Abstract
Membrane fusion induced by enveloped viruses proceeds through the actions of viral fusion proteins. Once activated, viral fusion proteins undergo large protein conformational changes to execute membrane fusion. Fusion is thought to proceed through a “hemifusion” intermediate in which the outer membrane leaflets of target and viral membranes mix (lipid mixing) prior to fusion pore formation, enlargement, and completion of fusion. Herpes simplex virus type 1 (HSV-1) requires four glycoproteins—glycoprotein D (gD), glycoprotein B (gB), and a heterodimer of glycoprotein H and L (gH/gL)—to accomplish fusion. gD is primarily thought of as a receptor-binding protein and gB as a fusion protein. The role of gH/gL in fusion has remained enigmatic. Despite experimental evidence that gH/gL may be a fusion protein capable of inducing hemifusion in the absence of gB, the recently solved crystal structure of HSV-2 gH/gL has no structural homology to any known viral fusion protein. We found that in our hands, all HSV entry proteins—gD, gB, and gH/gL—were required to observe lipid mixing in both cell-cell- and virus-cell-based hemifusion assays. To verify that our hemifusion assay was capable of detecting hemifusion, we used glycosylphosphatidylinositol (GPI)-linked hemagglutinin (HA), a variant of the influenza virus fusion protein, HA, known to stall the fusion process before productive fusion pores are formed. Additionally, we found that a mutant carrying an insertion within the short gH cytoplasmic tail, 824L gH, is incapable of executing hemifusion despite normal cell surface expression. Collectively, our findings suggest that HSV gH/gL may not function as a fusion protein and that all HSV entry glycoproteins are required for both hemifusion and fusion. The previously described gH 824L mutation blocks gH/gL function prior to HSV-induced lipid mixing.
Membrane fusion is an essential step during the entry process of enveloped viruses, such as herpes simplex virus (HSV), into target cells. The general pathway by which enveloped viruses fuse with target membranes through the action of fusion proteins is fairly well understood. Viral fusion proteins use the free energy liberated during their own protein conformational changes to draw the two membranes—viral and target—together. Fusion is thought to proceed through a “hemifusion” intermediate, in which the proximal leaflets of the two bilayers have merged but a viral pore has not yet formed and viral contents have not yet mixed with the cell cytoplasm (10, 38). Fusion proteins then drive the completion of fusion, which includes fusion pore formation, pore enlargement, and complete content mixing.
HSV, an enveloped neurotropic virus, requires four glycoproteins—glycoprotein B (gB), glycoprotein D (gD), glycoprotein H (gH), and glycoprotein L (gL)—to execute fusion (9, 57, 60). gB, gD, and gH are membrane bound; gL is a soluble protein which complexes with gH to form a heterodimer (gH/gL). HSV-1 gH is not trafficked to the cell or virion surface in the absence of gL (32, 52). The requirement of four entry glycoproteins sets HSV apart from other enveloped viruses, most of which induce fusion through the activity of a single fusion protein. Although the specific mode of HSV entry is cell type dependent—fusion with neurons and Vero cells occurs at the plasma membrane at neutral pH; fusion with HeLa and CHO cells involves pH-dependent endocytosis, and fusion with C10 cells involves pH-independent endocytosis (42, 45)—all routes of entry require gD, gB, and gH/gL. Furthermore, although some discrepancies between virus-cell and cell-cell fusion have been observed (8, 44, 55, 58), both generally require the actions of gD, gB, and gH/gL.
Much work has gone toward the understanding of how the required HSV entry glycoproteins work together to accomplish fusion, and many questions remain. After viral attachment, mediated by glycoprotein C and/or gB (54), the first step in HSV fusion is thought to be gD binding a host cell receptor (either herpesvirus entry mediator [HVEM], nectin-1, nectin-2, or heparan sulfate modified by specific 3-O-sulfotransferases) (56). The gD-receptor interaction induces a conformational change in gD (39) that is thought to trigger gD-gB and/or gD-gH/gL interactions that are required for the progression of fusion (1-4, 13, 18, 23, 49).
gB and gH/gL are considered the core fusion machinery of most herpesviruses. The HSV-1 gB structure revealed surprising structural homology to the postfusion structures of two known viral fusion proteins (31, 35, 51). This structural homology indicates that despite not being sufficient for HSV fusion, gB is likely a fusion protein. Although the gB cytoplasmic tail (CT) is not included in the solved structure, it acts as a regulator of fusion, as CT truncations can cause either hyperfusion or fusion-null phenotypes (5, 17). The gB CT has been proposed to bind stably to lipid membranes and negatively regulate membrane fusion (12). Another proposed regulator of gB function is gH/gL. Despite conflicting accounts of whether gD and a gD receptor are required for the interaction of gH/gL and gB (1, 3, 4), a recent study indicates that gH/gL and gB interact prior to fusion and that gB may interact with target membranes prior to an interaction with gH/gL (2). The gB-gH/gL interaction seems to be required for the progression of fusion.
Compared to the other required HSV entry glycoproteins, the role of gH/gL during fusion remains enigmatic. Mutational studies have revealed several regions of the gH ectodomain, transmembrane domain (TM), and CT that are required for its function (19, 25, 26, 30, 33). gH/gL of another herpesvirus, Epstein-Barr virus (EBV), have been shown to bind integrins during epithelial cell fusion, and soluble forms of HSV gH/gL have been shown to bind cells and inhibit viral entry in vitro (24, 46). However, the role of gH/gL binding to target cells in regard to the fusion process remains to be determined.
