEM, gD(285t) was more efficient at blocking entry than gD(306t) (50% blocking at 0.1 μM versus 10 μM, respectively, on CHO-HVEM cells). BTLAt and LTα(r) specifically blocked HSV entry into CHO-HVEM cells (Fig. 7A). BTLAt was slightly more efficient (50% blocking at 1 μM) than gD(306t) at blocking entry into CHO-HVEM cells, while LTα(r) blocked entry only as efficiently as gD(306t). The same hierarchy of blocking efficiency was observed on B78-HVEM cells (not shown), indicating that competition is not cell specific. However, since B78-HVEM cells express more HVEM, about 5-fold-higher concentrations of soluble proteins were required to achieve the levels of blocking observed in CHO-HVEM cells. Interestingly, although both BTLAt and LTα(r) have an affinity for HVEM that is similar to or higher than that of gD(285t), they were less efficient at blocking entry into HVEM-expressing cells. This suggests that the interactions of these natural ligands with HVEM are structurally different from that of gD. As expected, neither BTLAt nor LTα(r) blocked entry into cells expressing nectin-1, because they do not interact with this receptor (Fig. 7B).
FIG. 7.
Soluble BTLA and LTα block HSV entry into HVEM-expressing cells. CHO-HVEM (A) and CHO-nectin-1 (B) cells were preincubated with various concentrations of BTLAt, LTα(r) gD(285t), or gD(306t) for 1 h at 4°C. KOStk12 virus was then added (MOI = 5), and cells were shifted to 37°C for 6 h. Cells were lysed for quantitation of β-galactosidase activity. Data are represented as a percentage of β-galactosidase activity in the absence of added protein (100%).
DISCUSSION
HVEM is expressed mainly on cells of the immune system which are not targets of productive HSV infection or latency in vivo, yet clinical isolates of HSV retain the ability to use this receptor (32). However, HSV can enter T lymphocytes, and infection can be achieved by stimulation with mitogens (6, 51). Accordingly, viral replication appears to be blocked prior to DNA replication in T cells (26, 54). Activation may affect multiple steps of HSV infection, including levels of HVEM expression (43, 44, 58, 59) but it is not known whether the entry pathway is affected. Because gD can interfere with HVEM binding to its ligands, the interaction of HSV with HVEM may be important for regulating the immune response to the advantage of the virus rather than leading to productive infection. Previous studies showed that BTLA and gD bind to overlapping sites on HVEM (7, 12), suggesting that gD might interfere directly with the interaction between HVEM and some of its natural ligands. Using soluble versions of LTα, LIGHT, BTLA, and CD160 along with gD and HVEM (48, 77), we found that despite the fact that wild-type gD(306t) had the lowest affinity for HVEM, it can block the binding of BTLA, but not that of LTα or LIGHT, to HVEM. In this study, we also showed that gD as well as BTLA and LIGHT causes HVEM downregulation. Thus, gD binding to HVEM may interfere with the interaction of the natural HVEM ligands either by direct competition or by removing the receptor from the cell surface.
Receptor downregulation by gD and other ligands.
We previously showed that gD induced the internalization of its receptor nectin-1 during HSV endocytic entry and after transient trans-interaction at cell-cell contacts (65, 66). Here, we showed that gD also induces HVEM downregulation during entry and upon cell-cell contact. It is remarkable that despite the structural and functional differences between HVEM and nectin-1, HSV can use both receptors to access the endocytic pathway. After transient interaction at cell-cell contacts, gD induces HVEM downregulation in B78-HVEM cells and in human primary T cells. Downregulation of endogenous HVEM in resting T cells is achieved more efficiently by a high-affinity form of gD. This may be due to the smaller amount of HVEM at the surface of these cells or to a renewed expression of HVEM in lymphocytes that do not remain in constant contact with the effector cells. Upon exposure to gD, HVEM levels are reduced, suggesting an enhanced turnover of the receptor, similar to that of nectin-1 (65, 66). The natural rate of turnover of HVEM in human lymphocytes varies depending on the activation state of the cells (43, 45). Further experiments on HVEM expression in resting and activated T cells are required to correlate the effect of gD with the regulation of HVEM expression in lymphocytes.
HVEM downregulation has potentially important consequences for immune modulation, since the removal of HVEM from the cell surface would make it inaccessible for binding by any of its other ligands. Like direct competition by gD, this might interfere with the normal immune regulatory signals provided by HVEM and, in turn, may influence the immune response to HSV. Since gD is not likely to be found in a soluble form in its host but rather is expressed on virions and on the surface of infected cells, HVEM downregulation might be the preferred method of gD-mediated interference with the HVEM immunoregulatory pathways. Paradoxically, downregulation of HVEM on adjacent cells decreases the availability of receptors for cell-to-cell spread. However, uninfected cells continuously produce receptors appearing on the surface, which may either encounter cell-bound gD and be downregulated or encounter gD on egressing virions and lead to virus spread. It is not clear how downregulation affects the extent of spread, but the advantage of inducing HVEM downregulation appears to prevail over the inconvenience of a limitation of spread to T cells or monocytes.
HSV may have tapped into a normal mechanism used to regulate HVEM function, as BTLA and LIGHT also both induce HVEM downregulation (Fig. 2). Indeed, Morel et al. showed that LIGHT downregulates HVEM expression upon T cell activation (43). HVEM was redistributed to the inside of the cell and this effect could be blocked either by anti-LIGHT antibodies or by soluble HVEM-Fc. Thus, the costimulatory ligands of HVEM may be self-regulating so that the receptor is concurrently downregulated as cells are activated. The fact that BTLA induces downregulation of HVEM suggests that this mechanism could also hold true for coinhibitory ligands of HVEM. Therefore, we suggest that receptor downregulation has a broad effect; however, further studies are necessary to understand whether HSV gD mimics LIGHT or BTLA or both.
