Abstract
Rapid evolution and high intrahost sequence diversity are hallmarks of human and simian immunodeficiency virus (HIV/SIV) infection. Minor viral variants have important implications for drug resistance, receptor tropism, and immune evasion. Here, we used ultradeep pyrosequencing to sequence complete HIV/SIV genomes, detecting variants present at a frequency as low as 1%. This approach provides a more complete characterization of the viral population than is possible with conventional methods, revealing low-level drug resistance and detecting previously hidden changes in the viral population. While this work applies pyrosequencing to immunodeficiency viruses, this approach could be applied to virtually any viral pathogen.
The viral population within each human immunodeficiency virus (HIV)-infected individual is highly diverse and constantly evolving (2, 3). However, our understanding of the viral population is based largely on the consensus sequence of the dominant circulating virus because the full diversity of the viral population is extremely difficult to characterize. One recent study showed that despite viral fitness recovery in vitro, recovery was not correlated with changes observed in the consensus sequence of HIV. Instead, increased fitness correlated with general viral heterogeneity (5). This finding suggests that by limiting our studies to consensus sequences, we are missing many aspects of viral evolution that influence fitness, drug resistance, and immune evasion, among other characteristics. Studies that examined minor viral variants have provided new insights into HIV transmission and pathogenesis, with direct implications for HIV treatment (7, 13). Unfortunately, traditional techniques to identify rare variants, such as molecular cloning, single-genome amplification, or quantitative real-time (qRT)-PCR, are either labor intensive or restricted to the detection of single variants, limiting their widespread use (8, 11, 12, 14).
New second-generation technologies have radically altered DNA sequencing. Recent work by our group and others has employed pyrosequencing for targeted ultradeep sequencing of short regions of the viral genome, including CD8+ T-lymphocyte epitopes and regions of known drug resistance mutations, demonstrating a practical method to identify extremely low-frequency viral variants (4, 15). While sequencing short regions is appropriate in certain circumstances, the region of interest must be identified in advance, and the effect of mutations in that region on the remaining genome is ignored. Studying the heterogeneity of HIV across the entire genome may provide insights into interactions between minor variants, improve our understanding of HIV evolution, and ultimately provide insights into disease pathogenesis.
In this study, we combined pyrosequencing with a transposon-based fragmentation method to allow powerful ultradeep sequencing of the full-length HIV and simian immunodeficiency virus (SIV) genomes, demonstrating a new and highly practical approach to study the complexity of the viral population within a host and identify minor variants on a genome-wide scale. While this study applied pyrosequencing to immunodeficiency viruses, this approach could be applied to any viral pathogen.
Genome-wide pyrosequencing of SIV.
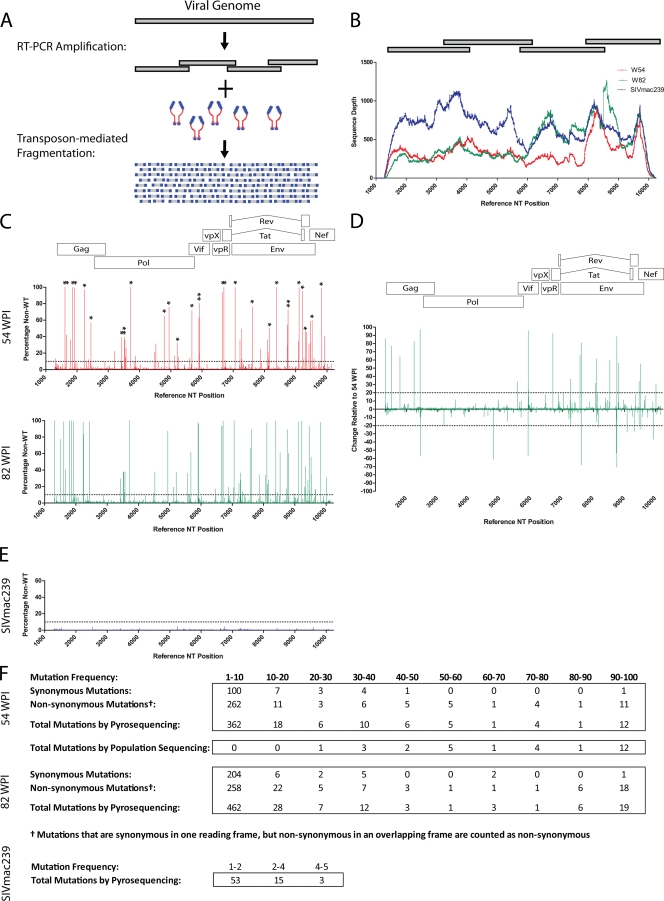
