Abstract
Deciphering immune events during early stages of human immunodeficiency virus type 1 (HIV-1) infection is critical for understanding the course of disease. We characterized the hierarchy of HIV-1-specific T-cell gamma interferon (IFN-γ) enzyme-linked immunospot (ELISPOT) assay responses during acute subtype C infection in 53 individuals and associated temporal patterns of responses with disease progression in the first 12 months. There was a diverse pattern of T-cell recognition across the proteome, with the recognition of Nef being immunodominant as early as 3 weeks postinfection. Over the first 6 months, we found that there was a 23% chance of an increased response to Nef for every week postinfection (P = 0.0024), followed by a nonsignificant increase to Pol (4.6%) and Gag (3.2%). Responses to Env and regulatory proteins appeared to remain stable. Three temporal patterns of HIV-specific T-cell responses could be distinguished: persistent, lost, or new. The proportion of persistent T-cell responses was significantly lower (P = 0.0037) in individuals defined as rapid progressors than in those progressing slowly and who controlled viremia. Almost 90% of lost T-cell responses were coincidental with autologous viral epitope escape. Regression analysis between the time to fixed viral escape and lost T-cell responses (r = 0.61; P = 0.019) showed a mean delay of 14 weeks after viral escape. Collectively, T-cell epitope recognition is not a static event, and temporal patterns of IFN-γ-based responses exist. This is due partly to viral sequence variation but also to the recognition of invariant viral epitopes that leads to waves of persistent T-cell immunity, which appears to associate with slower disease progression in the first year of infection.
For more than a decade, there has been a wealth of evidence to show that human immunodeficiency virus (HIV)-specific cytotoxic T-cell (CTL) responses play a role in the control of HIV-1 and simian immunodeficiency virus (SIV) infection. In humans, the first appearance of CTL in primary HIV-1 infection coincides with the decline of peak viremia (7, 27), while depletion of CD8+ T cells in SIV infection resulted in elevated viremia (45). Additionally, polymorphisms in HLA class I-restricted CTL responses are associated with differential HIV-1 disease outcomes (25), and the emergence of viral escape within CTL epitopes during acute and chronic SIV or HIV-1 infection demonstrates the effectiveness of CD8+ T cells to exert viral selection pressure (21). Dissecting the specificity of HIV-1-specific CD8+ T-cell responses that associate with the control of viral replication during acute/early infection is thought to be critical for the design of vaccines and potential immunotherapeutic strategies aimed at stimulating these responses.
Preferential targeting of class I-restricted CTL epitopes in Gag during early and chronic HIV-1 infection has been associated with lower viral loads (15, 25, 34, 48, 55), whereas Env- and Nef-specific CD8+ T-cell responses have been associated with higher viremia (15, 34, 55). Increasing evidence suggests that patterns of immunodominant HIV-specific CD8+ T-cell responses restricted by specific HLA alleles are major determinants of the viral set point (47). In addition, Goonetilleke et al. (17) have provided insight into the rapidity of early escape and the contribution of the first HIV-specific CD8+ T-cell responses to the transmitted/founder virus in control of acute viremia. The restriction of CTL epitopes by HLA-B*5801, for example, has also been associated with better viral control (16, 24). However, the temporal nature of epitope-specific responses that associate with viral control has not been explored. Recently, we found no association between the magnitude and breadth of gamma interferon (IFN-γ) enzyme-linked immunospot (ELISPOT) assay responses at a static 3-month time point with the viral set point at 12 months (22). The unpredictability of early T-cell responses with later viral control could be a result of HIV variability resulting in epitope escape from humoral and T-cell pressure (1, 8). For example, the impact of CTL pressure on shaping viral diversity at a human population level has been observed through HLA imprinting (6, 9, 44), and several studies have shown that certain selected escape mutations can compromise viral fitness (10, 29, 33, 39). Other studies have also demonstrated that the selection of escape variants in chronic HIV-1 and SIV infection can result in the loss of immune control and disease progression (3, 20). Assessing the nature of T-cell responses longitudinally and relating the patterns of contemporaneous viral recognition with viral diversity may represent alternative insights into factors associated with set point and disease progression.
As the global AIDS epidemic continues to expand in sub-Saharan Africa, and South Africa in particular, the need to implement a preventive vaccine through the public health sector remains paramount. To date, several prototype antibody and T-cell-based candidate vaccine trials have been completed worldwide (37), and the recent failure of a phase IIb Ad5-Gag-Pol-Nef HIV-1 vaccine trial has emphasized the challenge of producing an effective T-cell-based vaccine against HIV. Data from the recent ALVAC and AIDSVAX (RV144) trials in Thailand have provided modest efficacy of a vaccine regimen in reducing HIV infection (42), and while the immune mechanisms for this are as yet unclear, these findings have created a platform for identifying immune responses that correlate with protection.
The identification of the earliest targets of T cells during acute HIV-1 infection would be helpful in understanding the evolution of immunity when a host first encounters the virus and also would provide insight into the host-pathogen interplay when there is a rapidly changing target. We describe some of the earliest T-cell responses that occur during acute subtype C HIV-1 infection, how these change over time and associate with early disease progression, as well as the kinetics of these changes in relation to autologous viral escape.
MATERIALS AND METHODS
Study subjects.
The cohort consisted of 53 HIV-1 clade C-infected subjects enrolled as part of the Centre for AIDS Programme Research in South Africa (CAPRISA) 002 acute infection study in Durban, South Africa, as previously reported (52). The date of HIV infection was either estimated by a prospective RNA-positive/antibody-negative measurement or set as the midpoint between the last antibody-negative and first antibody-positive enzyme-linked immunosorbent assay (ELISA) test. Herein, we report on the data from events within the first 12 months of infection. Viral load was measured with the COBAS Amplicor HIV-1 monitor test, version 1.5 (Roche Diagnostics, Branchburg, NJ), and the CD4+ count was measured by flow cytometry. The studied subjects were estimated to have been infected for a median duration of 6 weeks, ranging from 2 to 15 weeks postinfection at enrollment (22). The median plasma viral load at enrollment was 75,600 HIV RNA copies/ml (range, 547 to 5,510,000), and the median CD4+ count was 494 cells/mm3 (range, 197 to 989). The University of KwaZulu-Natal, University of Witwatersrand, and University of Cape Town institutional review boards approved this study, and all the subjects provided written informed consent for participation in this study.
HLA typing.
High-resolution HLA class I genotyping was performed by sequencing of exons 2, 3, and 4 using Atria Allele SEQR kits (Abbott Diagnostics) and Assign SBT 3.5 software (Conexio Genomics) as described previously (10).
Synthetic subtype C HIV-1 peptides.
Peptide sets spanning the entire HIV-1 clade C proteome corresponding to gene products from the HIV-1 consensus C (Gag, Vif, Vpr, and Vpu), isolate Du151 (Pol, Nef, Tat, and Rev), and isolate Du179 (gp160 Env) were synthesized and dissolved in dimethyl sulfoxide (DMSO) at a concentration of 10 mg/ml as previously described (34). The peptide sets based on Du151 were sequences matching clade C vaccine candidates in clinical trials (54).
Cell preparations.
Peripheral blood mononuclear cells (PBMC) were isolated by standard Ficoll-Hypaque density gradient centrifugation (Amersham Pharmacia, Uppsala, Sweden) and used immediately in an ELISPOT assay, and the rest were cryopreserved in 90% heat-inactivated fetal bovine serum (FBS; Invitrogen, Paisley, United Kingdom) plus 10% DMSO and stored in liquid nitrogen until needed for further analysis.
ELISPOT assay.
HIV-1-specific T-cell responses were quantified by IFN-γ ELISPOT assay using a set of 432 overlapping peptides spanning the entire HIV-1 C proteome arranged in a pool matrix format as previously described (22, 34). Briefly, freshly isolated PBMC were stimulated with HIV-1 peptide pools (2 μg/ml), cytomegalovirus, Epstein-Barr virus, and influenza virus (CEF) pools (1 μg/ml), and phytohemagglutinin (4 μg/ml), incubated overnight at 37°C with 5% CO2, and developed as previously described. Confirmation of positive responses at the single-peptide level within peptide pools was undertaken in a second ELISPOT assay and monitored longitudinally for selected participants. The CTL epitopes within peptides showing responses were predicted from the published epitopes on the HIV immunology database based on the matched HLA from the participants.
Sequencing of autologous virus.