Differences in binding kinetics and affinities of HVEM ligands.
A variety of affinities for LIGHT, LTα, or BTLA binding to HVEM have been previously reported, but the different methods used made it difficult to compare them directly. Here, we used one system to produce the soluble ectodomain of each ligand and SPR to determine binding affinities and kinetics. We found that the trimeric TNF family ligands LTα and LIGHT had the highest affinities for HVEM (57 nM and 3 nM, respectively). The affinities of Ig family ligands BTLA and CD160 were approximately 4-fold lower than that of LTα and 70-fold lower than that of LIGHT (250 nM and 170 nM, respectively). BTLA and CD160 have similar affinities, but the binding kinetics of BTLA are very different from those of the other HVEM ligands. The BTLA affinity determined by SPR is approximately 10-fold lower than a previously reported measure of BTLA binding to HVEM-expressing cells (12). The difference in the method used for measurement likely accounts for the discrepancy in the determined affinity (12). The differences in affinities of the TNF and Ig family ligands for HVEM may be a result of their different functions and sites of binding. LIGHT and LTα, which are both trimers, are each thought to bind to a trimer of HVEM (3). Since TNFRs such as HVEM are believed to be dimers on the cell surface, high-affinity ligand binding may be needed to induce trimerization (8, 46). In contrast, BTLA-HVEM binding is known to be 1:1 (12), and this is also likely to be the case for CD160-HVEM. Similarly, the stoichiometry of gD-HVEM binding is also 1:1 (7), which is in accordance with its low affinity for HVEM. Overall, our studies have revealed a wide range of affinities, binding kinetics, and stoichiometries that likely reflect the structural differences among the ligands (Table 4).
TABLE 4.
Properties of HVEM ligands
| Ligand | Affinitya | Oligomerb | HVEM downregulationc |
|---|---|---|---|
| gD(306t) | + | 2 | ++d |
| gD(285t) | +++ | 2 | NAe |
| BTLA | ++ | 1 | +++ |
| LIGHT | ++++ | 3 | ++ |
| LTα | +++ | 3 | NA |
| CD160 | ++ | 1 | +/− |
| gD(W294A) | +++ | 2 | +++ |
KD summarized according to Table 2.
Number of protomers in the soluble proteins used in this study. On the cell surface, BTLA is likely a dimer and CD160 forms multimers (10, 21).
Determined in cocultures with HVEM ligands expressed on the surface of effector cells.
Downregulation by full-length wt gD.
NA, not applicable. gD285t and LTα(r) are soluble proteins.
Binding of gD relative to HVEM natural ligands.
Interestingly, the affinity of wild-type gD for HVEM is lower than that of all the natural ligands of HVEM. Although gD contains an Ig V domain like BTLA and CD160, this domain is not involved in the interaction of gD with HVEM. Instead, gD binds to HVEM through a loop formed by a flexible N-terminal extension (7, 12). Binding of HVEM to gD requires conformational changes in gD to displace the C terminus and form the N-terminal loop (14, 33). The presence of the gD C terminus slows complex formation with HVEM, and when it is removed or destabilized, the affinity increases 50- to 100-fold. Thus, the shorter gD(285t) has a higher affinity for the receptor because the HVEM binding site is more accessible. The HVEM binding site on BTLA is exposed, allowing a very fast interaction; however, the BTLA complex dissociates more rapidly from HVEM than gD despite sharing a similar binding site on HVEM.
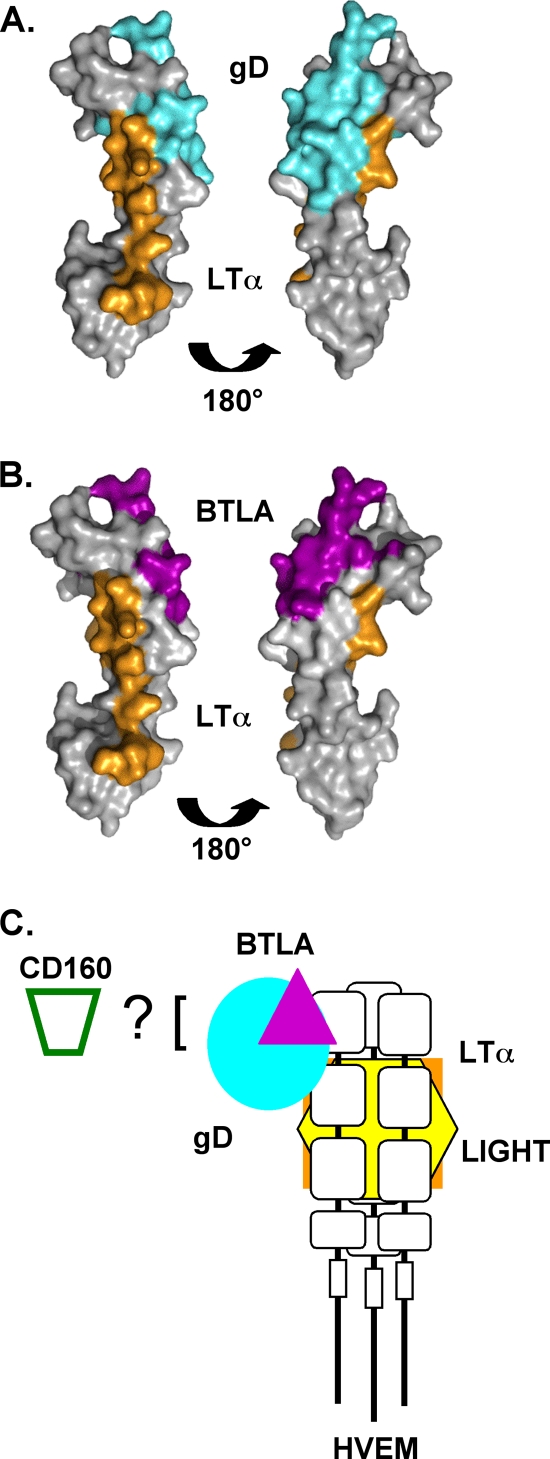
Indeed, despite having a lower binding affinity, wild-type gD(306t) inhibits the binding of BTLA to HVEM. As both gD and BTLA bind to an overlapping site on CRD1 of HVEM, competition for binding with BTLA is expected (Fig. 8). Neither gD nor BTLA competed with LTα or LIGHT binding to CRD2/3 on the opposite face of HVEM, which is unlikely to be masked by the Ig-like ligands. Interestingly, LIGHT and LTα both could block the binding of gD(306t) to HVEM but have no effect on the binding of either gD(285t), gD(W294A), or BTLA. By modeling the binding residues of LTα on HVEM based on its binding to TNFR1 (2), it is clear that the binding sites of gD and LTα may be less distinct than has been previously suggested (Fig. 8A). The inhibition of binding of all the ligands by the Y23A point mutation (Fig. 6) and the MAb CW3 (Table 3) also supports this idea. It is possible that trimerization caused by LTα and LIGHT binding changes the conformation of CRD2. This may be enough to prevent binding of the low-affinity gD(306t). The higher affinity of gD(285t) and gD(W294A) may allow these ligands to overcome this change. BTLA would likely not be affected, as it binds exclusively in CRD1. Alternatively, gD(306t), which is larger, could be more sensitive to steric hindrance. However, the fact that the mutant gD(W294A), which contains the C terminus but has a high affinity for HVEM, behaves like gD(285t) suggests that it is the higher affinity that allows them to overcome the block by LIGHT or LTα. Thus, our data support the formation of a ternary complex between HVEM, BTLA, and LTα as well as the previously reported complex between HVEM, BTLA, and LIGHT (22). Our data also indicate that a ternary complex could form between HVEM, gD, and LIGHT or LTα, but only if gD binds to HVEM first (Fig. 8C). In this configuration the effect of the TNF-like ligand may be modulated differently in the presence of BTLA or gD.