There are some lines of evidence that suggest that gH/gL is a fusion protein. The gH/gL complexes of VZV and CMV have been reported to independently execute some level of cell-cell fusion (14, 37). HSV-1 gH/gL has been reported to independently mediate membrane fusion during nuclear egress (15). In silico analyses and studies of synthetic HSV gH peptides have proposed that gH has fusogenic properties (20, 21, 25-28). Finally, of most importance to the work we report here, gH/gL has been shown to be sufficient for induction of hemifusion in the presence of gD and a gD receptor, further promoting the premise that gH/gL is a fusion protein (59). However, the recently solved crystal structure of HSV-2 gH/gL revealed a tight complex of gH/gL in a “boot-like” structure, which bears no structural homology to any known fusion proteins (11). The HSV-2 gH/gL structure and research demonstrating that gH/gL and gB interactions are critical to fusion (2) have together prompted a new model of HSV fusion in which gH/gL is required to either negatively or positively regulate the activity of gB through direct binding.
We wanted to investigate the ability of a previously reported gH CT mutant, 824L, to execute hemifusion. 824L gH contains a five-residue insertion at gH residue 824, just C-terminal of the TM domain. 824L is expressed on cell surfaces and incorporated into virions at levels indistinguishable from those of wild-type gH by either cell-based ELISA or immunoblotting, yet it is nonfunctional (33). We relied on a fusion assay capable of detecting hemifusion, developed by Subramanian et al. (59), which we modified to include an additional control for hemifusion or nonenlarging pore formation, glycosylphosphatidylinositol (GPI)-linked hemagglutinin (GPI-HA). GPI-HA is a variant of the influenza virus fusion protein, HA, that is known to stall the fusion process before enlarging fusion pores are formed.
We were surprised to find that in our hands, gD, a gD receptor, and gH/gL were insufficient for the induction of hemifusion or lipid mixing in both cell-based and virus-based fusion assays. We found that gD, gB, and gH/gL are all required to observe lipid mixing. Further, we found that gB, gD, gL, and 824L gH are insufficient for lipid mixing. Our findings support the emerging view, based on gH/gL structure, that the gH/gL complex does not function as a fusion protein and does not insert into target membranes to initiate the process of fusion through a hemifusion intermediate. Our findings also further demonstrate that mutations in the CT of gH can have a dramatic effect on the ability of gH/gL to function in fusion.
MATERIALS AND METHODS
Cells, plasmids, and viruses.
Cell lines used were CHO cells, CHO cells stably expressing human nectin-1 (22) (CHO-nectin-1), Vero cells, and Vero cells stably expressing HVEM via the expression plasmid pBG20 (Vero-BG20). The CHO lines were grown in Ham's F-12 medium supplemented with 10% fetal bovine serum (FBS). The Vero lines were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% FBS. CHO-nectin-1 and Vero-BG20 cells were maintained in 500 μg/ml G418. The HSV glycoprotein plasmids used were as previously published (50). The mutant influenza hemagglutinin (HA) expression plasmid used encodes GPI-linked HA (GPI-HA) in pCB6 (generously provided by J. White, University of Virginia). Virus isolates used were HSV-1 (KOS) tk-12 (wild type [WT]), HSV-1 (KOS) gBKO82 (gB-null), and HSV-1 (KOS) gH87 (gH-null) (7, 43, 61).
Cell-based hemifusion assay.
The cell-based hemifusion assay was performed as previously described (59) with some modifications. CHO cells (effectors) were seeded onto glass coverslips in six-well plates and grown to 70 to 80% confluence (approximately 1 × 106 cells per well). For all HSV samples, effector cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) with 400 ng plasmid expressing gB and 300 ng each of plasmid expressing gD, gH, gL, and green fluorescent protein (GFP), with empty vector (pCAGGs) substituted as indicated for 1.6 μg total DNA per sample. For GPI-HA samples, effector cells were transfected using Lipofectamine 2000 with 400 ng plasmid expressing GPI-HA, 300 ng GFP, and 900 ng pCAGGs. For all samples, cells were transfected for 5 h and then returned to serum-containing medium for 12 h. For GPI-HA samples, HA was cleaved with 5 μg/ml trypsin from bovine pancreas and TPCK treated (Sigma, St. Louis, MO) for 5 min at room temperature. Trypsin was then quenched by three washes in serum-containing medium. Vero BG20 cells (targets) were added to all samples as described previously (59). For GPI-HA samples, targets were allowed to find effectors at 37°C for 20 to 30 min. Fusion was triggered by warmed fusion buffer, pH 5.3 (10 mM MES [morpholineethanesulfonic acid], 10 mM HEPES, 2 mM MgCl2, 2 mM CaCl2 in phosphate-buffered saline [PBS]), for 2 min. Cells were washed and returned to serum-containing medium. For HSV samples, targets were added to effectors directly and fusion was not triggered by pH. For all samples, effectors and targets were coincubated at 37°C for 10 h, washed with PBS, and fixed with fresh 4% paraformaldehyde. Cells were washed with PBS, and 2 μg/ml tetramethyl rhodamine isocyanate (TRITC)-conjugated cholera toxin, beta subunit (CTX) (CTX-555; Molecular Probes, Eugene, OR), was added for 30 min at 37°C. Cells were washed with PBS, stained with DAPI (4′,6-diamidino-2-phenylindole), washed, and mounted with Prolong gold anti-fade mounting medium (Invitrogen, Carlsbad, CA). Images were taken on a Zeiss LSM 510 Meta UV confocal microscope. Quantification of fusion and hemifusion was performed by counting GFP-positive and GFP/TRITC-double-positive cells.
Cell surface glycoprotein staining.
Cell surface expression of glycoproteins was probed using mouse monoclonal antibodies (MAbs) III-174 (gD), II-105 (gB), and 53S (gH/gL). All MAbs were diluted 1:500. Secondary goat anti-mouse IgG Texas Red was added at a 1:100 dilution (Invitrogen, Carlsbad, CA). Nonspecific binding was blocked by blocking buffer (10% donkey serum, 0.01% sodium azide, 0.1% Triton X-100).