Viral sequencing was carried out as previously described (10). RNA isolated from plasma samples using the MagNA Pure compact nucleic extractor (Roche) was reverse transcribed using the Invitrogen ThermoScript reverse transcription kit (Invitrogen) and the primers Gag D reverse (5′-AAT TCC TCC TAT CAT TTT TGG-3′; HXB position 2382 to 2402) and Nef O reverse (5′-AGG CAA GCT TTA TTG AGG-3′; HXB position 9608 to 9625) for nef. Limiting-dilution nested PCR was carried out by serial endpoint dilution of the cDNA (43). The first-round PCR primers were Gag D forward (5′-TCT CTA GCA GTG GCG CCC G-3′; HXB position 626 to 644) and Gag D reverse (5′-AAT TCC TCC TAT CAT TTT TGG-3′; HXB position 2382 to 2402). The second-round PCR primers were Gag A forward (5′-CTC TCG ACG CAG GAC TCG GCT T-3′; HXB position 683 to 704) and Gag C reverse (5′-TCT TCT AAT ACT GTA TCA TCT GC-3′; HXB position 2334 to 2356). For nef, the first-round PCR primers were SQ15FC (5′-GAG AGC GGT GGA ACT TCT-3′; HXB position 8561 to 8578) and Nef O reverse. The second-round PCR primers were Nef forward (5′-CCT AGA AGA ATA AGA CAG GGC TT-3′; HXB position 8754 to 8776) and Nef reverse (5′-CCT GGA ACG CCC CAG TGG-3′; HXB position 9443 to 9461). PCR products were either directly sequenced or cloned using the pGEM-T Easy vector system (Promega). Sequencing was carried out using an ABI Prism dye terminator cycle sequencing kit (Applied Biosysytems) and the primers Gag A forward, Gag A reverse (5′-ACA TGG GTA TCA CTT CTG GGC T-3′; HXB position 1282 to 1303), Gag B forward (5′-CCA TAT CAC CTA GAA CTT TGA AT-3′; HXB position 1226 to 1246), Gag B reverse (5′-CTC CCT GAC ATG CTG TCA TCA T-3′; HXB position 1825 to 1846), Gag C forward (5′-CCT TGT TGG TCC AAA ATG CGA-3′; HXB position 1748 to 1768), and Gag C reverse for direct sequencing. Sequences were assembled using ChromasPro and aligned using ClustalW (with default settings) (49).
Statistical analysis.
Statistical analysis and graphical presentation were performed using InStat, GraphPad Prism version 3.0 software, and SAS version 9.1.3 (SAS Institute, Inc., Cary, NC). Data are expressed as medians ± ranges and analyzed by the use of nonparametric statistics. Statistical analysis of significance was based on two-tailed t tests using either Mann-Whitney or Kruskal-Wallis analysis of variance (Dunn's test) for multiple comparisons. The frequency of recognition was modeled using generalized estimating equations (GEE), using a binomial distribution and a logit link, effectively modeling a logistic regression model and adjusting for repeated measures. The magnitudes of responses were not normally distributed, and due to the overdispersion in the data, a Poisson regression model was not appropriate; hence, using the GEE approach, a negative binomial model was used to model the rate of change of the responses over weeks postinfection (13, 36). A P value of <0.05 was considered statistically significant.
RESULTS
Evolving frequency of HIV-1-specific T-cell recognition and associations with viral load.
We first wished to identify the evolving frequency of recognition to regions of HIV after acute infection, using consensus and consensus-like overlapping peptides. Figure 1 A shows that Nef, Pol, and Gag were the most frequently targeted proteins from as early as 3 to 8 weeks postinfection, rising from more than 50% recognition at 12 weeks to 100% of individuals reactive to one or more of the Nef peptide pools at 6 months postinfection. Between 3 to 8 weeks postinfection, when the earliest measurements were made, the immunodominance of Nef appeared to be established, with more than 75% of individuals recognizing the Nef region, followed by Pol (50%), Gag and Env (47%), Vif (38%), Vpr (32%), and Rev, Vpu, and Tat (12%). Using a GEE (see Materials and Methods) to fit the frequency of recognition, we found that over the first 6 months there was a 23% chance of an increased response to Nef for every week postinfection (P = 0.0024), followed by a nonsignificant increased response to both Pol (4.6%) and Gag (3.2%). Responses to Env and the regulatory proteins remained stable over the first 6 months postinfection. Collectively, these data show a distinct hierarchy of the rate of responses during acute infection, with responses to Nef evolving most rapidly. When using the GEE model to determine whether a response to a specific region was associated with increased viral loads, there was a significant (P = 0.0042) odds ratio of 1.86 for responses to Env for every log10 increase in viral load. This translates into an 86% increase in the chance of having an Env response for every log10 increase in viral load. Conversely, there was a 35% decreased chance of having a Gag response for each log10 viral load increase (P = 0.0796) (Fig. 1B). These results mean that although responses to Env appear to remain stable over the first 6 months of infection, the recognition of this region relates to a higher rate of viral load increase, and the recognition of Gag relates to a lower rate of viral load increase. Of note, responses to Nef and Pol, the earliest and most immunodominant regions recognized during acute infection, were not associated with any increased or decreased rates of viral load.
FIG. 1.
Frequency of T-cell recognition across the expressed subtype C HIV-1 proteome. (A) Dotted lines represent response frequencies to Nef, Pol, Gag, Env, Vif, Vpr, Rev, Vpu, and Tat at 6, 12, and 24 weeks postinfection (ranges of weeks postinfection for each time point are indicated in parentheses). The two horizontal dotted lines represent cutoffs for immunodominant (>50%) and subdominant (<25%) responses. (B) Association of viral load with the magnitudes of HIV-1-specific T-cell responses across the entire expressed genome using a generalized estimating equation model. This model was fitted to the data using a binomial distribution with a logit link, adjusted for repeated measurements (with an unstructured covariance structure). The horizontal solid line represents an odds ratio of 1.
Evolving magnitudes of HIV-1-specific T-cell responses.
In terms of magnitude, the immunodominance of Nef is further highlighted in Fig. 2 A, where a longitudinal analysis shows Nef to have a significantly higher magnitude of response over time than any other protein. Responses directed against Nef made up more than 30% of the total cumulative T-cell response relative to other proteins and were proportionally higher in magnitude than those directed against other regions at any time point analyzed (Fig. 2B). We have previously reported on specific Nef epitopes recognized at 12 weeks postinfection (22), and when responses were normalized, taking into account the amino acid length of the protein for each responder, Nef was the most immunogenic protein at all time points, corroborating findings of Streeck et al. (48) and Masemola et al. (34). The highest density of targeted peptides was embedded within the conserved central region of Nef between amino acids 52 to 171 (22). As the magnitudes of responses to each peptide region were not normally distributed, a negative binomial model was used (see Materials and Methods) to model the rate of change in the magnitude of each response over weeks postinfection, while also adjusting for repeated measures. Figure 2C shows that the magnitude of Gag recognition was the only response which had a significant positive slope (P = 0.0096), where a unit increase in weeks postinfection led to a log 0.0364 increase in the slope of responses to Gag in the first 6 months postinfection. There was a nonsignificant increase in the response to Nef (P = 0.0943), with a log 0.0218 increase in the slope of responses to Nef for every week postinfection. The magnitudes of responses to other regions showed declines or increased responses over time, but none showed a significant rate of change over time. Taken together, although Nef responses were highly immunodominant and increased over 6 months, neither the frequency nor the magnitude had any impact on the rate of viral load changes. However, the later emergence of responses to Gag over time, both in frequency and magnitude, was associated with lower rates of viral load increases in this cohort. These data provide a unique insight into the dynamic nature of T-cell responses during acute infection and suggest that neither a static measurement nor the magnitude of response alone is enough to determine the course of viremia (22).
FIG. 2.
Magnitude, proportions, and hierarchy of IFN-γ ELISPOT responses. (A) Cumulative magnitudes of IFN-γ ELISPOT responses across the HIV-1 proteome, expressed as spot-forming units (SFU)/106 PBMC, at 5 (range, 3 to 8), 13 (range, 12 to 15), and 22 (range, 20 to 26) weeks postinfection. Each symbol represents a response per participant, and the solid vertical lines represent the upper and lower ranges, with the median responses as short horizontal bars. (B) Pie charts depicting the relative contributions of Nef, Gag, Pol, Env, and VVTRV (Vif, Vpr, Tat, Rev, Vpu) to the total magnitude of HIV-1-specific T-cell responses at 5 (range, 3 to 8), 13 (range, 12 to 15), and 22 (range, 20 to 26) weeks postinfection. (C) Rate of change in the magnitudes of HIV-1-specific T-cell responses across the entire expressed genome over weeks postinfection.