FIG. 8.
Crystal structures of HVEM with footprint of contact residues for ligands. Contact residues for BTLA and gD were determined from the crystal structures of the complexes (7, 12). For LTα, the contact residues of LTα on TNFR1 were determined based on the crystal structure (2). Based on a sequence alignment of TNFR1 and HVEM, the homologous residues in HVEM were determined. (A) Comparison of gD binding site (cyan) and modeled LTα binding site (orange) on HVEM. (B) Comparison of BTLA binding site (purple) and modeled LTα binding site (orange) on HVEM. (C) Model of relative spacing of ligand interactions with HVEM. LTα, orange; LIGHT, yellow; BTLA, purple; gD, cyan.
Soluble BTLA and LTα block HSV entry.
Soluble gD is known to block HSV entry into cells expressing either HVEM or nectin-1 due to occupation of the receptor or downregulation from the cell surface (16, 77). We found that soluble BTLAt and LTα(r) could specifically block HSV entry into cells expressing HVEM. This result correlates with the direct blocking of wild-type gD [i.e., gD(306t)] binding to HVEM by BTLAt and LTα(r) in the competition studies. LTα is a soluble ligand for HVEM that could possibly interfere directly with infection mediated by HVEM. Interestingly, despite having a 50-fold-higher affinity, LTα is not more efficient than gD(306t) at blocking virus entry. This may be because LTα does not bind to the same site as virion gD and therefore might block indirectly through a change in conformation. It is currently unknown whether LTα has any effect on HSV infection in vivo by this mechanism.
Implications for HSV infection.
What are the implications for the ability of wild-type gD to compete with BTLA for binding to HVEM? HVEM acts as a molecular switch that helps to regulate the immune response through both costimulatory and coinhibitory signaling (5, 18, 44, 45, 74). Blocking the binding of BTLA or CD160 would likely hinder inhibitory signaling which could potentially augment the immune response. Since both LIGHT and LTα can bind to other receptors to promote costimulatory signaling (19, 64), it has been proposed that inhibitory signaling is the most important function of HVEM (73).
It is also possible that gD evolved to use the binding site of a natural ligand of HVEM to induce signaling itself. Several studies have indicated that gD induces NF-κB activation upon HVEM binding (11, 39, 57). NF-κB activation is advantageous for virus replication but may also augment T or B cell responses. BTLA and gD binding to HVEM share the most similarity regarding structure, binding site, downregulation, and, to some extent, affinity. It is possible that gD mimics some functional aspects of BTLA as well. In fact, both gD and BTLA binding to HVEM in trans induced survival signals in the HVEM-expressing T cells (11). The interaction with HVEM has the opposite outcome for the BTLA-expressing cells, which receive an inhibitory signal (22, 58). Thus, gD binding to HVEM both induces a T cell survival signal and eliminates the possibility that BTLA can send an inhibitory signal by removing HVEM from the cell surface. This could potentially enhance a T cell response. It is unclear why it would be advantageous for HSV to enhance the immune response through the gD-HVEM interaction. We suggest that this may be a means of activating an immune evasion mechanism similar to the T cell fratricide model, in which HSV-infected cytotoxic T lymphocytes (CTLs) kill other virus-specific T cells (50). Alternatively, it may be advantageous for the virus to locally stimulate an immune response as a way to keep itself in check and cause only mild local replication so as to not overwhelm its host. In fact, several recent studies have shown that HSV-specific T cells are found near latently infected trigeminal ganglia (30, 61). These T cells may play a role in controlling reactivation of the virus (31, 60). Since these immune cells likely express HVEM, as well as the other ligands, the interaction with gD at the sites of virus exit from neurons may locally stimulate the immune response to keep the virus in check and prevent it from spreading to the central nervous system.