We first applied this approach to sequence virus from an Indian rhesus macaque experimentally infected with SIVmac239. We designed four overlapping reverse transcription PCR amplicons of approximately 2.5-kb each to span nucleotides 1269 to 10235 of the SIVmac239 genome, which includes all coding regions (Fig. 1A and B). We then performed reverse transcription PCR on plasma viral RNA isolated at two time points in chronic-stage infection, weeks 54 and 82. The reverse transcription PCR products were randomly fragmented using modified transposons (Nextera; Epicenter Biotechnology) and were then pyrosequenced. As a control, we sequenced an SIVmac239 viral stock. Sample preparation and pyrosequencing are described in greater detail at the following URL: https://xnight.primate.wisc.edu:8443/labkey/files/WNPRC/WNPRC_Laboratories/oconnor/public/publications/%40files/2010%20JV%20Bimber-Dudley%20et%20al%20Supplemental%20Material.pdf?renderAs=DEFAULT.
FIG. 1.
Genome-wide pyrosequencing of SIV. (A) Schematic representation of SIV amplification strategy and fragmentation. (B) Sequence coverage obtained for each SIV genome. Gray bars above graph denote RT-PCR amplicons. Sequence position is based on the SIVmac239 consensus reference sequence. (C) Graphs indicate the percentage of nucleotides that are non-wild-type at each position at 54 (top) and 82 (bottom) weeks postinfection. Asterisks above the graph for 54 wpi indicate mutations detected by conventional Sanger sequencing at that time point. The dotted lines represent mutations identified at a frequency of less than 10%. (D) Changes in mutation frequency between 54 and 82 weeks postinfection. (E) Graph of mutation frequency for each position of an SIVmac239 stock. (F) Summary of nonsynonymous and synonymous mutation frequencies from 54- and 82-wpi genomes. A mutation summary is also shown for the SIVmac239 stock. NT, nucleotide.
Intrahost viral diversity.
We obtained an average of 41,826 sequence reads per SIV genome. This provided an average coverage depth of 380 sequences across an 8.9-kb segment spanning the coding region (Fig. 1B). This deep coverage creates a high-resolution view of the viral population, not only revealing a large number of mutations but also capturing the frequency of each mutation within the population (Fig. 1C to F). The frequencies of each mutation in our SIV genomes varied widely, with only a small number of mutations approaching fixation, demonstrating the extensive heterogeneity of the viral population (Fig. 1C). Changes in the relative frequencies of variants may provide information about the fitness cost associated with a mutation or selective pressures of the host. Note that the majority of detected mutations are present in <10% of the viral population. Many of these mutations may be selectively neutral or nearly so and thus subject to genetic drift, although they contribute to overall genetic diversity and their presence may enable the virus to respond more rapidly to changes in selection pressure. Areas with sustained low levels of nonsynonymous mutations may also serve to identify regions of the virus that are under selective pressure but are unable to escape due to functional constraints.
As a control, we compared the pattern of mutation detected by pyrosequencing with mutations detected by bulk population sequencing (https://xnight.primate.wisc.edu:8443/labkey/files/WNPRC/WNPRC_Laboratories/oconnor/public/publications/%40files/2010%20JV%20Bimber-Dudley%20et%20al%20Supplemental%20Material.pdf?renderAs=DEFAULT). All mutations detected by bulk population sequencing were also detected by pyrosequencing (Fig. 1C, asterisks), supporting the accuracy of the technique, although mutations present below 50% were reliably detected only by pyrosequencing (Fig. 1F). We also sequenced an SIVmac239 stock, identifying only low levels of variation (Fig. 1E and F).