Recognition patterns of HIV-1-specific T-cell responses over time.
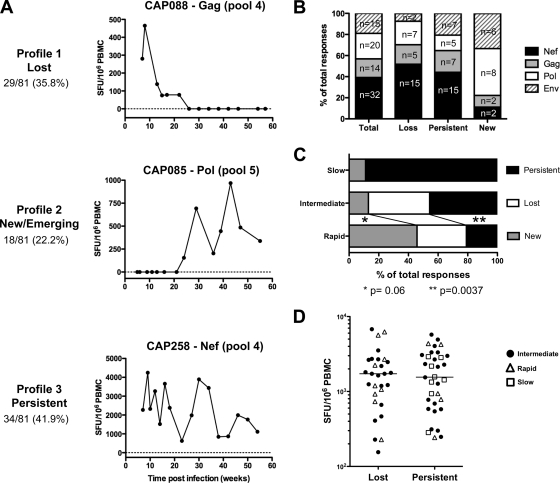
We investigated a subset of 34 individuals at more intensive time points immediately after infection and identified three distinct response profiles: (i) lost responses were defined as a drop of 80% or more in the magnitude of the peak response over two consecutive time points, (ii) new or emerging responses were defined as the appearance of detectable IFN-γ ELISPOT responses to a peptide pool which were not present at two previous consecutive time points, and (iii) persistent responses were defined as those responses that persisted over time and were characterized by fluctuating recognition profiles. Figure 3 A shows each profile, with 36% of individuals having profile 1 responses, characterized by a rapid loss of HIV-1-specific IFN-γ T-cell responses soon after primary infection; 22% having profile 2 responses, characterized by responses that were new or emerging over time; and 42% having profile 3 responses, characterized by responses that persisted over time. Although the majority of HIV-infected individuals showed persistent responses, virtually all these responses showed waxing and waning reactivities over time. To discount for the possibility of fluctuating responses being due to assay variability, we determined the coefficient of variation against CEF peptides in quality control samples to be 24% and within two standard deviations of the mean IFN-γ ELISPOT response (data not shown). The average percentage of fluctuations over time across all persistent responses was well above the assay coefficient of variation and was 43%. Of note, oscillations over time in the magnitudes of non-HIV-specific responses (i.e., CEF, PHA) were also observed for both HIV-infected and noninfected individuals. Moreover, in most infected participants, the fluctuation pattern of PHA and CEF responses mirrored those observed for HIV-specific responses (data not shown). Taken together, these data show that variation of HIV-specific responses over time is likely to be a biological phenomenon and suggest that fluctuations are a natural physiological phenomenon, reflecting the flexible, nonstatic nature of T-cell responses that lead to waves of immunodominance (51).
FIG. 3.
Longitudinal characterization of HIV-1-specific T-cell responses over the first year of infection. (A) Tracking of HIV-1 specific T-cell responses over time showing three distinct profiles of T-cell recognition; (B) proportion of IFN-γ ELISPOT responses that are lost, persistent, or new to Nef, Gag, Pol, and Env when followed longitudinally over 1 year; (C) proportion of lost, persistent, and new IFN-γ ELISPOT responses in rapid progressors (n = 10), intermediate progressors (n = 33), and slow progressors (n = 8); (D) peak magnitudes of lost or persistent IFN-γ ELISPOT responses.
The fluidity of recognition profiles that constitute an apparent stable response at the overall protein level is shown in Fig. 3B, which illustrates the simultaneous distribution of lost, persistent, and new responses toward Gag, Pol, Env, and Nef. There were no significant differences between these patterns. In total, we observed the emergence of 18 new responses with the concomitant loss of 29 different responses. Although there were no statistically significant differences between protein regions, there was a trend toward more responses to Nef peptides either persisting or being lost and more new responses to Pol and Env peptides. These data also underscore that new or lost T-cell recognition is not confined to a specific protein region and that HIV-specific T-cell responses are dynamic and fluid in nature.
The dynamic nature of T-cell recognition patterns and early disease progression.
We next aimed to identify whether lost, new, or persistent responses had any impact on early disease progression in the first 12 months postinfection. Study participants were grouped according to defined disease progression categories (22), and the numbers of lost, new, or persistent responses were compared between the three groups (Fig. 3C). Rapid progressors were defined as study participants who had CD4 counts below 350 cells/ml and viral loads above 100,000 copies/ml on two consecutive measurements between 10 to 15 months postinfection, slow progressors were defined as participants having CD4 counts above 350 cells/ml and viral loads below 2,000 RNA copies/ml on two consecutive measurements between 10 to 15 months postinfection, and intermediate progressors were defined as those who fitted neither the rapid nor the slow category. Individuals with rapid progression (n = 9) possessed significantly fewer (P = 0.0037) persistent responses (profile 3) and a greater number (P = 0.06) of new responses (profile 2) than those with intermediate (or typical) progression (n = 20). The frequencies of lost responses (profile 1) were equal between the intermediate and rapid progressors. No loss of IFN-γ ELISPOT responses was detected for individuals showing early slow disease progression (n = 5), where these subjects overwhelmingly possessed persistent responses (89%). These data suggest that persistent responses over time, recognizing invariant epitopes, may provide advantageous immune responses leading to sustained viral suppression and/or maintenance of CD4 counts. The loss of response, which was invariably associated with epitope escape, appeared to provide no advantage to the host, while new responses which emerged over time are probably driven by antigen load and therefore appear deleterious during early HIV infection.
To understand whether a lost response may be due to the loss of a weak subdominant response, we compared the peak magnitudes of responses between lost and persistent responses (Fig. 3D) and found that there was no preferential disappearance of subdominant responses. Furthermore, the magnitudes of peak responses for each recognition profile were unrelated to disease categories of rapid, intermediate, and slow (Fig. 3D). Collectively, these data suggest that single-peptide responses that are lost, emerge as new responses, or persist during the first year of infection are not associated with high or low initial peak magnitudes of responses during acute infection.
Recognition patterns associated with autologous viral escape.
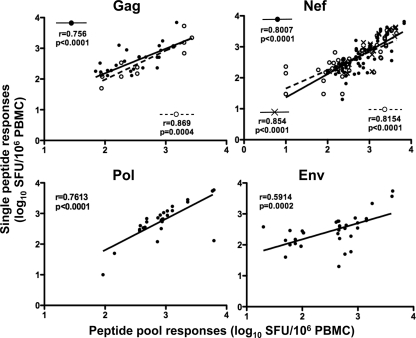
Our approach to identify lost, persistent, or new IFN-γ responses was made using peptide pools. After deconvoluting the peptide-matrix pools in the ELISPOT assay (34) and confirming single-peptide responses in follow-up assays, we were able to verify that many of the pool responses were derived from the recognition of one immunodominant peptide response. Figure 4 shows the correlation between the peptide pool responses initially identified and the single peptides used to confirm the responses. For Gag and Nef, there were highly significant correlations between peptide pool responses and confirmed single peptides, where the peptide confirmations falling into profile 1 showed r values of 0.869 (P = 0.0004) and 0.8154 (P < 0.0001) for Gag and Nef, respectively, and profile 2 showed an r value of 0.854 (P < 0.0001) for Nef. The correlation of overall peptide pool responses with single peptides (regardless of the profile of response) showed significant correlations for Gag (r = 0.756; P < 0.0001), Nef (r = 0.8007; P < 0.0001), Pol (r = 0.7613; P < 0.0001), and Env (r = 0.5914; P < 0.0002) (Fig. 4). Tables 1 and 2 show the confirmed peptides that constitute the responses identified in the peptide pool responses for Gag and Nef, identifying the likely epitopes within the peptide being targeted and those which are associated with lost, new, or persistent responses.
FIG. 4.
Correlations between peptide pool responses and confirmed single peptides. Spearman correlations between IFN-γ ELISPOT responses derived from the peptide pool versus single confirmed peptides within the pool for Gag, Nef, Pol, and Env. The black circles represent peptide pools versus single peptides for all responses, open circles represent peptide pools versus single peptides for peptides that showed evidence for autologous viral sequence change, and crosses represent peptide pools versus single peptides for peptides that showed persistent IFN-γ ELISPOT responses.
TABLE 1.