The competition and downregulation data suggest that HSV gD mimics some aspects of BTLA binding to HVEM to induce receptor downregulation which would prevent further interaction of HVEM with natural ligands. In addition to BTLA, other HVEM ligands interfere with HSV infection by blocking access to HVEM. Thus, gD is at the center of the interplay between the various types of ligands that bind to HVEM. Further studies are necessary to determine how the ability of gD to interfere with binding of HVEM to its ligands translates to immunomodulation during infection in vivo.
Acknowledgments
This investigation was supported by Public Health Service grants AI-073384 to C.K., AI-18289 to G.H.C., and AI-056045 and AI-076231 to R.J.E. from the National Institute of Allergy and Infectious Diseases. K.M.S. was supported by NIH training grant T32-AI07324. C.K. was supported by the Joseph and Josephine Rabinowitz award for Research Excellence at the University of Pennsylvania School of Dental Medicine.
We are grateful to Patricia G. Spear for the gD-null KOSgDβ virus, David C. Johnson for VD60 cells, Gordon Freeman for CHO-CD160 cells and CD160 antibodies, and Marcio Lazaro and Hildegund Ertl for the pLIGHT plasmid. We thank Ali Zekavat and Bruce Shenker for FACS processing at the Flow Cytometry Facility of the University of Pennsylvania School of Dental Medicine. We thank Sarah Connolly for plasmids and the B78-HVEM and B78-HVEM-Y23A cells. We are grateful to the members of the Cohen-Eisenberg and Krummenacher laboratories for suggestions and helpful discussions.
Footnotes
Published ahead of print on 8 September 2010.
REFERENCES
- 1.Anumanthan, A., A. Bensussan, L. Boumsell, A. D. Christ, R. S. Blumberg, S. D. Voss, A. T. Patel, M. J. Robertson, L. M. Nadler, and G. J. Freeman. 1998. Cloning of BY55, a novel Ig superfamily member expressed on NK cells, CTL, and intestinal intraepithelial lymphocytes. J. Immunol. 161:2780-2790. [PubMed] [Google Scholar]
- 2.Banner, D. W., A. D'Arcy, W. Janes, R. Gentz, H. J. Schoenfeld, C. Broger, H. Loetscher, and W. Lesslauer. 1993. Crystal structure of the soluble human 55 kd TNF receptor-human TNF beta complex: implications for TNF receptor activation. Cell 73:431-445. [DOI] [PubMed] [Google Scholar]
- 3.Bodmer, J. L., P. Schneider, and J. Tschopp. 2002. The molecular architecture of the TNF superfamily. Trends Biochem. Sci. 27:19-26. [DOI] [PubMed] [Google Scholar]
- 4.Cai, G., A. Anumanthan, J. A. Brown, E. A. Greenfield, B. Zhu, and G. J. Freeman. 2008. CD160 inhibits activation of human CD4+ T cells through interaction with herpesvirus entry mediator. Nat. Immunol. 9:176-185. [DOI] [PubMed] [Google Scholar]
- 5.Cai, G., and G. J. Freeman. 2009. The CD160, BTLA, LIGHT/HVEM pathway: a bidirectional switch regulating T-cell activation. Immunol. Rev. 229:244-258. [DOI] [PubMed] [Google Scholar]
- 6.Calistri, A., C. Parolin, M. Pizzato, P. Calvi, I. Giaretta, and G. Palu. 1999. Herpes simplex virus chronically infected human T lymphocytes are susceptible to HIV-1 superinfection and support HIV-1 pseudotyping. J. Acquir. Immune Defic. Syndr. 21:90-98. [PubMed] [Google Scholar]
- 7.Carfi, A., S. H. Willis, J. C. Whitbeck, C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169-179. [DOI] [PubMed] [Google Scholar]
- 8.Chan, F. K., H. J. Chun, L. Zheng, R. M. Siegel, K. L. Bui, and M. J. Lenardo. 2000. A domain in TNF receptors that mediates ligand-independent receptor assembly and signaling. Science 288:2351-2354. [DOI] [PubMed] [Google Scholar]
- 9.Cheung, T. C., I. R. Humphreys, K. G. Potter, P. S. Norris, H. M. Shumway, B. R. Tran, G. Patterson, R. Jean-Jacques, M. Yoon, P. G. Spear, K. M. Murphy, N. S. Lurain, C. A. Benedict, and C. F. Ware. 2005. Evolutionarily divergent herpesviruses modulate T cell activation by targeting the herpesvirus entry mediator cosignaling pathway. Proc. Natl. Acad. Sci. U. S. A. 102:13218-13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung, T. C., L. M. Oborne, M. W. Steinberg, M. G. Macauley, S. Fukuyama, H. Sanjo, C. D'Souza, P. S. Norris, K. Pfeffer, K. M. Murphy, M. Kronenberg, P. G. Spear, and C. F. Ware. 2009. T cell intrinsic heterodimeric complexes between HVEM and BTLA determine receptivity to the surrounding microenvironment. J. Immunol. 183:7286-7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung, T. C., M. W. Steinberg, L. M. Oborne, M. G. Macauley, S. Fukuyama, H. Sanjo, C. D'Souza, P. S. Norris, K. Pfeffer, K. M. Murphy, M. Kronenberg, P. G. Spear, and C. F. Ware. 2009. Unconventional ligand activation of herpesvirus entry mediator signals cell survival. Proc. Natl. Acad. Sci. U. S. A. 106:6244-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Compaan, D. M., L. C. Gonzalez, I. Tom, K. M. Loyet, D. Eaton, and S. G. Hymowitz. 2005. Attenuating lymphocyte activity: the crystal structure of the BTLA-HVEM complex. J. Biol. Chem. 280:39553-39561. [DOI] [PubMed] [Google Scholar]