Pyrosequencing data provide a unique opportunity to study the composition of the viral population. As expected, over the 7 months of this study the viral population diverged from SIVmac239 (see Table 3 at https://xnight.primate.wisc.edu:8443/labkey/files/WNPRC/WNPRC_Laboratories/oconnor/public/publications/%40files/2010%20JV%20Bimber-Dudley%20et%20al%20Supplemental%20Material.pdf?renderAs=DEFAULT). Concurrent with this divergence, there was a reduction in the relative frequency of nonsynonymous single nucleotide polymorphisms (SNPs) and a lower level of genetic diversity of nonsynonymous SNPs compared to that of synonymous SNPs (P < 0.001) (see Table 4 at https://xnight.primate.wisc.edu:8443/labkey/files/WNPRC/WNPRC_Laboratories/oconnor/public/publications/%40files/2010%20JV%20Bimber-Dudley%20et%20al%20Supplemental%20Material.pdf?renderAs=DEFAULT). These observations are consistent with purifying selection and may be at least partially explained by the size of the viral population. At 54 weeks postinfection (wpi), the animal had a comparatively small viral population, 6.8-e4 copies/ml, which increased to 7.5-e6 copies/ml by 82 wpi. Random mutations are constantly introduced into the viral population. The majority of mutations that introduce nonsynonymous mutations into coding regions are at least slightly deleterious. With a larger population size, purifying selection becomes more efficient, resulting in improved removal of deleterious mutations and creating a more homogeneous population (10). Supporting the model of more efficient purifying selection, we found a significant reduction over time in presumably deleterious mutations such as insertions, deletions, and premature stop codons but not in more neutral synonymous mutations (see Table 5 at https: //xnight.primate.wisc.edu:8443/labkey/files/WNPRC/WNPRC _Laboratories/oconnor/public/publications/%40files/2010 %20JV%20Bimber-Dudley%20et%20al%20Supplemental %20Material.pdf?renderAs=DEFAULT). We cannot rule out other potential explanations for these observations, such as changes in the nature of selective pressure over the course of infection. The overall heterogeneity of the viral population has been suggested to influence viral persistence, yet heterogeneity has been difficult to quantify (5). The ability to measure intrahost viral diversity may provide novel insights into disease pathogenesis.
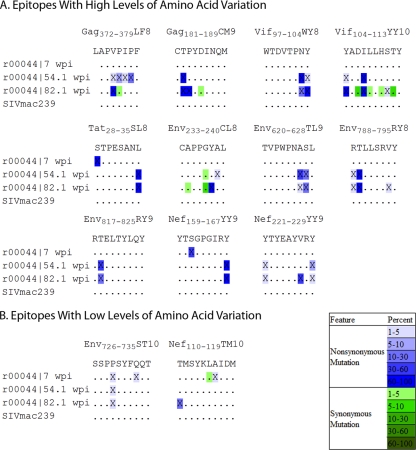
Cytotoxic T-lymphocyte (CTL) escape is one of the major forces driving intrahost evolution of SIV/HIV. The animal in this study expressed the well-characterized major histocompatibility complex (MHC) class I alleles Mamu-A1*001 and Mamu-A1*002 (formerly published as Mamu-A*01 and Mamu-A*02, respectively). We examined the pattern of CTL escape in 31 epitopes restricted by these alleles, using three time points, 7, 54, and 82 wpi (Fig. 2). Comparable with previously published work, we identified 11 epitopes with high levels of escape by week 82; however, the number of variants and the pattern of escape detected using pyrosequencing, including earlier kinetics of escape and the preservation of minor variants within the epitope, are considerably more complex than normally appreciated (Fig. 2A). We also detected sustained low-frequency mutations in 2 additional epitopes that would not otherwise be detected (Fig. 2B). This finding suggests that the CTL response may leave a much greater footprint on the genome than previously detected and that areas of low-frequency mutation may serve to identify putative CTL epitopes.
FIG. 2.
Cytotoxic T-lymphocyte escape. (A) Sequences of epitopes restricted by Mamu-A1*001 and Mamu-A1*002 with high levels of mutation. An SIVmac239 stock is included as a control. Mutations are color coded by frequency. (B) Sequences of epitopes restricted by Mamu-A1*001 and Mamu-A1*002 with low levels of mutation.