Virus generation.
The titer of HSV-1 (KOS) tk-12 was determined on Vero cells and transiently complemented HSV-1 (KOS) gBKO82 (genotype gB-null), and the titers of HSV-1 (KOS) gH87 (genotype gH-null) virus stocks were determined on complementing cells lines Vero-VgB and Vero-VgH4, respectively, by blue plaque assay as previously described (59). Vero cells were infected at a multiplicity of infection (MOI) of 20 with tk-12, gBKO82, and gH87 viruses or mock infected to generate wild-type or phenotypically glycoprotein-null viruses as previously described (59). gB-complemented gB-null virus was generated as previously described (40). All viral supernatants were concentrated as previously described (59).
Western blot analysis.
Virus supernatants were boiled in loading buffer (2% SDS, 175 mM β-mercaptoethanol [BME]) and separated by electrophoresis on 4 to 20% gradient gels (Bio-Rad, Hercules, CA). Gels were transferred to polyvinylidene difluoride (PVDF) membranes and probed for VP5 using HSV ICP5 MAb (East Coast Bio, North Berwick, ME) diluted 1:3,000, anti-gB antiserum R74 diluted 1:10,000, and anti-gH-gL antiserum R137 diluted 1:10,000 (47) (generously provided by G. Cohen and R. Eisenberg, University of Pennsylvania). For sequential probing with different antibodies, membranes were stripped (100 mM BME, 62.5 Tris-HCl, pH 6.8, 20% SDS), washed, and reprobed. Blots were visualized by enhanced chemiluminescence, and densitometry was determined by ImageJ.
Virus-based fusion assay.
The virus-based hemifusion assay was performed as previously described (59). Briefly, concentrated virus-containing supernatants generated from Vero cell infections were added to 80% confluent CHO-nectin-1 cells labeled with 5 μM Celltracker blue 7-amino-4-chloromethylcoumarin (CMAC) (Molecular Probes) grown on coverslips in six-well plates. Cells were incubated with virus for 2 h at 37°C at an approximate MOI of 50. Where indicated, the HSV-neutralizing anti-gB antibody H1838 (48) (generously provided by Gary H. Cohen and Roselyn J. Eisenberg) was preincubated with virus at a 1:100 dilution for 1 h at 37°C. Cells were then treated with 0.1 M sodium citrate buffer (pH 3.0), washed, and fixed with fresh 4% paraformaldehyde. Then, 2 μg/ml TRITC-conjugated cholera toxin, beta subunit (CTX) (CTX-555; Molecular Probes, Eugene, OR), was added for 30 min at 37°C before cells were washed and mounted with Prolong gold anti-fade mounting medium (Invitrogen, Carlsbad, CA). Images were taken on a Zeiss LSM 510 Meta UV confocal microscope.
RESULTS
HSV-induced cell-cell lipid mixing requires all HSV entry glycoproteins.
Based on a model of HSV-induced membrane fusion in which gH/gL is capable of executing hemifusion in the presence of gD and a gD receptor (59), we aimed to test a gH mutant generated in a prior study (33) for hemifusion function. We utilized cell- and virus-based hemifusion assays based on previous work by others (59). The cell-based hemifusion assay detects lipid mixing or hemifusion during HSV-induced cell-cell fusion and differentiates it from content mixing or full fusion. This assay utilizes the absence of the common lipid ganglioside GM1 in the membrane of CHO cells (29) and the abundant presence of GM1 on Vero cells. Vero cells stably expressing the gD receptor HVEM (Vero-BG20) were used as target cells. Lipid mixing, measured by GM1 transfer from Vero-BG20 (target) cells to CHO (effector) cells, is measured using TRITC-conjugated cholera toxin β-subunit (CTX), which binds directly to GM1. Transient transfection of effector cells with GFP and HSV glycoproteins allows for differentiation of effector (GFP+/GM1−) and target (GFP−/GM1+) cells. Full fusion results in GM1 and GFP double-positive (GFP+/GM1+) multinucleated cells. Hemifusion results in GFP+/GM1+ single-nucleated cells. As a positive control for full fusion, effectors were transfected with GFP and the four required HSV entry glycoproteins—gD, gB, gH, and gL—and mixed with target cells.
We adapted the cell-based hemifusion assay to include an additional positive control for hemifusion or nonenlarging pore formation, glycosylphosphatidylinositol (GPI)-linked hemagglutinin (HA). Researchers have shown that replacing the transmembrane (TM) domain and cytoplasmic tail of the influenza virus fusion protein, HA, with GPI (GPI-HA) causes induction of the fusion process, which then stalls prior to transfer of soluble contents. Originally, GPI-HA was thought to stall HA-induced fusion at the hemifusion intermediate step (36). More recently it was shown that GPI-HA is capable of progressing the fusion process beyond hemifusion to nonenlarging pore formation (41). For our purposes, GPI-HA expressed in effector cells served as a positive control for lipid mixing in the absence of full fusion by transfecting effector cells with GFP and GPI-HA prior to mixing with GM1+ target cells.