- 13.Connolly, S. A., D. J. Landsburg, A. Carfi, J. C. Whitbeck, Y. Zuo, D. C. Wiley, G. H. Cohen, and R. J. Eisenberg. 2005. Potential nectin-1 binding site on herpes simplex virus glycoprotein D. J. Virol. 79:1282-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connolly, S. A., D. J. Landsburg, A. Carfi, D. C. Wiley, G. H. Cohen, and R. J. Eisenberg. 2003. Structure-based mutagenesis of herpes simplex virus glycoprotein D defines three critical regions at the gD/HveA interface. J. Virol. 77:8127-8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connolly, S. A., D. J. Landsburg, A. Carfi, D. C. Wiley, R. J. Eisenberg, and G. H. Cohen. 2002. Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA (HVEM). J. Virol. 76:10894-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Connolly, S. A., J. C. Whitbeck, A. H. Rux, C. Krummenacher, S. van Drunen Littel-van den Hurk, G. H. Cohen, and R. J. Eisenberg. 2001. Glycoprotein D homologues in herpes simplex virus type 1, pseudorabies virus, and bovine herpes virus type 1 bind directly to human HveC (nectin-1) with different affinities. Virology 280:7-18. [DOI] [PubMed] [Google Scholar]
- 17.Dean, H. J., S. S. Terhune, M. T. Shieh, N. Susmarski, and P. G. Spear. 1994. Single amino acid substitutions in gD of herpes simplex virus 1 confer resistance to gD-mediated interference and cause cell-type-dependent alterations in infectivity. Virology 199:67-80. [DOI] [PubMed] [Google Scholar]
- 18.del Rio, M. L., C. L. Lucas, L. Buhler, G. Rayat, and J. I. Rodriguez-Barbosa. 2010. HVEM/LIGHT/BTLA/CD160 cosignaling pathways as targets for immune regulation. J. Leukoc. Biol. 87:223-235. [DOI] [PubMed] [Google Scholar]
- 19.De Trez, C., and C. F. Ware. 2008. The TNF receptor and Ig superfamily members form an integrated signaling circuit controlling dendritic cell homeostasis. Cytokine Growth Factor Rev. 19:277-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geraghty, R. J., C. Krummenacher, R. J. Eisenberg, G. H. Cohen, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 21.Giustiniani, J., A. Bensussan, and A. Marie-Cardine. 2009. Identification and characterization of a transmembrane isoform of CD160 (CD160-TM), a unique activating receptor selectively expressed upon human NK cell activation. J. Immunol. 182:63-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez, L. C., K. M. Loyet, J. Calemine-Fenaux, V. Chauhan, B. Wranik, W. Ouyang, and D. L. Eaton. 2005. A coreceptor interaction between the CD28 and TNF receptor family members B and T lymphocyte attenuator and herpesvirus entry mediator. Proc. Natl. Acad. Sci. U. S. A. 102:1116-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gray, P. W., B. B. Aggarwal, C. V. Benton, T. S. Bringman, W. J. Henzel, J. A. Jarrett, D. W. Leung, B. Moffat, P. Ng, L. P. Svedersky, et al. 1984. Cloning and expression of cDNA for human lymphotoxin, a lymphokine with tumour necrosis activity. Nature 312:721-724. [DOI] [PubMed] [Google Scholar]
- 24.Handler, C. G., G. H. Cohen, and R. J. Eisenberg. 1996. Crosslinking of glycoprotein oligomers during herpes simplex virus type 1 entry. J. Virol. 70:6076-6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harrop, J. A., P. C. McDonnell, M. Brigham-Burke, S. D. Lyn, J. Minton, K. B. Tan, K. Dede, J. Spampanato, C. Silverman, P. Hensley, R. DiPrinzio, J. G. Emery, K. Deen, C. Eichman, M. Chabot-Fletcher, A. Truneh, and P. R. Young. 1998. Herpesvirus entry mediator ligand (HVEM-L), a novel ligand for HVEM/TR2, stimulates proliferation of T cells and inhibits HT29 cell growth. J. Biol. Chem. 273:27548-27556. [DOI] [PubMed] [Google Scholar]
- 26.Hayward, A., M. Laszlo, M. Turman, A. Vafai, and D. Tedder. 1988. Non-productive infection of human newborn blood mononuclear cells with herpes simplex virus: effect on T cell activation, IL-2 production and proliferation. Clin. Exp. Immunol. 74:196-200. [PMC free article] [PubMed] [Google Scholar]
- 27.Heldwein, E. E., and C. Krummenacher. 2008. Entry of herpesviruses into mammalian cells. Cell Mol. Life Sci. 65:1653-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu, H., I. Solovyev, A. Colombero, R. Elliott, M. Kelley, and W. J. Boyle. 1997. ATAR, a novel tumor necrosis factor receptor family member, signals through TRAF2 and TRAF5. J. Biol. Chem. 272:13471-13474. [DOI] [PubMed] [Google Scholar]