Confirmed variant peptide responses in 17 study participants in Gag and Nef, showing the profile of response over time, disease category, region of viral variation within the putative epitope, peak IFN-γ ELISPOT response, time to autologous viral epitope escape, and subsequent time to loss of peptide-specific responses
| Participant identifier | Protein | Confirmed peptide responseb | HLA background and restricting allelec | Profile | Disease categoryd | Viral variant positione | Peak response (SFU/106 PBMC) | No. of weeks to autologous viral escape | No. of weeks to lost response |
|---|---|---|---|---|---|---|---|---|---|
| CAP8 | p17 | 9GKKHYMLKHLVWASREL26 | A*2301/-, B*0801/15:10, C*0701/1601 | 1 | R | E | 2,695 | 11.5 | 30 |
| CAP37 | Nef | 6SKSSIVGWPAVRERIRRTE24 | A*2301/2402, B*0702/5301, C*1701/- | 1 | R | F | 2,243 | 13 | |
| CAP37 | Nef | 129PGPGVRYPLTFGWCFFKLVP147 | A*2301/2402, B*0702/5301, C*1701/- | 1 | R | F+E | 5,653 | 9.5 | 15 |
| CAP85 | Nef | 113WVYHTQGYFPDWQNYTPGP131 | A*3002/-, B*0801/4501, C*0701/1601 | 1 | I | F | 1,638 | 18.5 | 16 |
| CAP88a | p15 | 357SHKARVLAEAMSQANSA374 | A*2902/6601, B*4501/5802, C*0602/- | 1 | I | F | 465 | 10 | 26 |
| CAP129 | Nef | 65EVGFPVRPQVPLRPMTYKA93 | A*2601/8001, B*1801/8101, C*0202/0401 | 1 | I | E | 155 | 2 | 13 |
| CAP129 | Nef | 101IHSKRRQDILDLWVYYHTQG119 | A*2601/8001, B*1801/8101 C*0202/0401 | 1 | I | F+E | 1,775 | 2 | 13 |
| CAP217 | p24 | 234SDIAGTTSTLQEQIAWMTSNPPVPV2 | A*0202/2901, B*1503/5801, C*0210/0602 | 1 | I | E | 2,190 | 14.5 | 19 |
| CAP217 | Nef | 77RPMTYKAAFDLSFFLKEKG95 | A*0202/2901, B*1503/5801, C*0210/0602 | 1 | I | E | 3,528 | 17.5 | 19 |
| CAP225a | Nef | 125QNYTPGPGVRYPLTF139 | A*0101/3001, B*4202/8101, C*1701/1801 | 1 | I | E | 2,468 | 1.5 | 29 |
| CAP225a | Nef | 161NNCLLHPMSQHGMEDADRE179 | A*0101/3001, B*4202/8101, C*1701/1801 | 1 | I | E | 1,248 | 28.5 | 33 |
| CAP229a | p24 | 234SDIAGTTSTLQEQIAWMTSNPPVPV2 | A*0123/-, B*5801/-, C*0602/- | 1 | I | F | 1,188 | 18.5 | 34 |
| CAP255 | Nef | 129PGPGVRYPLTFGWCFFKLVP147 | A*0301/8001, B*0801/1801, C*0202/0702 | 1 | I | E | 2,688 | 24.5 | 51 |
| CAP264 | Nef | 77RPMTYKAAFDLSFFLKEKG95 | A*3601/6802, B*1510/5301, C*0401/0804 | 1 | I | E | 2,725 | 17.5 | 20 |
| CAP8 | Nef | 101IHSKRRQDILDLWVYYHTQG119 | A*2301/-; B*0801/1510; Cw*0701/1601 | 2 | R | F+E | 2,495 | 11.5 | |
| CAP256 | Nef | 77RPMTYKAAFDLSFFLKEKG95 | A*2901/6601; B*1503/5802; Cw*0401/0602 | 2 | R | E | 2,935 | 21 | |
| CAP30 | Nef | 77RPMTYKAAFDLSFFLKEKG95 | A*0201/3402; B*4403/4501; Cw*0401/1601 | 3 | I | E | 313 | 8 | |
| CAP244 | Nef | 129PGPGVRYPLTFGWCFFKLVP147 | A*2301/3004; B*4403/5802; Cw*0401/0602 | 3 | I | F | 5,735 | 52 | |
| CAP255 | Nef | 101IHSKRRQDILDLWVYYHTQG119 | A*0301/8001; B*0801/1801; Cw*0202/0702 | 3 | I | F+E | 4,045 | ||
| CAP258 | Nef | 129PGPGVRYPLTFGWCFFKLVP147 | A*2301/2902; B*4101/4201; Cw*1701/- | 3 | R | E | 4,245 | 19.5 | |
| CAP261 | Nef | 101IHSKRRQDILDLWVYYHTQG119 | A*2911/4301; B*1302/1503; Cw*0602/- | 3 | I | F+E | 300 | 5 | |
| CAP262a | Nef | 65EVGFPVRPQVPLRPMTYKA93 | A*0101/6602; B*4201/8101; Cw*06/1701 | 3 | S | E | 935 | 14 | |
| CAP268a | Nef | 101IHSKRRQDILDLWVYYHTQG119 | A*0205/2601; B*0705/5801; Cw*0701/0702 | 3 | I | F+E | 1,543 | 4 | |
| Mean ± SD | 2,313 ± 1,563 | 14 ± 11 | 24 ± 11 |
This study participant showed a simultaneous profile of lost and persistent peptide-specific responses.
Bold and underlining represent the putative optimal epitope.
Bold and underlining represent the putative restricting HLA. Hyphens refer to the same HLA allele and depict a homozygous locus.
R, rapid progressor; I, intermediate progressor; S, slow progressor.
E, mutation occurs within the targeted epitope; F, mutation occurs within the flanking region of the targeted epitope.
TABLE 2.
Confirmed invariant peptide responses in 13 study participants in Gag and Nef, showing the profile of response over time, disease category, and peak IFN-γ ELISPOT responses
| Participant identifier | Protein | Confirmed peptide responseb | HLA background and restricting allelec | Profile | Disease categoryd | Peak response (SFU/106 PBMC) | No. of weeks to autologous viral escape |
|---|---|---|---|---|---|---|---|
| CAP174 | Nef | 73QVPLRPMTYKAAFDL87 | A*0301/7401; B*4901/5802; Cw*0602/0701 | 1 | R | 923 | 11 |
| CAP229a | Nef | 77RPMTYKAAFDLSFFLKEKG95 | A*0123/-; B*5801/-; Cw*0602/- | 1 | I | 1,808 | 15 |
| CAP210 | p15 | 290KEPFRDYVDRFFKTLRAEQATQD321 | A*6802/-; B*1510/-; Cw*0304/- | 2 | R | 2,068 | |
| CAP228 | p24 | 163AFSPEVIPMFTALSEGA179 | A*2301/2638; B*4403/5101; Cw*0303/0701 | 2 | S | 320 | |
| CAP45 | Nef | 129PGPGVRYPLTFGWCFFKLVP147 | A*2301/2902; B*1510/4501; Cw*0602/1601 | 3 | S | 2,178 | |
| CAP88a | Nef | 113WVYHTQGYFPDWQNYTPGP131 | A*2902/6601; B*4501/5802; Cw*0602/- | 3 | I | 2,677 | |
| CAP206 | Nef | 129PGPGVRYPLTFGWCFFKLVP147 | A*0301/3201; B*0702/4403; Cw*0210/0702 | 3 | R | 243 | |
| CAP222 | Nef | 65EVGFPVRPQVPLRPMTYKA93 | A*3001/3303; B*5301/8101; Cw*0401/- | 3 | S | 2,838 | |
| CAP222 | p24 | 178GATPQDLNTMLNTVGGH194 | A*3001/3303; B*5301/8101; Cw*0401/- | 3 | S | 1,568 | |
| CAP225a | p24 | 178GATPQDLNTMLNTVGGH194 | A*0101/3001; B*4202/8101; Cw*1701/1801 | 3 | I | 3,327 | |
| CAP228 | Nef | 129PGPGVRYPLTFGWCFFKLVP147 | A*2301/2638; B*4403/5101; Cw*0303/0701 | 3 | S | 2,915 | |
| CAP257 | p17 | 9GKKHYMLKHLVWASREL26 | A*2301/2902; B*4202/4403; Cw*1701/- | 3 | I | 775 | |
| CAP257 | Nef | 129PGPGVRYPLTFGWCFFKLVP147 | A*2301/2902; B*4202/4403; Cw*1701/- | 3 | I | 4,928 | |
| CAP262a | p24 | 178GATPQDLNTMLNTVGGH194 | A*0101/6602; B*4201/8101; Cw*06/1701 | 3 | S | 1,335 | |
| CAP268a | p24 | 235DIAGTTSTLQEQIAWMTSNPPVPV258 | A*0205/2601; B*0705/5801; Cw*0701/0702 | 3 | I | 2,325 | |
| Mean ± SD | 2,015 ± 1,245 |
This study participant showed a simultaneous profile of lost and persistent peptide-specific responses.