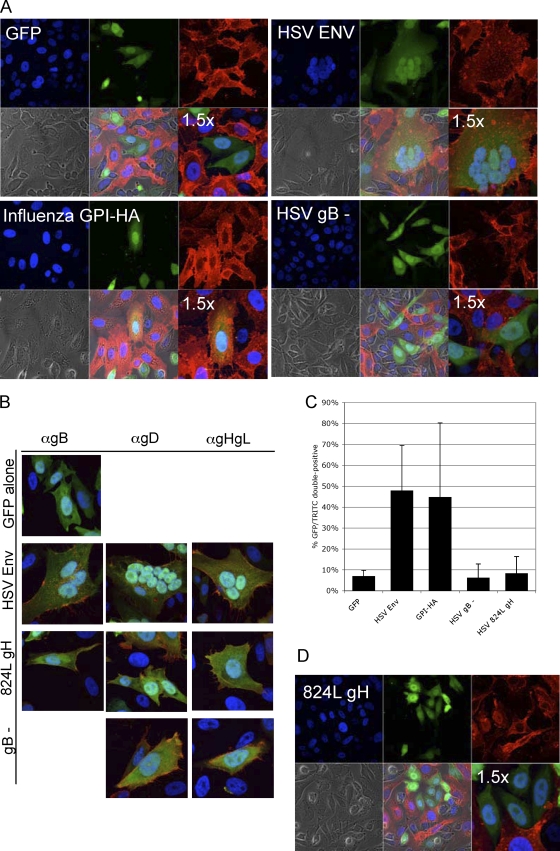
As expected, our positive control for full fusion, HSV envelope (gB, gD, gH, gL), produced GFP+/GM1+ multinucleated cells (Fig. 1 A). The average number of nuclei per syncytium was 4 (range, 2 to 10) (data not shown). Our positive control for lipid mixing in the absence of full fusion, GPI-HA, produced GFP+/GM1+ single-nucleated cells (Fig. 1A). Based on the work of Subramanian et al. (59), we expected HSV samples lacking gB, but not gH, to execute lipid mixing. However, we found little evidence of lipid mixing in samples lacking gB (Fig. 1A), despite gD, gH, and gL expression on the effector cell surface detectable by monoclonal antibody staining (Fig. 1B). The images shown are of a representative experiment. These data suggest that both gH/gL and gB are required for hemifusion or nonenlarging pore formation.
FIG. 1.
(A) Lipid mixing does not occur in the absence of gB. CHO cells (GM1−) were transfected with either GFP alone (top left), GFP and HSV gB, gD, and gH/gL (top right), GFP and GPI-HA (bottom left), or GFP and gD and gH/gL (bottom right). Transfected cells (green) were overlaid with Vero-BG20 cells (GM1+), fixed, and stained with TRITC-conjugated CTX to detect GM1 (red). Nuclei were stained with DAPI. Images were taken with a 63× objective on a Zeiss LSM 510 UV Meta laser scanning confocal microscope. Images representative of three independent experiments are shown. For clarity, a 1.5× magnification is also shown. (B) Cell surface glycoprotein expression. Glycoprotein expression in the cell-based hemifusion assay was confirmed by monoclonal antibodies. Mouse anti-gB (II-105), anti-gD (III-174), and anti-gH/gL (53S) antibodies were added to fixed, transfected effector cells in the presence of blocking buffer. Goat anti-mouse IgG conjugated to Texas Red detected primary monoclonal antibodies. Images were taken with a 63× objective on a Zeiss LSM 510 UV Meta laser scanning confocal microscope. (C) Quantification of cell-based hemifusion results. A minimum of 10 fields of view (FOV) per sample were imaged in each independent experiment. In each FOV, the numbers of GFP+ cells and GFP+/GM1+ cells were counted. The total number of GFP+/GM1+ cells was divided by the total number of GFP+ cells per experiment to determine the percentage of GFP+/GM1+ cells. Mean values of three independent experiments are shown, with standard deviations between results of experiments. (D) 824L gH is incapable of executing lipid mixing. Cells transfected with GFP, gB, gD, gL, and 824L gH were overlaid with Vero-BG20 cells (GM1+), fixed, and stained with TRITC-conjugated CTX to detect GM1. Images were taken with a 63× objective on a Zeiss LSM 510 UV Meta laser scanning confocal microscope. Images representative of results of two independent experiments are shown.
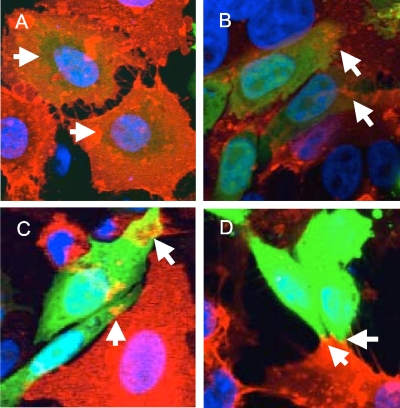
When quantifying the percentage of GFP+/GM1+ cells, we noted that the area of double-positive signal ranged from the whole cell to small areas of the cell (Fig. 2). We counted any area of double-positive signaling as evidence of lipid mixing. Cells with double-positive signals in a small portion of the cell were still scored as GFP+/GM1+. However, only two cells in all of the HSV gB fields of view quantified displayed the whole-cell double-positive signal that was typical of GPI-HA samples. We quantified our cell-based hemifusion results by randomly imaging multiple fields of view for each sample in multiple independent experiments and counting the number of GFP single-positive (GFP+/GM1−) and GFP+/GM1+ cells (Table 1). The average percentage of GFP+/GM1+ cells for HSV samples lacking gB (6.2%) was similar to that for the GFP-alone negative control (7.0%), both of which differed significantly from the average percentage of GFP+/GM1+ cells for HSV envelope (gD, gB, gH, gL) (48.0%) and GPI-HA (44.8%) (Fig. 1C). We conclude that our cell-based hemifusion assay is capable of detecting lipid mixing, as evidenced by the whole-cell GFP/GM1 double-positive signal induced by GPI-HA, and full fusion, detected by the GFP+/GM1+ signal within the syncytia of HSV envelope samples. Despite a liberally defined scoring scheme for GFP+/GM1+ cells, we did not observe significant lipid mixing in HSV samples lacking gB. We conclude that gH/gL is not sufficient for inducing lipid mixing, a process that appears to require the presence of both gH/gL and gB.