- 29.Hung, S. L., Y. Y. Cheng, Y. H. Wang, K. W. Chang, and Y. T. Chen. 2002. Expression and roles of herpesvirus entry mediators A and C in cells of oral origin. Oral Microbiol. Immunol. 17:215-223. [DOI] [PubMed] [Google Scholar]
- 30.Khanna, K. M., R. H. Bonneau, P. R. Kinchington, and R. L. Hendricks. 2003. Herpes simplex virus-specific memory CD8+ T cells are selectively activated and retained in latently infected sensory ganglia. Immunity 18:593-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knickelbein, J. E., K. M. Khanna, M. B. Yee, C. J. Baty, P. R. Kinchington, and R. L. Hendricks. 2008. Noncytotoxic lytic granule-mediated CD8+ T cell inhibition of HSV-1 reactivation from neuronal latency. Science 322:268-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krummenacher, C., F. Baribaud, M. Ponce De Leon, I. Baribaud, J. C. Whitbeck, R. Xu, G. H. Cohen, and R. J. Eisenberg. 2004. Comparative usage of herpesvirus entry mediator A and nectin-1 by laboratory strains and clinical isolates of herpes simplex virus. Virology 322:286-299. [DOI] [PubMed] [Google Scholar]
- 33.Krummenacher, C., V. M. Supekar, J. C. Whitbeck, E. Lazear, S. A. Connolly, R. J. Eisenberg, G. H. Cohen, D. C. Wiley, and A. Carfi. 2005. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J. 24:4144-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwon, B. S., K. B. Tan, J. Ni, K. O. Oh, Z. H. Lee, K. K. Kim, Y. J. Kim, S. Wang, R. Gentz, G.-L. Yu, J. Harrop, S. D. Lyn, C. Silverman, T. G. Porter, A. Truneh, and P. R. Young. 1997. A newly identified member of the tumor necrosis factor receptor superfamily with a wide tissue distribution and involvement in lymphocyte activation. J. Biol. Chem. 272:14272-14276. [DOI] [PubMed] [Google Scholar]
- 35.Ligas, M. W., and D. C. Johnson. 1988. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by β-galactosidase sequences binds to but is unable to penetrate into cells. J. Virol. 62:1486-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Locksley, R. M., N. Killeen, and M. J. Lenardo. 2001. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell 104:487-501. [DOI] [PubMed] [Google Scholar]
- 37.Marsters, S. A., T. M. Ayres, M. Skubatch, C. L. Gray, M. Rothe, and A. Ashkenazi. 1997. Herpes virus entry mediator, a member of the tumor necrosis factor receptor (TNFR) family, interacts with members of the TNFR-associated factor family and activates the transcription factors NF-κB and AP-1. J. Biol. Chem. 272:14029-14032. [DOI] [PubMed] [Google Scholar]
- 38.Mauri, D. N., R. Ebner, K. D. Kochel, R. I. Montgomery, T. C. Cheung, G.-L. Yu, M. Murphy, R. J. Eisenberg, G. H. Cohen, P. G. Spear, and C. F. Ware. 1998. LIGHT, a new member of the TNF superfamily, and lymphotoxin (LT) α are ligands for herpesvirus entry mediator (HVEM). Immunity 8:21-30. [DOI] [PubMed] [Google Scholar]
- 39.Medici, M. A., M. T. Sciortino, D. Perri, C. Amici, E. Avitabile, M. Ciotti, E. Balestrieri, E. De Smaele, G. Franzoso, and A. Mastino. 2003. Protection by herpes simplex virus glycoprotein D against Fas-mediated apoptosis: role of nuclear factor kappaB. J. Biol. Chem. 278:36059-36067. [DOI] [PubMed] [Google Scholar]
- 40.Miller, C. G., C. Krummenacher, R. J. Eisenberg, G. H. Cohen, and N. W. Fraser. 2001. Development of a syngenic murine B16 cell line-derived melanoma susceptible to destruction by neuroattenuated HSV-1. Mol. Ther. 3:160-168. [DOI] [PubMed] [Google Scholar]
- 41.Milne, R. S., A. V. Nicola, J. C. Whitbeck, R. J. Eisenberg, and G. H. Cohen. 2005. Glycoprotein D receptor-dependent, low-pH-independent endocytic entry of herpes simplex virus type 1. J. Virol. 79:6655-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 43.Morel, Y., J. M. Schiano de Colella, J. Harrop, K. C. Deen, S. D. Holmes, T. A. Wattam, S. S. Khandekar, A. Truneh, R. W. Sweet, J. A. Gastaut, D. Olive, and R. T. Costello. 2000. Reciprocal expression of the TNF family receptor herpes virus entry mediator and its ligand LIGHT on activated T cells: LIGHT down-regulates its own receptor. J. Immunol. 165:4397-4404. [DOI] [PubMed] [Google Scholar]
- 44.Murphy, K. M., C. A. Nelson, and J. R. Sedy. 2006. Balancing co-stimulation and inhibition with BTLA and HVEM. Nat. Rev. Immunol. 6:671-681. [DOI] [PubMed] [Google Scholar]
- 45.Murphy, T. L., and K. M. Murphy. 2010. Slow down and survive: enigmatic immunoregulation by BTLA and HVEM. Annu. Rev. Immunol. 28:389-411. [DOI] [PubMed] [Google Scholar]
- 46.Naismith, J. H., T. Q. Devine, B. J. Brandhuber, and S. R. Sprang. 1995. Crystallographic evidence for dimerization of unliganded tumor necrosis factor receptor. J. Biol. Chem. 270:13303-13307. [DOI] [PubMed] [Google Scholar]