Bold and underlining represent the putative optimal epitope.
Bold and underlining represent the putative restricting HLA. Hyphens refer to the same HLA allele and depict a homozygous locus.
R, rapid progressor; I, intermediate progressor; S, slow progressor.
When we overlaid the recognition profile with autologous viral sequence changes, it was evident that profile 1 (lost responses) were mostly observed concurrently with fixed viral escape over time (87.5% [14 of 16]) (Fig. 5 A), where escape occurred either in the putative epitope (E) or in the flanking region (F). Table 1 shows the peak T-cell response in 14 of the T-cell responses with profile 1 at the single-peptide level and the timing of viral epitope escape. Fifty percent of profile 2 responses (new/emerging responses) were associated with autologous sequence variation; however, the low number of individuals (n = 4) presenting with new responses to Gag or Nef did not allow us to draw defined conclusions (Fig. 5B). Profile 3 responses (persistent responses) were least associated with autologous viral escape, with 37% (7 of 19) showing epitope variation, or almost two-thirds of participants with invariant sequences. In the representative example shown in Fig. 5A, study participant CAP217 possessed a peak IFN-γ ELISPOT response of 2,190 spot-forming units (SFU)/106 PBMC at 12 weeks postinfection, recognizing the HLA-B*5801-restricted TW10 (TSTLQEQIAW) epitope in p24 Gag (Table 1). By 17 weeks postinfection, a fixed T→N mutation occurred at position 3, and by 19 weeks postinfection, the response fell below 80% of the peak response (Table 1). For CAP88, there was a relatively weaker peak IFN-γ ELISPOT response of 465 SFU/106 PBMC at 7 weeks postinfection, recognizing a probable HLA-B*4501-restricted AS9 (AEAMSQANS) epitope in p15 Gag. There was a subsequent transient escape in the flanking region detected by 13 weeks postinfection that was associated with reduction in responses to 138 SFU/106 PBMC. It was interesting to note that although there was a reversion back from valine to alanine at amino acid position 374 at some point between 26 and 54 weeks postinfection, there was no detectable IFN-γ ELISPOT response during this time (Fig. 5A). CAP256, showing a profile 2 pattern, possessed a late peak IFN-γ ELISPOT response (2,935 SFU/106 PBMC) at 34 weeks postinfection. It was evident from the autologous sequence change within the recognized epitopic region (probable HLA-Cw*0602-restricted AL9 [AAFDLSFFL] [Table 1]) that the initial nonrecognition of the peptide used in the assay was due to sequence variation in the flanking region and within the epitope (Fig. 5B). By 30 weeks postinfection, the infecting virus sequence (measured at 6 weeks postinfection) changed to match the peptide sequence. Of note in this individual, more than 80% of the peak IFN-γ ELISPOT response was lost, regained, and then lost despite there being no further variations in the autologous sequence (Fig. 5B). CAP210 showed a profile very similar to that of CAP256, where there were two putative epitopes being recognized: HLA-B*1510-restricted YVDRFFKTL and HLA Cw*0304-restricted RAEQATQD in p15 Gag (Table 2). However, in contrast to CAP256, CAP210 showed no detectable autologous sequence variation despite showing a peak IFN-γ ELISPOT response of 2,068 SFU/106 PBMC (Table 2) at 30 weeks postinfection (Fig. 5B). Both CAP225 and CAP257 represented individuals with a profile 3 pattern, showing persistent IFN-γ ELISPOT responses in the absence of autologous sequence variation, despite high-magnitude peak responses of several thousand SFU/106 PBMC (Table 2 and Fig. 5C). Collectively, when we assessed the Shannon entropy scores for the peptides falling into either profile 1 or profile 3, we found there to be no significant difference.
FIG. 5.
Profiles of T-cell recognition in relation to autologous sequence variations in Gag and Nef. Representative examples of the three T-cell recognition profiles in relation to autologous sequence variation, shown as the magnitudes of IFN-γ ELISPOT responses over weeks postinfection. Each example is a representative individual within the cohort, showing the high-resolution HLA background for A, B, and C alleles. The sequences of the autologous virus at different weeks postinfection are shown underneath each graph. The gray shaded areas indicate the putative targeted epitope. The boxes with dashed borders on the graphs show the windows of time in which autologous viral escape occurred. “E” corresponds to a mutation within the target epitope, and “F” corresponds to a mutation in the flanking region of the targeted epitope.
Decay of IFN-γ ELISPOT responses after epitope escape.
Assessing the tempo of lost IFN-γ ELISPOT responses in relation to autologous viral escape, we were able to show a strong association between the estimated time of viral escape and the time to disappearance of the IFN-γ ELISPOT response (P = 0.019; r = 0.61). For the 10 individuals and 14 epitopes showing escape (Table 1), we estimated the time taken for the IFN-γ ELISPOT response to decay after the target epitope varied (Fig. 6) to be a mean 14 weeks (range, 4.4 to 24 weeks, 95% confidence interval). These data indicate that once the presenting epitope was removed, there was likely no requirement for the continued survival of IFN-γ-producing effector T cells, which dissipated between 1 and 6 months after epitope escape.
FIG. 6.
Relationship between the time of loss of IFN-γ responses and the median time of viral escape or autologous sequence variation. Linear regression showing a linear correlation between the estimated median time of autologous epitope escape and the time of IFN-γ ELISPOT response loss. The dashed lines indicate the 95% confidence intervals (c.i.) above and below the mean regression line. The estimated time of escape was calculated as the median time between the last time point measured where the autologous sequence was wild type and the first time point where the variant sequence was detected.
DISCUSSION
In a cohort of 53 subjects infected with subtype C HIV-1, we recently reported that the breadth and magnitude of HIV-1-specific IFN-γ ELISPOT T-cell responses at 3 months postinfection were unrelated to the viral set point at 12 months postinfection and that there was a dominant, narrow, and focused recognition of Nef at 3 months postinfection (22). In the current paper, we investigated the dynamics and kinetics of IFN-γ ELISPOT T-cell responses over time in this cohort. We found a strong and diverse pattern of T-cell recognition across the entire HIV-1 proteome, with immunodominant responses emerging as early as 3 weeks postinfection. Consistent with previously reported immunodominance patterns of T-cell recognition, Nef, Pol, and Gag were the most frequently recognized regions during the first 6 months of infection (22, 31, 34, 48), with a distinct hierarchy of evolving responses. In this cohort, Nef responses were the most rapid to emerge over time, being dominant at all time points analyzed, in agreement with other publications (31, 34, 35, 48). However, despite the vigorous targeting of Nef, we found no association between anti-Nef-specific T-cell responses and viral load. This contrasts with studies that have demonstrated a direct correlation between Nef-specific T-cell responses and high viremia in early and chronic HIV-1 infection (25, 34, 55). However, our analysis takes into account the temporal patterns of response, making this analysis unique from previous studies. Of note, of the responses to increase over time, anti-Gag-specific T cells emerged the slowest.
In agreement with previous findings (15, 25, 34, 48, 55), the frequencies of Gag- and Env-specific T-cell responses were found to be associated with lower and higher viremia, respectively, supporting the hypothesis that the ability to target Gag in vivo contributes to viral control. Further support for the importance of Gag is when mutations in key epitopes result in a fitness cost to the virus. For example, in this cohort, we have previously found that lower viremia was noted for HIV-infected individuals after viral transmission with viruses containing B*57/58 escape mutations to HLA-B*57/B58-negative recipients, suggesting that mutations in the ISW9/TW10 Gag epitopes are associated with a transmitted viral fitness cost (10, 11).