FIG. 2.
Sampling of the range of GFP+/GM1+ signal observed in our cell-based hemifusion assay. Each image includes two cells scored as GFP+/GM1+. (A) A representative GPI-HA sample with two GFP+/GM1+ cells. The TRITC signal surrounds the entirety of the GFP+ cells. (B and C) Representative samples lacking gB (GFP, gD, gH/gL) displaying two GFP+/GM1+ cells each. Arrows indicate regions of the cell with GFP/TRITC overlap. The area of TRITC/GFP overlap is limited; however, using liberal scoring, these cells were counted as positive for lipid transfer. (D) A representative 824L gH sample with two GFP+/GM1+ cells. Despite the limited lipid transfer, these cells were scored as positive for lipid transfer.
TABLE 1.
Quantification of cells
| Cell parameter | Result |
SD | |||
|---|---|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | Avg for two or more experiments | ||
| GFP-alone control (GFP, empty vector) | |||||
| No. of GFP+ cells | 112 | 108 | 52 | ||
| No. of GFP+ GM1+ cells | 10 | 9 | 2 | ||
| % GFP+ GM1+ | 8.9 | 8.3 | 3.8 | 7.0 | 2.8 |
| HSV envelope (GFP, gB, gD, gH, gL) | |||||
| No. of GFP+ cells | 41 | 22 | 89 | ||
| No. of GFP+ GM1+ cells | 14 | 16 | 33 | ||
| % GFP+ GM1+ | 34.1 | 72.7 | 37.1 | 48.0 | 21.5 |
| HSV gB− (GFP, gD, gH, gL) | |||||
| No. of GFP+ cells | 121 | 58 | 216 | ||
| No. of GFP+ GM1+ cells | 2 | 8 | 7 | ||
| % GFP+ GM1+ | 1.7 | 13.8 | 3.2 | 6.2 | 6.6 |
| 824L gH (GFP, gD, gB, gL, 824L gH) | |||||
| No. of GFP+ cells | 64 | 71 | |||
| No. of GFP+ GM1+ cells | 9 | 2 | |||
| % GFP+ GM1+ | 14.0 | 2.8 | 8.4 | 8.0 | |
| Influenza GPI-HA (GPI-HA, GFP, empty vector) | |||||
| No. of GFP+ cells | 76 | 22 | 129 | ||
| No. of GFP+ GM1+ cells | 20 | 18 | 29 | ||
| % GFP+ GM1+ | 26.3 | 81.7 | 22.5 | 44.8 | 35.0 |
gH-null and gB-null viruses do not mediate lipid transfer.
Herpesvirus cell-cell and virus-cell fusion are related but distinct processes (8, 44, 55, 58). In order to establish the minimal repertoire of HSV glycoproteins required for virus-cell lipid mixing, we used a virus-based hemifusion assay. Virus particles lacking gB or gH were generated by infecting Vero cells with genotypically gB- or gH-null viruses that had been propagated on complementing cell lines. Phenotypically complemented virus particles are capable of infecting Vero cells, undergoing replication, and producing GM1+ noninfectious virus lacking the indicated glycoprotein. Vero cells were infected at an MOI of 20 with either wild-type HSV-1 KOS tk-12 or complemented gB- or gH-null virus. In addition, Vero-VgB cells that express gB were infected with gB-null virus. After a 2-day incubation, virus was isolated from culture supernatants and concentrated.
Wild-type, transiently complemented, or null virus was added to CHO target cells at an approximate MOI of 50, based on wild-type virus titers. Because the titers of the null viruses could not be determined, null virus MOI was estimated based on Western blot densitometry of viral capsid bands (anti-VP5) relative to wild-type virus. After a 2-h infection, virus was neutralized with citrate buffer, and target cells were fixed. GM1 transfer from virus to CHO target cell expressing nectin-1 (CHO-nectin-1) was assessed using TRITC-conjugated CTX, as in the cell-based assay.
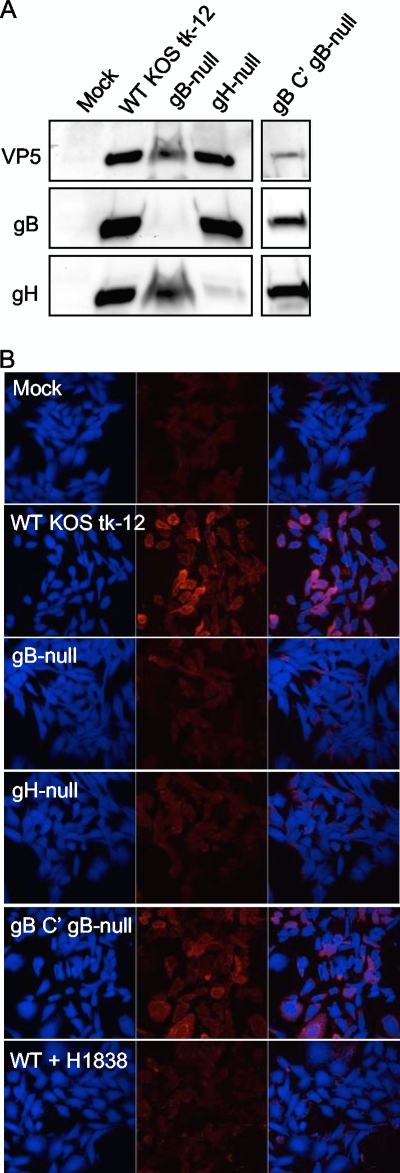
The concentrated virus-containing supernatants were probed with VP5, gB, and gH antibodies to confirm that null viruses lacked the appropriate glycoprotein and that relative levels of VP5 were similar between wild-type and null viruses (Fig. 3 A). Prior to incubation with virus, CHO-nectin-1 cells were stained with Celltracker blue, which exhibits blue fluorescence in the cytoplasm of live cells at physiological pH. As shown in Fig. 3B, CHO-nectin-1 cells incubated with HSV-1 KOS tk-12, but not gB- or gH-null virus, contained GM1 in their membranes. The specificity of viral membrane transfer was assessed by preincubating wild-type virus with an anti-gB virus-neutralizing antibody, H1838. Preincubation with H1838 abrogated GM1 transfer (Fig. 3B). To ensure that the lack of lipid transfer in gB-null samples was due to the absence of gB specifically, gB-null virus was phenotypically complemented with gB (Fig. 3A) and incubated with target cells. Complementation of gB-null virus with gB restored the virus-to-cell lipid transfer (Fig. 3B). We conclude that in both cell and viral fusion, both gB and gH/gL are required for induction of lipid mixing.