- 47.Nicola, A. V., A. M. McEvoy, and S. E. Straus. 2003. Roles for endocytosis and low pH in herpes simplex virus entry into HeLa and Chinese hamster ovary cells. J. Virol. 77:5324-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicola, A. V., S. H. Willis, N. N. Naidoo, R. J. Eisenberg, and G. H. Cohen. 1996. Structure-function analysis of soluble forms of herpes simplex virus glycoprotein D. J. Virol. 70:3815-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otsuki, N., Y. Kamimura, M. Hashiguchi, and M. Azuma. 2006. Expression and function of the B and T lymphocyte attenuator (BTLA/CD272) on human T cells. Biochem. Biophys. Res. Commun. 344:1121-1127. [DOI] [PubMed] [Google Scholar]
- 50.Raftery, M. J., C. K. Behrens, A. Muller, P. H. Krammer, H. Walczak, and G. Schonrich. 1999. Herpes simplex virus type 1 infection of activated cytotoxic T cells: induction of fratricide as a mechanism of viral immune evasion. J. Exp. Med. 190:1103-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rinaldo, C. R., Jr., B. S. Richter, P. H. Black, R. Callery, L. Chess, and M. S. Hirsch. 1978. Replication of herpes simplex virus and cytomegalovirus in human leukocytes. J. Immunol. 120:130-136. [PubMed] [Google Scholar]
- 52.Rooney, I. A., K. D. Butrivich, A. A. Glass, S. Borboroglu, C. A. Benedict, J. C. Whitbeck, G. H. Cohen, R. J. Eisenberg, and C. F. Ware. 2000. The lymphotoxin-β receptor is necessary and sufficient for LIGHT-mediated apoptosis of tumor cells. J. Biol. Chem. 275:14307-14315. [DOI] [PubMed] [Google Scholar]
- 53.Rux, A. H., S. H. Willis, A. V. Nicola, W. Hou, C. Peng, H. Lou, G. H. Cohen, and R. J. Eisenberg. 1998. Functional region IV of glycoprotein D from herpes simplex virus modulates glycoprotein binding to the herpes virus entry mediator. J. Virol. 72:7091-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarmiento, M., and E. S. Kleinerman. 1990. Innate resistance to herpes simplex virus infection. Human lymphocyte and monocyte inhibition of viral replication. J. Immunol. 144:1942-1953. [PubMed] [Google Scholar]
- 55.Sarrias, M. R., J. C. Whitbeck, I. Rooney, C. F. Ware, R. J. Eisenberg, G. H. Cohen, and J. D. Lambris. 2000. The three HveA receptor ligands, gD, LT-alpha and LIGHT bind to distinct sites on HveA. Mol. Immunol. 37:665-673. [DOI] [PubMed] [Google Scholar]
- 56.Schneider, K., K. G. Potter, and C. F. Ware. 2004. Lymphotoxin and LIGHT signaling pathways and target genes. Immunol. Rev. 202:49-66. [DOI] [PubMed] [Google Scholar]
- 57.Sciortino, M. T., M. A. Medici, F. Marino-Merlo, D. Zaccaria, M. Giuffre-Cuculletto, A. Venuti, S. Grelli, and A. Mastino. 2008. Involvement of HVEM receptor in activation of nuclear factor kappaB by herpes simplex virus 1 glycoprotein D. Cell. Microbiol. 10:2297-2311. [DOI] [PubMed] [Google Scholar]
- 58.Sedy, J. R., M. Gavrieli, K. G. Potter, M. A. Hurchla, R. C. Lindsley, K. Hildner, S. Scheu, K. Pfeffer, C. F. Ware, T. L. Murphy, and K. M. Murphy. 2005. B and T lymphocyte attenuator regulates T cell activation through interaction with herpesvirus entry mediator. Nat. Immunol. 6:90-98. [DOI] [PubMed] [Google Scholar]
- 59.Shaikh, R. B., S. Santee, S. W. Granger, K. Butrovich, T. Cheung, M. Kronenberg, H. Cheroutre, and C. F. Ware. 2001. Constitutive expression of LIGHT on T cells leads to lymphocyte activation, inflammation, and tissue destruction. J. Immunol. 167:6330-6337. [DOI] [PubMed] [Google Scholar]
- 60.Sheridan, B. S., T. L. Cherpes, J. Urban, P. Kalinski, and R. L. Hendricks. 2009. Reevaluating the CD8 T-cell response to herpes simplex virus type 1: involvement of CD8 T cells reactive to subdominant epitopes. J. Virol. 83:2237-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sheridan, B. S., K. M. Khanna, G. M. Frank, and R. L. Hendricks. 2006. Latent virus influences the generation and maintenance of CD8+ T cell memory. J. Immunol. 177:8356-8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shukla, D., J. Liu, P. Blaiklock, N. W. Shworak, X. Bai, J. D. Esko, G. H. Cohen, R. J. Eisenberg, R. D. Rosenberg, and P. G. Spear. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99:13-22. [DOI] [PubMed] [Google Scholar]