Genome-wide pyrosequencing of HIV.
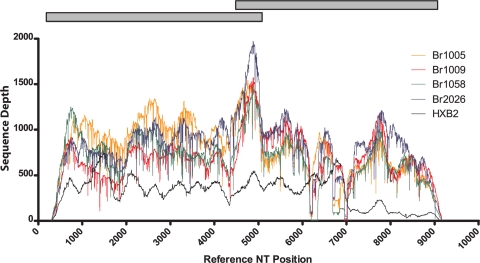
We next extended these studies to examine HIV. A challenge when sequencing HIV from patient isolates is the considerable sequence heterogeneity between and within patient samples. The approach we describe for SIV sequencing has considerable advantages for the sequencing of HIV because it requires relatively few virus-specific primers and a small amount of input material. Using methods similar to those described for SIV, we sequenced the coding regions from 11 HIV-positive patients, along with an HIV plasmid (pHXBnPLAP-IRES-N+). We aligned reads to the HXB2 reference strain and obtained full or nearly full coverage with libraries created from 50 ng of DNA (Fig. 3; see also Fig. 1 at https://xnight.primate.wisc.edu:8443/labkey/files/WNPRC/WNPRC_Laboratories/oconnor/public/publications/%40files/2010%20JV%20Bimber-Dudley%20et%20al%20Supplemental%20Material.pdf?renderAs=DEFAULT). We obtained an average of 29,000 sequence reads per genome, with a sequencing depth range of 208 to 846. More information about the PCR and sequencing methods may be found at https://xnight.primate.wisc.edu:8443/labkey/files/WNPRC/WNPRC_Laboratories/oconnor/public/publications/%40files/2010%20JV%20Bimber-Dudley%20et%20al%20Supplemental%20Material.pdf?renderAs=DEFAULT.
FIG. 3.
Genome-wide pyrosequencing of HIV. Sequence coverage obtained across the genome of a representative set of four HIV genomes and an HIV plasmid is shown. Gray bars represent RT-PCR amplicons used to amplify the HIV coding region for pyrosequencing. Sequence position is based on the HXB2 consensus reference sequence. BR1005, BR1009, etc., are numbers used to identify patients in the study.
Characterization of HIV genomes.
Using our methodology, we characterized several clinically relevant aspects of our patient viruses from a single sequencing run per patient. First, we created a consensus sequence representing the coding region of each patient and used the Rega HIV subtyping tool to determine the predominant subtype of each patient (6). As expected based on the origin of our patient samples (United States and Brazil), all of our samples were predicted as subtype B across the coding region, with a bootstrap value of 100%.
Next, we extracted all envelope sequences and utilized the Geno2Pheno [454] tool to determine the tropism of the virus infecting each patient (Table 1) (9). Tropism determined by hundreds of sequences per patient allows greater confidence in predicting the prevalence of CXCR4-tropic viruses than the use of a consensus sequence or sequences derived from multiple clones. We found evidence of CXCR4 tropism in 3 of 11 patients (Table 1) infected for >9 years. As expected, 100% of the sequences from our HXB2 plasmid control were CXCR4 tropic. Because pyrosequencing provides a clonal sequence, we were able to identify distinct variants within the V3 loop. While many patients had a fairly homogenous viral population in this region, in patients 115 and 116 we identified multiple distinct major variants circulating (Fig. 4). While the ability to resolve the linkage of mutations is limited to the length of a sequence read (∼400 bp), the sequence of short diagnostic regions may help distinguish patients coinfected with multiple distinct strains.
TABLE 1.