FIG. 3.
(A) Generation of WT KOS tk-12, gB-null, gH-null, and gB-complemented (C′) gB-null virus. The volume of concentrated virus-containing supernatants used in each virus-based hemifusion assay was examined by SDS-PAGE. Samples were separated on a 4 to 20% gradient gel, transferred to a PVDF membrane, and incubated with anti-VP5, anti-gB, and anti-gH antibodies. The blot shown is from a single gel transfer that was incubated with anti-VP5 antibody, stripped, and reprobed with anti-gB and then anti-gH antibody. This blot is representative of two separate virus preparations. (B) HSV-1 gB and gH are required for virus-cell lipid mixing. Celltracker blue-labeled CHO-nectin-1 cells were inoculated with virus-containing supernatants depicted in panel A at an approximate MOI of 50 for 2 h. Where indicated, virus was preincubated with anti-gB virus neutralizing antibody H1838. Cells were treated with citrate buffer, fixed, and stained with TRITC-CTX (red). Images were taken with a 40× objective on a Zeiss LSM 510 UV Meta laser scanning confocal microscope. Images representative of results of three independent experiments are shown.
Wild-type gH cytoplasmic tail is required for HSV-induced lipid mixing.
We next wanted to examine whether a gH cytoplasmic tail mutant, 824L, was capable of inducing lipid mixing in the presence of gD, gB, and gL. 824L gH contains a five-residue insertion mutation at residue 824L, within the short gH cytoplasmic tail. 824L gH is expressed at the cell surface and incorporated into virions normally (33). Mutations in the gH TM and CT, such as 824L, can dramatically reduce cell-cell and viral HSV fusion (6, 33, 34, 62).
We used the cell-based hemifusion assay to test the ability of 824L gH to execute hemifusion. Effector cells were transfected with gB, gD, gL, and 824L gH prior to mixing with target cells. We did not observe significant lipid mixing in 824L gH samples (Fig. 1D), despite cell surface expression of gH/gL detectable by anti-gH/gL monoclonal antibody (Fig. 1B). The average percentage of GFP+/GM1+ cells in 824L gH samples (8.4%) was similar to that of the GFP-alone negative control (7.0%) (Table 1; Fig. 1C). Thus, an alteration in the cytoplasmic tail of gH can abrogate gB- and gH/gL-mediated HSV lipid mixing.
DISCUSSION
At least two models of HSV-1 fusion with experimental support have been proposed. In one, the sequential model, gD binds receptor and triggers a fusion cascade in which gH/gL prompts hemifusion and gB supports the completion of fusion (23, 59). In another, termed either the regulatory, structural, or complex-formation model, once gD binds receptor and triggers fusion, gB is activated and receives signals or structural support from gH/gL, and the fusion process progresses to completion (2, 31, 39). Neither model can be considered comprehensive, as gB and gH/gL also have receptor-binding roles (24, 53), various routes of viral entry have different pH requirements, and each entry glycoprotein likely fulfills several required roles to accomplish fusion.
Our findings support the emerging view, based largely on the recently solved crystal structure of HSV-2 gH/gL, that despite being a required part of the HSV fusion core, gH/gL is not an HSV fusion protein. In our hands, gD, gB, and gH/gL were all required for lipid mixing, an intermediate stage of the fusion process. These results support the regulatory/structural/complex model of HSV fusion over the sequential model. Lipid mixing in the absence of full fusion was observed with GPI-HA, an HA variant known to stall fusion at nonenlarging pore formation. Both lipid mixing and full fusion were observed in the cell-based fusion assay when gD, gB, and gH/gL were expressed. Similarly, neither gB- nor gH-null HSV-1 was capable of transferring virus lipid membrane components into target membranes.
We noted relatively high deviations in the overall percentage of GFP+/GM1+ cells in HSV envelope and GPI-HA samples between experiments (Table 1). Variability between experiments may be simply the result of variable transfection efficiency but could alternatively highlight a weakness of this hemifusion detection strategy. In these cell- and virus-based assays the kinetics of hemifusion and fusion are not measured and the detection of both processes is not directly quantifiable. Hence, we used a liberally defined scoring scheme for GFP+/GM1+ cells that increased our confidence in our conclusions. In future studies, single-particle detection of HSV fusion using techniques similar to those developed to study influenza fusion (16) will be necessary for determining the kinetics of fusion and further unraveling the molecular steps required for HSV fusion.