- 63.Sisk, W. P., J. D. Bradley, R. J. Leipold, A. M. Stoltzfus, M. Ponce de Leon, M. Hilf, C. Peng, G. H. Cohen, and R. J. Eisenberg. 1994. High-level expression and purification of secreted forms of herpes simplex virus type 1 glycoprotein gD synthesized by baculovirus-infected insect cells. J. Virol. 68:766-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Steinberg, M. W., J. W. Shui, C. F. Ware, and M. Kronenberg. 2009. Regulating the mucosal immune system: the contrasting roles of LIGHT, HVEM, and their various partners. Semin. Immunopathol. 31:207-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stiles, K. M., and C. Krummenacher. 2010. Glycoprotein D actively induces rapid internalization of two nectin-1 isoforms during herpes simplex virus entry. Virology 399:109-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stiles, K. M., R. S. Milne, G. H. Cohen, R. J. Eisenberg, and C. Krummenacher. 2008. The herpes simplex virus receptor nectin-1 is down-regulated after trans-interaction with glycoprotein D. Virology 373:98-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tamada, K., K. Shimozaki, A. I. Chapoval, Y. Zhai, J. Su, S. F. Chen, S. L. Hsieh, S. Nagata, J. Ni, and L. Chen. 2000. LIGHT, a TNF-like molecule, costimulates T cell proliferation and is required for dendritic cell-mediated allogeneic T cell response. J. Immunol. 164:4105-4110. [DOI] [PubMed] [Google Scholar]
- 68.Tan, K. B., J. Harrop, M. Reddy, P. Young, J. Terrett, J. Emery, G. Moore, and A. Truneh. 1997. Characterization of a novel TNF-like ligand and recently described TNF ligand and TNF receptor superfamily genes and their constitutive and inducible expression in hematopoietic and non-hematopoietic cells. Gene 204:35-46. [DOI] [PubMed] [Google Scholar]
- 69.Terry-Allison, T., R. I. Montgomery, J. C. Whitbeck, R. Xu, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. HveA (herpesvirus entry mediator A), a coreceptor for herpes simplex virus entry, also participates in virus-induced cell fusion. J. Virol. 72:5802-5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tessier, D. C., D. Y. Thomas, H. E. Khouri, F. Laliberte, and T. Vernet. 1991. Enhanced secretion from insect cells of a foreign protein fused to the honeybee melittin signal peptide. Gene 98:177-183. [DOI] [PubMed] [Google Scholar]
- 71.Tiwari, V., C. D. O'Donnell, M. J. Oh, T. Valyi-Nagy, and D. Shukla. 2005. A role for 3-O-sulfotransferase isoform-4 in assisting HSV-1 entry and spread. Biochem. Biophys. Res. Commun. 338:930-937. [DOI] [PubMed] [Google Scholar]
- 72.Tiwari, V., S. Y. Shukla, B. Y. Yue, and D. Shukla. 2007. Herpes simplex virus type 2 entry into cultured human corneal fibroblasts is mediated by herpesvirus entry mediator. J. Gen. Virol. 88:2106-2110. [DOI] [PubMed] [Google Scholar]
- 73.Wang, Y., S. K. Subudhi, R. A. Anders, J. Lo, Y. Sun, S. Blink, J. Wang, X. Liu, K. Mink, D. Degrandi, K. Pfeffer, and Y. X. Fu. 2005. The role of herpesvirus entry mediator as a negative regulator of T cell-mediated responses. J. Clin. Invest. 115:711-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ware, C. F. 2008. Targeting lymphocyte activation through the lymphotoxin and LIGHT pathways. Immunol. Rev. 223:186-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Watanabe, N., M. Gavrieli, J. R. Sedy, J. Yang, F. Fallarino, S. K. Loftin, M. A. Hurchla, N. Zimmerman, J. Sim, X. Zang, T. L. Murphy, J. H. Russell, J. P. Allison, and K. M. Murphy. 2003. BTLA is a lymphocyte inhibitory receptor with similarities to CTLA-4 and PD-1. Nat. Immunol. 4:670-679. [DOI] [PubMed] [Google Scholar]
- 76.Whitbeck, J. C., S. A. Connolly, S. H. Willis, W. Hou, C. Krummenacher, M. Ponce de Leon, H. Lou, I. Baribaud, R. J. Eisenberg, and G. H. Cohen. 2001. Localization of the gD-binding region of the human herpes simplex virus receptor, HveA. J. Virol. 75:171-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Whitbeck, J. C., C. Peng, H. Lou, R. Xu, S. H. Willis, M. Ponce de Leon, T. Peng, A. V. Nicola, R. I. Montgomery, M. S. Warner, A. M. Soulika, L. A. Spruce, W. T. Moore, J. D. Lambris, P. G. Spear, G. H. Cohen, and R. J. Eisenberg. 1997. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the TNFR superfamily and a mediator of HSV entry. J. Virol. 71:6083-6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Willis, S. H., C. Peng, M. Ponce de Leon, A. V. Nicola, A. H. Rux, G. H. Cohen, and R. J. Eisenberg. 1998. Expression and purification of secreted forms of HSV glycoproteins from baculovirus-infected insect cells. Methods Mol. Med. 10:131-156. [DOI] [PubMed] [Google Scholar]
- 79.Willis, S. H., A. H. Rux, C. Peng, J. C. Whitbeck, A. V. Nicola, H. Lou, W. Hou, L. Salvador, G. H. Cohen, and R. J. Eisenberg. 1998. Examination of the kinetics of herpes simplex virus glycoprotein D binding to the herpesvirus entry mediator, using surface plasmon resonance. J. Virol. 72:5937-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wittels, M., and P. G. Spear. 1991. Penetration of cells by herpes simplex virus does not require a low pH-dependent endocytic pathway. Virus Res. 18:271-290. [DOI] [PubMed] [Google Scholar]