HIV tropisms predicted by the Geno2Pheno[454] typing tool, drug resistance mutations, and the corresponding drugs HIV-positive patients in this study have been exposed to previously or are currently takinga
| Patient | No. of yr infectedb | % CXCR4 tropic | Drug resistance mutation(s) | % sequences with mutation(s)c | Coverage depth at DR sited | Drugs taken by patient that may influence mutationse | Patient currently taking drugsb |
|---|---|---|---|---|---|---|---|
| 108 | >14 | 94.8 | K103N | 1.4 | 497 | EFV, NVP | No |
| G333E | 99.4 | 664 | 3TC | No | |||
| 113 | 20 | 0 | M41L | 23.2 | 283 | D4T, ddI | No |
| D67N | 4.1 | 276 | D4T, ddI | No | |||
| K219Q | 1.2 | 244 | D4T | No | |||
| 114 | >10 | 4.1 | D67N | 2.4 | 403 | TDF | No |
| K219Q | 2.7 | 333 | None | N/A | |||
| 115 | 9 | 7.1 | K65R | 1.3 | 73 | 3TC, FTC, TDF | Yes |
| F77L | 2.9 | 69 | ZDV | No | |||
| 116 | 1 | 0 | K219Q | 7.9 | 178 | None | N/A |
| G333E | 36.1 | 242 | None | N/A | |||
| 121 | 9 | 0 | L74I | 74 | 231 | None | N/A |
| A98G | 1.7 | 236 | EFV | No | |||
| K103N | 95.9 | 224 | EFV | No | |||
| V108I | 8.9 | 235 | EFV | No | |||
| M184V | 98.9 | 294 | 3TC | No | |||
| P225H | 92.3 | 234 | ZDV, 3TC | No | |||
| G333E | 100 | 346 | ZDV, 3TC | No | |||
| Br1005 | 2 | 0 | None | ||||
| Br2026 | 4 | 0 | None | ||||
| Br1058 | 2 | 0 | None | ||||
| Br1009 | <2 mo | 0 | D67N | 1.3 | 798 | None | N/A |
| Br1048 | <1 mo | 0 | D67N | 1.5 | 350 | None | N/A |
| HXB2_1 | N/A | 100 | None | ||||
| HXB2_2 | N/A | 100 | None |
The false-positive rate for predictions with the Geno2Pheno[454] typing tool was 1%.
N/A, not applicable.
Boldface type indicates that the background mutation was observed in the HBX2 vector control. Underlining indicates a low level of sequence coverage.
DR, drug resistance.
EFV, efavirenz; NVP, nevirapine; 3TC, lamivudine; D4T, stavudine; ddI, dideoxyinosine; TDF, tenofovir; FTC, emtricitabine; ZDV, zidovudine.
FIG. 4.
Variation within the Env V3 loop. Consensus sequences representing the major variants spanning the envelope V3 loop in patient 114 (A) and patient 115 (B) are shown. Nonsynonymous mutations are in yellow. Synonymous mutations are in green.
Lastly, we characterized drug resistance to reverse transcriptase and protease inhibitors. As shown in Table 1, we identified drug resistance mutations in eight patient samples. Four of eight patients were previously exposed to all the drugs for which they harbored resistance mutations, while the remaining patients had resistance mutations despite being antiretroviral therapy (ART)-naïve to those drugs. Despite the lack of current drug selection pressure in most patients, several mutants remained at very high percentages in the viral population.
Many minor and intermediate variants that may be missed by traditional sequencing methods were also found. Until recently, the ability to study minor drug resistance variants has been limited (<20%), but preliminary studies suggest that these variants may contribute to drug failure after initiation of ART (1, 13). With the expanded use of ART, it is especially important to detect minor drug resistance variants in ART-naïve individuals and to characterize the duration and frequency of resistance mutations prior to the failure of ART. Because detection of minor variants is sensitive to experimental artifacts, probing these mutations may require new analysis tools that take this into account (see the supplemental material at https://xnight.primate.wisc.edu:8443/labkey/files/WNPRC/WNPRC_Laboratories/oconnor/public/publications/%40files/2010%20JV%20Bimber-Dudley%20et%20al%20Supplemental%20Material.pdf?renderAs=DEFAULT).