If we presume for the time being that HSV gH/gL does not act as a viral fusion protein, we are left with many unanswered questions about its function during fusion. For some herpesviruses such as varicella-zoster virus (VZV) and cytomegalovirus (CMV), gH/gL appears capable of executing fusion in the absence of gB, yet HSV gH/gL cannot (14, 37). Are the roles of HSV and VZV or CMV gH/gL truly divergent, or do variable experimental conditions highlight differences that are not physiologically relevant? It seems unlikely that the structure of HSV-2 gH/gL would differ dramatically from the as-yet-unsolved gH/gL structures of other herpesviruses. It remains possible that the structurally unique HSV gH/gL still contains some fusogenic properties, a premise supported by the finding that HSV gH/gL is capable of executing fusion during nuclear egress in the absence of gB (15).
Our initial intent in this study was to screen a gH mutant for hemifusion function based on the premise that gH/gL alone is sufficient to induce hemifusion. After finding that in our hands, both gB and gH/gL were required for hemifusion, we aimed to determine whether a gH mutant that is not functional in full fusion could facilitate hemifusion in the presence of gB and thus differentiate between gH/gL functional domains required for fusion versus hemifusion. We found that a five-residue insertion at residue 824L in the CT of gH prevented lipid mixing when coexpressed with wild-type gB, gD, and gL in the cell-based hemifusion assay. 824L gH abrogated lipid mixing despite normal cell surface expression as detected by an anti-gH/gL monoclonal antibody. Regions outside the ectodomain of gH/gL and within its TM region and CT have also been shown previously to be crucial for its function (6, 33, 34, 62). The data we present here suggest that an intact gH/gL ectodomain is insufficient for triggering gB and prompting fusion. However, it is possible that the 824L gH mutation is affecting the gH ectodomain indirectly in such a way that disrupts either gB-gH/gL interaction or other functions of the gH ectodomain.
The recently described crystal structure of HSV-2 gH/gL and our work presented here both suggest that gH/gL may not play a direct role in mediating fusion. Rather, gH/gL may be required to either negatively or positively regulate the activities of gB. Finally, it will be of great interest to determine how mutations in the CT and TM can disrupt HSV-induced lipid mixing, a process that requires gD, gB, and gH/gL.
Acknowledgments
We thank Judith White for the generous gift of the pCB6 GPI-HA expression plasmid. We thank Robert Geraghty for the generous gift of Vero-BG20 cells. We thank Gary H. Cohen and Roselyn J. Eisenberg for the generous gift of the R137, R74, and H1838 antibodies and Nanette Susmarski for providing cells and her cell line expertise. We thank the members of the Longnecker, Spear, and Jardetzky laboratories as well as members of the Northwestern University Cell Imaging Facility for help and support.
This research was supported by CA117794 (R.L.). Imaging work was performed at the Northwestern University Cell Imaging Facility, generously supported by NCI CCSG P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center.
Footnotes
Published ahead of print on 15 September 2010.
REFERENCES
- 1.Atanasiu, D., J. C. Whitbeck, T. M. Cairns, B. Reilly, G. H. Cohen, and R. J. Eisenberg. 2007. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc. Natl. Acad. Sci. U. S. A. 104:18718-18723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atanasiu, D., J. C. Whitbeck, M. P. de Leon, H. Lou, B. P. Hannah, G. H. Cohen, and R. J. Eisenberg. 2010. Bimolecular complementation defines functional regions of herpes simplex virus gB that are involved with gH/gL as a necessary step leading to cell fusion. J. Virol. 84:3825-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avitabile, E., C. Forghieri, and G. Campadelli-Fiume. 2007. Complexes between herpes simplex virus glycoproteins gD, gB, and gH detected in cells by complementation of split enhanced green fluorescent protein. J. Virol. 81:11532-11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avitabile, E., C. Forghieri, and G. Campadelli-Fiume. 2009. Cross talking among the glycoproteins involved in herpes simplex virus entry and fusion: the interaction between gB and gH/gL does not necessarily require gD. J. Virol. 83:10752-10760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baghian, A., L. Huang, S. Newman, S. Jayachandra, and K. G. Kousoulas. 1993. Truncation of the carboxy-terminal 28 amino acids of glycoprotein B specified by herpes simplex virus type 1 mutant amb1511-7 causes extensive cell fusion. J. Virol. 67:2396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browne, H. M., B. C. Bruun, and A. C. Minson. 1996. Characterization of herpes simplex virus type 1 recombinants with mutations in the cytoplasmic tail of glycoprotein H. J. Gen. Virol. 77(pt. 10):2569-2573. [DOI] [PubMed] [Google Scholar]
- 7.Cai, W. H., B. Gu, and S. Person. 1988. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J. Virol. 62:2596-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cairns, T. M., R. S. Milne, M. Ponce-de-Leon, D. K. Tobin, G. H. Cohen, and R. J. Eisenberg. 2003. Structure-function analysis of herpes simplex virus type 1 gD and gH-gL: clues from gDgH chimeras. J. Virol. 77:6731-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campadelli-Fiume, G., M. Amasio, E. Avitabile, A. Cerretani, C. Forghieri, T. Gianni, and L. Menotti. 2007. The multipartite system that mediates entry of herpes simplex virus into the cell. Rev. Med. Virol. 17:313-326. [DOI] [PubMed] [Google Scholar]
- 10.Chernomordik, L. V., and M. M. Kozlov. 2005. Membrane hemifusion: crossing a chasm in two leaps. Cell 123:375-382. [DOI] [PubMed] [Google Scholar]