One of the major factors limiting the effectiveness of virus-specific CTL is the ability of HIV-1 to evade CTL responses through sequence variation or viral escape (1, 19). Longitudinal studies of CTL escape suggest that CTL responses represent a major driving force of HIV, SIV, and hepatitis C virus evolution, with more than 50% of sequence variation across the genome attributed to CTL-mediated pressure in SIV and HIV (1, 28, 40). In this paper, we show that the magnitude of HIV-specific T-cell responses fluctuates widely over the first year of infection, highlighting the rapidity of T-cell evolution and the often unpredictable nature of T-cell recognition patterns. Three distinct profiles of T-cell recognition were identified, characterized by either lost, new, or persistent responses, and there was a tendency of lost and persistent responses to be clustered in Nef and emerging new responses to be clustered in Pol and Env. Almost 90% of lost T-cell recognition (profile 1) was coincidental with autologous sequence variation in either the putative or the flanking region of epitopes and was consistent with CTL immune pressure. In support of this, CTL-driven immune escape has been demonstrated in the acute phase of SIV and HIV infection, suggesting a rapid selection or evasion of virus-specific CTL responses during the first few weeks of infection (1, 5, 8, 17, 30). Using single-genome amplification (SGA), IFN-γ ELISPOT assay, and algorithms to identified sites of positive selection across the HIV genome, Goonetilleke et al. (17) identified the first T-cell responses to the transmitted/founder virus and showed that these T-cell responses rapidly select escape mutations concurrent with a decrease in viral load in acute HIV-1 infection. These data provide insight into the contribution of CTL in the resolution of acute-phase viremia. There is also strong inferential evidence supporting HLA class 1-driven viral evolution across the HIV-1 genome in both subtype B and C HIV infection (4, 9, 14, 23, 44, 50, 53), where a high density of HLA-associated mutation sites can be found in Nef relative to Pol, Gag, and other regulatory proteins, suggesting that CTL recognition exerts differential selection pressures in selected regions (9, 44, 53). This also may explain the high proportion of lost responses in Nef relative to Gag, Pol, and Env that we observed. Toggling selection sites, defined as a model of positive selection associated with immune escape and reversion, have also been observed for Nef, consistent with the high density of HLA-associated polymorphisms (12), supporting the evidence of CTL-driven immune selective pressure in shaping HIV diversity. It is noteworthy, however, that the use of consensus and consensus-like peptides in our study might underestimate or limit the potential to detect HIV-1-specific CTL responses, especially to those highly variable regions of the HIV-1 genome. Ideally, the use of autologous sequence-based peptide sets may have improved our detection of T-cell responses in more variable regions of HIV-1 proteins than previously reported (2) and would have provided a more comprehensive profile of the evolution of T-cell responses from acute to chronic HIV infection. It is probable that the use of autologous peptides based on the transmitted infecting virus would avoid the potential underestimation of epitope escape.
Similar to Turnbull et al. (51), we found a relatively low proportion of autologous sequence variation within epitopic regions being targeted by persisting HIV-specific T cells in this study, suggesting that the fluctuations we observed for many of the persisting responses were due to mechanisms other than viral escape. Several mechanisms have been postulated for the temporal fluctuation of CTL responses, including (i) fluctuations in antigen load, (ii) divergence of the CD8+ T-cell receptor (TCR) repertoire, and (iii) T-cell exhaustion (16, 38, 51). Other studies have reported perturbations of the T-cell receptor repertoire during primary HIV infection and hypothesized that this may be influenced by the clonal expansion and exhaustion of virus-specific CTL clones during the initial burst of viremia during acute infection (18, 41, 46). It has also been hypothesized that persistence of some epitopes are due to insufficient antiviral pressure from the corresponding CTL responses (26). In addition, conserved epitopes have been shown to elicit subdominant CTL responses throughout HIV-1 infection (32). However, in our study, there were no differences between subdominant or dominant T-cell responses in persistent or lost epitopes, which appeared to have no bearing on disease progression in our study. These data are in agreement with the findings of Lui et al. (33), who found no differences in the peak magnitudes of CTL responses between evolving and persistent epitopes. Based on the fact that persistent T-cell responses tended to target peptides preferentially with low variability, we could hypothesize that persistent immune responses are directed against highly conserved epitopes (those with low entropy) and that the three different profiles of responses are a likely result of the structurally constrained nature of the epitopes targeted. However, no differences were observed in the average entropy scores of peptides within the differing profiles, making it difficult to associate the likelihood of epitope change with the profile of the T-cell response. Our data also suggest that the temporal nature of T-cell fluctuations means that a static measurement in time is not a predictable or reliable marker of recognition and may have little meaning for disease progression. The significance and novelty of these results lie at several levels: (i) T-cell epitope recognition is not a static event and there are temporal patterns of IFN-γ-based responses, (ii) persistent T-cell responses to invariant epitopes are enriched in individuals with slowly progressing viremia, and (iii) it takes an average of 14 weeks for IFN-γ-based T-cell responses to dissipate once the target epitope has escaped. Collectively, the broader interpretation of our data is that continual invariant viral epitope presentation is required for persistent T-cell immunity, which in turn appears necessary for the control of early disease progression.
Acknowledgments
We thank the participants and the clinical and laboratory staff at CAPRISA for the specimens.
This work was funded by the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), U.S. Department of Health and Human Services grants U19 A151794 (S.A.K.) and 5R01AI078936-02 (C.W.). R.N. also received a training grant from the Rwandan government.
Footnotes
Published ahead of print on 8 September 2010.
REFERENCES
- 1.Allen, T. M., M. Altfeld, S. C. Geer, E. T. Kalife, C. Moore, K. M. O'Sullivan, I. Desouza, M. E. Feeney, R. L. Eldridge, E. L. Maier, D. E. Kaufmann, M. P. Lahaie, L. Reyor, G. Tanzi, M. N. Johnston, C. Brander, R. Draenert, J. K. Rockstroh, H. Jessen, E. S. Rosenberg, S. A. Mallal, and B. D. Walker. 2005. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J. Virol. 79:13239-13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altfeld, M., M. M. Addo, R. Shankarappa, P. K. Lee, T. M. Allen, X. G. Yu, A. Rathod, J. Harlow, K. O'Sullivan, M. N. Johnston, P. J. Goulder, J. I. Mullins, E. S. Rosenberg, C. Brander, B. Korber, and B. D. Walker. 2003. Enhanced detection of human immunodeficiency virus type 1-specific T-cell responses to highly variable regions by using peptides based on autologous virus sequences. J. Virol. 77:7330-7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 4.Berger, C. T., J. M. Carlson, C. J. Brumme, K. L. Hartman, Z. L. Brumme, L. M. Henry, P. C. Rosato, A. Piechocka-Trocha, M. A. Brockman, P. R. Harrigan, D. Heckerman, D. E. Kaufmann, and C. Brander. 2010. Viral adaptation to immune selection pressure by HLA class I-restricted CTL responses targeting epitopes in HIV frameshift sequences. J. Exp. Med. 207:61-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernardin, F., D. Kong, L. Peddada, L. A. Baxter-Lowe, and E. Delwart. 2005. Human immunodeficiency virus mutations during the first month of infection are preferentially found in known cytotoxic T-lymphocyte epitopes. J. Virol. 79:11523-11528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharya, T., M. Daniels, D. Heckerman, B. Foley, N. Frahm, C. Kadie, J. Carlson, K. Yusim, B. McMahon, B. Gaschen, S. Mallal, J. I. Mullins, D. C. Nickle, J. Herbeck, C. Rousseau, G. H. Learn, T. Miura, C. Brander, B. Walker, and B. Korber. 2007. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science 315:1583-1586. [DOI] [PubMed] [Google Scholar]
- 7.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-211. [DOI] [PubMed] [Google Scholar]
- 9.Brumme, Z. L., C. J. Brumme, D. Heckerman, B. T. Korber, M. Daniels, J. Carlson, C. Kadie, T. Bhattacharya, C. Chui, J. Szinger, T. Mo, R. S. Hogg, J. S. Montaner, N. Frahm, C. Brander, B. D. Walker, and P. R. Harrigan. 2007. Evidence of differential HLA class I-mediated viral evolution in functional and accessory/regulatory genes of HIV-1. PLoS Pathog. 3:e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chopera, D. R., Z. Woodman, K. Mlisana, M. Mlotshwa, D. P. Martin, C. Seoighe, F. Treurnicht, D. de Assis Rosa, W. Hide, S. A. Karim, C. M. Gray, and C. Williamson. 2008. Transmission of HIV-1 CTL escape variants provides HLA-mismatched recipients with a survival advantage. PLoS Pathog. 4:e1000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crawford, H., W. Lumm, A. Leslie, M. Schaefer, D. Boeras, J. G. Prado, J. Tang, P. Farmer, T. Ndung'u, S. Lakhi, J. Gilmour, P. Goepfert, B. D. Walker, R. Kaslow, J. Mulenga, S. Allen, P. J. Goulder, and E. Hunter. 2009. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J. Exp. Med. 206:909-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delport, W., K. Scheffler, and C. Seoighe. 2008. Frequent toggling between alternative amino acids is driven by selection in HIV-1. PLoS Pathog. 4:e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diggle, P. J., P. Heagerty, K.-Y. Liang, and S. L. Zeger. 2002. Analysis of longitudinal data. Oxford University Press, Cary, NC.