Together, the SIV and HIV data demonstrate a practical method providing unparalleled resolution of the viral population on a genome-wide scale. The ability to readily identify and quantify viral variants should permit unprecedented assessment of viral diversity that can be applied to many viral pathogens.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants R21 AI068488-02, R21AI073230-02, and R01 AI077376 and by Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP) grant 04/15856-9. This publication was made possible in part by grants (P51 RR000167 and P40 RR019995) from the National Center for Research Resources (NCRR), a component of the NIH, to the Wisconsin National Primate Research Center (WNPRC), University of Wisconsin—Madison. This work was also funded by University of Wisconsin School of Medicine and Public Health Medical Education and Research Committee/Wisconsin Partnership Program grant 233KA28. This research was conducted in part at a facility constructed with support from Research Facilities Improvement Program grants RR15459-01 and RR020141-01. Analysis of data performed by Austin L. Hughes was funded in part by grant GM-43940, provided by the NIH.
We thank the WNPRC Virology Core for preparing the SIVmac239 inocula used in this study and the WNPRC animal care staff. We thank Chris Wright and the University of Illinois Sequencing Center. We thank Dawn Boh for assistance with recruiting patients and performing blood draws. We also thank members of the O'Connor lab for their careful reviews of the manuscript.
Footnotes
Published ahead of print on 15 September 2010.
REFERENCES
- 1.Abegaz, W. E., Z. Grossman, D. Wolday, et al. 2008. Threshold survey evaluating transmitted HIV drug resistance among public antenatal clinic clients in Addis Ababa, Ethiopia. Antivir. Ther. 13(Suppl. 2):89-94. [PubMed] [Google Scholar]
- 2.Allen, T. M., D. H. O'Connor, P. Jing, et al. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 3.Barouch, D. H., and N. L. Letvin. 2004. HIV escape from cytotoxic T lymphocytes: a potential hurdle for vaccines? Lancet 364:10-11. [DOI] [PubMed] [Google Scholar]
- 4.Bimber, B. N., B. J. Burwitz, S. O'Connor, et al. 2009. Ultradeep pyrosequencing detects complex patterns of CD8+ T-lymphocyte escape in simian immunodeficiency virus-infected macaques. J. Virol. 83:8247-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bordería, A. V., R. Lorenzo-Redondo, M. Pernas, et al. 2010. Initial fitness recovery of HIV-1 is associated with quasispecies heterogeneity and can occur without modifications in the consensus sequence. PLoS One 5:e10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Oliveira, T., K. Deforche, S. Cassol, et al. 2005. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics 21:3797-3800. [DOI] [PubMed] [Google Scholar]
- 7.Halvas, E. K., A. Wiegand, V. F. Boltz, et al. 2010. Low frequency nonnucleoside reverse-transcriptase inhibitor-resistant variants contribute to failure of efavirenz-containing regimens in treatment-experienced patients. J. Infect. Dis. 201:672-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keele, B. F., E. E. Georgi, J. F. Salazar-Gonzalez, et al. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lengauer, T., O. Sander, S. Sierra, et al. 2007. Bioinformatics prediction of HIV coreceptor usage. Nat. Biotechnol. 25:1407-1410. [DOI] [PubMed] [Google Scholar]
- 10.Li, W. H. 1978. Maintenance of genetic variability under the joint effect of mutation, selection and random drift. Genetics 90:349-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loh, L., and S. J. Kent. 2008. Quantification of simian immunodeficiency virus cytotoxic T lymphocyte escape mutant viruses. AIDS Res. Hum. Retroviruses 24:1067-1072. [DOI] [PubMed] [Google Scholar]
- 12.Loh, L., J. Petravic, C. J. Batten, et al. 2008. Vaccination and timing influence SIV immune escape viral dynamics in vivo. PLoS Pathog. 4:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macleod, I. J., C. F. Rowley, I. Thior, et al. 2010. Minor resistant variants in nevirapine-exposed infants may predict virologic failure on nevirapine-containing ART. J. Clin. Virol. 48:162-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peyerl, F. W., D. H. Barouche, H. S. Bazick, et al. 2005. Use of molecular beacons for rapid, real-time, quantitative monitoring of cytotoxic T-lymphocyte epitope mutations in simian immunodeficiency virus. J. Clin. Microbiol. 43:4773-4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simen, B. B., J. F. Simons, K. H. Hullsiek, et al. 2009. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J. Infect. Dis. 199:693-701. [DOI] [PubMed] [Google Scholar]