- 14.Duda, A., L. Lee-Turner, J. Fox, N. Robinson, S. Dustan, S. Kaye, H. Fryer, M. Carrington, M. McClure, A. R. McLean, S. Fidler, J. Weber, R. E. Phillips, and A. J. Frater. 2009. HLA-associated clinical progression correlates with epitope reversion rates in early human immunodeficiency virus infection. J. Virol. 83:1228-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geldmacher, C., J. R. Currier, E. Herrmann, A. Haule, E. Kuta, F. McCutchan, L. Njovu, S. Geis, O. Hoffmann, L. Maboko, C. Williamson, D. Birx, A. Meyerhans, J. Cox, and M. Hoelscher. 2007. CD8 T-cell recognition of multiple epitopes within specific Gag regions is associated with maintenance of a low steady-state viremia in human immunodeficiency virus type 1-seropositive patients. J. Virol. 81:2440-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geldmacher, C., C. Gray, M. Nason, J. R. Currier, A. Haule, L. Njovu, S. Geis, O. Hoffmann, L. Maboko, A. Meyerhans, J. Cox, and M. Hoelscher. 2007. A high viral burden predicts the loss of CD8 T-cell responses specific for subdominant gag epitopes during chronic human immunodeficiency virus infection. J. Virol. 81:13809-13815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goonetilleke, N., M. K. Liu, J. F. Salazar-Gonzalez, G. Ferrari, E. Giorgi, V. V. Ganusov, B. F. Keele, G. H. Learn, E. L. Turnbull, M. G. Salazar, K. J. Weinhold, S. Moore, N. Letvin, B. F. Haynes, M. S. Cohen, P. Hraber, T. Bhattacharya, P. Borrow, A. S. Perelson, B. H. Hahn, G. M. Shaw, B. T. Korber, and A. J. McMichael. 2009. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J. Exp. Med. 206:1253-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorochov, G., A. U. Neumann, A. Kereveur, C. Parizot, T. Li, C. Katlama, M. Karmochkine, G. Raguin, B. Autran, and P. Debre. 1998. Perturbation of CD4+ and CD8+ T-cell repertoires during progression to AIDS and regulation of the CD4+ repertoire during antiviral therapy. Nat. Med. 4:215-221. [DOI] [PubMed] [Google Scholar]
- 19.Goulder, P. J., C. Brander, Y. Tang, C. Tremblay, R. A. Colbert, M. M. Addo, E. S. Rosenberg, T. Nguyen, R. Allen, A. Trocha, M. Altfeld, S. He, M. Bunce, R. Funkhouser, S. I. Pelton, S. K. Burchett, K. McIntosh, B. T. Korber, and B. D. Walker. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412:334-338. [DOI] [PubMed] [Google Scholar]
- 20.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 21.Goulder, P. J., and D. I. Watkins. 2004. HIV and SIV CTL escape: implications for vaccine design. Nat. Rev. Immunol. 4:630-640. [DOI] [PubMed] [Google Scholar]
- 22.Gray, C. M., M. Mlotshwa, C. Riou, T. Mathebula, D. de Assis Rosa, T. Mashishi, C. Seoighe, N. Ngandu, F. van Loggerenberg, L. Morris, K. Mlisana, C. Williamson, and S. A. Karim. 2009. Human immunodeficiency virus-specific gamma interferon enzyme-linked immunospot assay responses targeting specific regions of the proteome during primary subtype C infection are poor predictors of the course of viremia and set point. J. Virol. 83:470-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawashima, Y., K. Pfafferott, J. Frater, P. Matthews, R. Payne, M. Addo, H. Gatanaga, M. Fujiwara, A. Hachiya, H. Koizumi, N. Kuse, S. Oka, A. Duda, A. Prendergast, H. Crawford, A. Leslie, Z. Brumme, C. Brumme, T. Allen, C. Brander, R. Kaslow, J. Tang, E. Hunter, S. Allen, J. Mulenga, S. Branch, T. Roach, M. John, S. Mallal, A. Ogwu, R. Shapiro, J. G. Prado, S. Fidler, J. Weber, O. G. Pybus, P. Klenerman, T. Ndung'u, R. Phillips, D. Heckerman, P. R. Harrigan, B. D. Walker, M. Takiguchi, and P. Goulder. 2009. Adaptation of HIV-1 to human leukocyte antigen class I. Nature 458:641-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiepiela, P., A. J. Leslie, I. Honeyborne, D. Ramduth, C. Thobakgale, S. Chetty, P. Rathnavalu, C. Moore, K. J. Pfafferott, L. Hilton, P. Zimbwa, S. Moore, T. Allen, C. Brander, M. M. Addo, M. Altfeld, I. James, S. Mallal, M. Bunce, L. D. Barber, J. Szinger, C. Day, P. Klenerman, J. Mullins, B. Korber, H. M. Coovadia, B. D. Walker, and P. J. Goulder. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769-775. [DOI] [PubMed] [Google Scholar]
- 25.Kiepiela, P., K. Ngumbela, C. Thobakgale, D. Ramduth, I. Honeyborne, E. Moodley, S. Reddy, C. de Pierres, Z. Mncube, N. Mkhwanazi, K. Bishop, M. van der Stok, K. Nair, N. Khan, H. Crawford, R. Payne, A. Leslie, J. Prado, A. Prendergast, J. Frater, N. McCarthy, C. Brander, G. H. Learn, D. Nickle, C. Rousseau, H. Coovadia, J. I. Mullins, D. Heckerman, B. D. Walker, and P. Goulder. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13:46-53. [DOI] [PubMed] [Google Scholar]
- 26.Koibuchi, T., T. M. Allen, M. Lichterfeld, S. K. Mui, K. M. O'Sullivan, A. Trocha, S. A. Kalams, R. P. Johnson, and B. D. Walker. 2005. Limited sequence evolution within persistently targeted CD8 epitopes in chronic human immunodeficiency virus type 1 infection. J. Virol. 79:8171-8181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuntzen, T., J. Timm, A. Berical, L. L. Lewis-Ximenez, A. Jones, B. Nolan, J. Schulze zur Wiesch, B. Li, A. Schneidewind, A. Y. Kim, R. T. Chung, G. M. Lauer, and T. M. Allen. 2007. Viral sequence evolution in acute hepatitis C virus infection. J. Virol. 81:11658-11668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leslie, A. J., K. J. Pfafferott, P. Chetty, R. Draenert, M. M. Addo, M. Feeney, Y. Tang, E. C. Holmes, T. Allen, J. G. Prado, M. Altfeld, C. Brander, C. Dixon, D. Ramduth, P. Jeena, S. A. Thomas, A. St. John, T. A. Roach, B. Kupfer, G. Luzzi, A. Edwards, G. Taylor, H. Lyall, G. Tudor-Williams, V. Novelli, J. Martinez-Picado, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282-289. [DOI] [PubMed] [Google Scholar]
- 30.Li, B., A. D. Gladden, M. Altfeld, J. M. Kaldor, D. A. Cooper, A. D. Kelleher, and T. M. Allen. 2007. Rapid reversion of sequence polymorphisms dominates early human immunodeficiency virus type 1 evolution. J. Virol. 81:193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lichterfeld, M., X. G. Yu, D. Cohen, M. M. Addo, J. Malenfant, B. Perkins, E. Pae, M. N. Johnston, D. Strick, T. M. Allen, E. S. Rosenberg, B. Korber, B. D. Walker, and M. Altfeld. 2004. HIV-1 Nef is preferentially recognized by CD8 T cells in primary HIV-1 infection despite a relatively high degree of genetic diversity. AIDS 18:1383-1392. [DOI] [PubMed] [Google Scholar]
- 32.Liu, Y., J. McNevin, M. Rolland, H. Zhao, W. Deng, J. Maenza, C. E. Stevens, A. C. Collier, M. J. McElrath, and J. I. Mullins. 2009. Conserved HIV-1 epitopes continuously elicit subdominant cytotoxic T-lymphocyte responses. J. Infect. Dis. 200:1825-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, Y., J. McNevin, H. Zhao, D. M. Tebit, R. M. Troyer, M. McSweyn, A. K. Ghosh, D. Shriner, E. J. Arts, M. J. McElrath, and J. I. Mullins. 2007. Evolution of human immunodeficiency virus type 1 cytotoxic T-lymphocyte epitopes: fitness-balanced escape. J. Virol. 81:12179-12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masemola, A., T. Mashishi, G. Khoury, P. Mohube, P. Mokgotho, E. Vardas, M. Colvin, L. Zijenah, D. Katzenstein, R. Musonda, S. Allen, N. Kumwenda, T. Taha, G. Gray, J. McIntyre, S. A. Karim, H. W. Sheppard, and C. M. Gray. 2004. Hierarchical targeting of subtype C human immunodeficiency virus type 1 proteins by CD8+ T cells: correlation with viral load. J. Virol. 78:3233-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mashishi, T., S. Loubser, W. Hide, G. Hunt, L. Morris, G. Ramjee, S. Abdool-Karim, C. Williamson, and C. M. Gray. 2001. Conserved domains of subtype C Nef from South African HIV type 1-infected individuals include cytotoxic T lymphocyte epitope-rich regions. AIDS Res. Hum. Retroviruses 17:1681-1687. [DOI] [PubMed] [Google Scholar]
- 36.McCullagh, P., and J. A. Nelder. 1989. Generalized linear models, 2nd ed. Chapman and Hall/CRC, Boca Raton, FL.
- 37.McMichael, A. J., P. Borrow, G. D. Tomaras, N. Goonetilleke, and B. F. Haynes. 2010. The immune response during acute HIV-1 infection: clues for vaccine development. Nat. Rev. Immunol. 10:11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer-Olson, D., K. W. Brady, M. T. Bartman, K. M. O'Sullivan, B. C. Simons, J. A. Conrad, C. B. Duncan, S. Lorey, A. Siddique, R. Draenert, M. Addo, M. Altfeld, E. Rosenberg, T. M. Allen, B. D. Walker, and S. A. Kalams. 2006. Fluctuations of functionally distinct CD8+ T-cell clonotypes demonstrate flexibility of the HIV-specific TCR repertoire. Blood 107:2373-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miura, T., M. A. Brockman, A. Schneidewind, M. Lobritz, F. Pereyra, A. Rathod, B. L. Block, Z. L. Brumme, C. J. Brumme, B. Baker, A. C. Rothchild, B. Li, A. Trocha, E. Cutrell, N. Frahm, C. Brander, I. Toth, E. J. Arts, T. M. Allen, and B. D. Walker. 2009. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphocyte [corrected] recognition. J. Virol. 83:2743-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Connor, D. H., A. B. McDermott, K. C. Krebs, E. J. Dodds, J. E. Miller, E. J. Gonzalez, T. J. Jacoby, L. Yant, H. Piontkivska, R. Pantophlet, D. R. Burton, W. M. Rehrauer, N. Wilson, A. L. Hughes, and D. I. Watkins. 2004. A dominant role for CD8+-T-lymphocyte selection in simian immunodeficiency virus sequence variation. J. Virol. 78:14012-14022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pantaleo, G., H. Soudeyns, J. F. Demarest, M. Vaccarezza, C. Graziosi, S. Paolucci, M. Daucher, O. J. Cohen, F. Denis, W. E. Biddison, R. P. Sekaly, and A. S. Fauci. 1997. Evidence for rapid disappearance of initially expanded HIV-specific CD8+ T cell clones during primary HIV infection. Proc. Natl. Acad. Sci. U. S. A. 94:9848-9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rerks-Ngarm, S., P. Pitisuttithum, S. Nitayaphan, J. Kaewkungwal, J. Chiu, R. Paris, N. Premsri, C. Namwat, M. de Souza, E. Adams, M. Benenson, S. Gurunathan, J. Tartaglia, J. G. McNeil, D. P. Francis, D. Stablein, D. L. Birx, S. Chunsuttiwat, C. Khamboonruang, P. Thongcharoen, M. L. Robb, N. L. Michael, P. Kunasol, and J. H. Kim. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209-2220. [DOI] [PubMed] [Google Scholar]
- 43.Rodrigo, A. G., P. C. Goracke, K. Rowhanian, and J. I. Mullins. 1997. Quantitation of target molecules from polymerase chain reaction-based limiting dilution assays. AIDS Res. Hum. Retroviruses 13:737-742. [DOI] [PubMed] [Google Scholar]
- 44.Rousseau, C. M., M. G. Daniels, J. M. Carlson, C. Kadie, H. Crawford, A. Prendergast, P. Matthews, R. Payne, M. Rolland, D. N. Raugi, B. S. Maust, G. H. Learn, D. C. Nickle, H. Coovadia, T. Ndung'u, N. Frahm, C. Brander, B. D. Walker, P. J. Goulder, T. Bhattacharya, D. E. Heckerman, B. T. Korber, and J. I. Mullins. 2008. HLA class I-driven evolution of human immunodeficiency virus type 1 subtype C proteome: immune escape and viral load. J. Virol. 82:6434-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 46.Soudeyns, H., G. Campi, G. P. Rizzardi, C. Lenge, J. F. Demarest, G. Tambussi, A. Lazzarin, D. Kaufmann, G. Casorati, L. Corey, and G. Pantaleo. 2000. Initiation of antiretroviral therapy during primary HIV-1 infection induces rapid stabilization of the T-cell receptor beta chain repertoire and reduces the level of T-cell oligoclonality. Blood 95:1743-1751. [PubMed] [Google Scholar]
- 47.Streeck, H., J. S. Jolin, Y. Qi, B. Yassine-Diab, R. C. Johnson, D. S. Kwon, M. M. Addo, C. Brumme, J. P. Routy, S. Little, H. K. Jessen, A. D. Kelleher, F. M. Hecht, R. P. Sekaly, E. S. Rosenberg, B. D. Walker, M. Carrington, and M. Altfeld. 2009. Human immunodeficiency virus type 1-specific CD8+ T-cell responses during primary infection are major determinants of the viral set point and loss of CD4+ T cells. J. Virol. 83:7641-7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Streeck, H., M. Lichterfeld, G. Alter, A. Meier, N. Teigen, B. Yassine-Diab, H. K. Sidhu, S. Little, A. Kelleher, J. P. Routy, E. S. Rosenberg, R. P. Sekaly, B. D. Walker, and M. Altfeld. 2007. Recognition of a defined region within p24 Gag by CD8+ T cells during primary human immunodeficiency virus type 1 infection in individuals expressing protective HLA class I alleles. J. Virol. 81:7725-7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Treurnicht, F. K., C. Seoighe, D. P. Martin, N. Wood, M. R. Abrahams, D. de Assis Rosa, H. Bredell, Z. Woodman, W. Hide, K. Mlisana, S. A. Karim, C. M. Gray, and C. Williamson. 2010. Adaptive changes in HIV-1 subtype C proteins during early infection are driven by changes in HLA-associated immune pressure. Virology 396:213-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turnbull, E. L., M. Wong, S. Wang, X. Wei, N. A. Jones, K. E. Conrod, D. Aldam, J. Turner, P. Pellegrino, B. F. Keele, I. Williams, G. M. Shaw, and P. Borrow. 2009. Kinetics of expansion of epitope-specific T cell responses during primary HIV-1 infection. J. Immunol. 182:7131-7145. [DOI] [PubMed] [Google Scholar]
- 52.van Loggerenberg, F., K. Mlisana, C. Williamson, S. C. Auld, L. Morris, C. M. Gray, Q. Abdool Karim, A. Grobler, N. Barnabas, I. Iriogbe, and S. S. Abdool Karim. 2008. Establishing a cohort at high risk of HIV infection in South Africa: challenges and experiences of the CAPRISA 002 acute infection study. PLoS One 3:e1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, Y. E., B. Li, J. M. Carlson, H. Streeck, A. D. Gladden, R. Goodman, A. Schneidewind, K. A. Power, I. Toth, N. Frahm, G. Alter, C. Brander, M. Carrington, B. D. Walker, M. Altfeld, D. Heckerman, and T. M. Allen. 2009. Protective HLA class I alleles that restrict acute-phase CD8+ T-cell responses are associated with viral escape mutations located in highly conserved regions of human immunodeficiency virus type 1. J. Virol. 83:1845-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williamson, C., L. Morris, M. F. Maughan, L. H. Ping, S. A. Dryga, R. Thomas, E. A. Reap, T. Cilliers, J. van Harmelen, A. Pascual, G. Ramjee, G. Gray, R. Johnston, S. A. Karim, and R. Swanstrom. 2003. Characterization and selection of HIV-1 subtype C isolates for use in vaccine development. AIDS Res. Hum. Retroviruses 19:133-144. [DOI] [PubMed] [Google Scholar]
- 55.Zuniga, R., A. Lucchetti, P. Galvan, S. Sanchez, C. Sanchez, A. Hernandez, H. Sanchez, N. Frahm, C. H. Linde, H. S. Hewitt, W. Hildebrand, M. Altfeld, T. M. Allen, B. D. Walker, B. T. Korber, T. Leitner, J. Sanchez, and C. Brander. 2006. Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. J. Virol. 80:3122-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]