- 11.Chowdary, T. K., T. M. Cairns, D. Atanasiu, G. H. Cohen, R. J. Eisenberg, and E. E. Heldwein. 2010. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat. Struct. Mol. Biol. 17:882-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chowdary, T. K., and E. E. Heldwein. 2010. Syncytial phenotype of C-terminally truncated herpes simplex virus type 1 gB is associated with diminished membrane interactions. J. Virol. 84:4923-4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cocchi, F., D. Fusco, L. Menotti, T. Gianni, R. J. Eisenberg, G. H. Cohen, and G. Campadelli-Fiume. 2004. The soluble ectodomain of herpes simplex virus gD contains a membrane-proximal pro-fusion domain and suffices to mediate virus entry. Proc. Natl. Acad. Sci. U. S. A. 101:7445-7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole, N. L., and C. Grose. 2003. Membrane fusion mediated by herpesvirus glycoproteins: the paradigm of varicella-zoster virus. Rev. Med. Virol. 13:207-222. [DOI] [PubMed] [Google Scholar]
- 15.Farnsworth, A., T. W. Wisner, M. Webb, R. Roller, G. Cohen, R. Eisenberg, and D. C. Johnson. 2007. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc. Natl. Acad. Sci. U. S. A. 104:10187-10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Floyd, D. L., J. R. Ragains, J. J. Skehel, S. C. Harrison, and A. M. van Oijen. 2008. Single-particle kinetics of influenza virus membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 105:15382-15387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster, T. P., J. M. Melancon, and K. G. Kousoulas. 2001. An alpha-helical domain within the carboxyl terminus of herpes simplex virus type 1 (HSV-1) glycoprotein B (gB) is associated with cell fusion and resistance to heparin inhibition of cell fusion. Virology 287:18-29. [DOI] [PubMed] [Google Scholar]
- 18.Fusco, D., C. Forghieri, and G. Campadelli-Fiume. 2005. The pro-fusion domain of herpes simplex virus glycoprotein D (gD) interacts with the gD N terminus and is displaced by soluble forms of viral receptors. Proc. Natl. Acad. Sci. U. S. A. 102:9323-9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galdiero, M., A. Whiteley, B. Bruun, S. Bell, T. Minson, and H. Browne. 1997. Site-directed and linker insertion mutagenesis of herpes simplex virus type 1 glycoprotein H. J. Virol. 71:2163-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galdiero, S., A. Falanga, M. Vitiello, M. D'Isanto, C. Collins, V. Orrei, H. Browne, C. Pedone, and M. Galdiero. 2007. Evidence for a role of the membrane-proximal region of herpes simplex virus type 1 glycoprotein H in membrane fusion and virus inhibition. Chembiochem 8:885-895. [DOI] [PubMed] [Google Scholar]
- 21.Galdiero, S., A. Falanga, M. Vitiello, L. Raiola, R. Fattorusso, H. Browne, C. Pedone, C. Isernia, and M. Galdiero. 2008. Analysis of a membrane interacting region of herpes simplex virus type 1 glycoprotein H. J. Biol. Chem. 283:29993-30009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 23.Gianni, T., M. Amasio, and G. Campadelli-Fiume. 2009. Herpes simplex virus gD forms distinct complexes with fusion executors gB and gH/gL through the C-terminal profusion. J. Biol. Chem. 284:17370-17382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gianni, T., A. Cerretani, R. Dubois, S. Salvioli, S. S. Blystone, F. Rey, and G. Campadelli-Fiume. 2010. Herpes simplex virus glycoproteins H/L bind to cells independently of αVβ3 integrin and inhibit virus entry, and their constitutive expression restricts infection. J. Virol. 84:4013-4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gianni, T., R. Fato, C. Bergamini, G. Lenaz, and G. Campadelli-Fiume. 2006. Hydrophobic alpha-helices 1 and 2 of herpes simplex virus gH interact with lipids, and their mimetic peptides enhance virus infection and fusion. J. Virol. 80:8190-8198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gianni, T., P. L. Martelli, R. Casadio, and G. Campadelli-Fiume. 2005. The ectodomain of herpes simplex virus glycoprotein H contains a membrane alpha-helix with attributes of an internal fusion peptide, positionally conserved in the Herpesviridae family. J. Virol. 79:2931-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gianni, T., L. Menotti, and G. Campadelli-Fiume. 2005. A heptad repeat in herpes simplex virus 1 gH, located downstream of the alpha-helix with attributes of a fusion peptide, is critical for virus entry and fusion. J. Virol. 79:7042-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gianni, T., A. Piccoli, C. Bertucci, and G. Campadelli-Fiume. 2006. Heptad repeat 2 in herpes simplex virus 1 gH interacts with heptad repeat 1 and is critical for virus entry and fusion. J. Virol. 80:2216-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giraudo, C. G., C. Hu, D. You, A. M. Slovic, E. V. Mosharov, D. Sulzer, T. J. Melia, and J. E. Rothman. 2005. SNAREs can promote complete fusion and hemifusion as alternative outcomes. J. Cell Biol. 170:249-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harman, A., H. Browne, and T. Minson. 2002. The transmembrane domain and cytoplasmic tail of herpes simplex virus type 1 glycoprotein H play a role in membrane fusion. J. Virol. 76:10708-10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heldwein, E. E., H. Lou, F. C. Bender, G. H. Cohen, R. J. Eisenberg, and S. C. Harrison. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217-220. [DOI] [PubMed] [Google Scholar]
- 32.Hutchinson, L., H. Browne, V. Wargent, N. Davis-Poynter, S. Primorac, K. Goldsmith, A. C. Minson, and D. C. Johnson. 1992. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J. Virol. 66:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson, J. O., E. Lin, P. G. Spear, and R. Longnecker. 2010. Insertion mutations in herpes simplex virus 1 glycoprotein H reduce cell surface expression, slow the rate of cell fusion, or abrogate functions in cell fusion and viral entry. J. Virol. 84:2038-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones, N. A., and R. J. Geraghty. 2004. Fusion activity of lipid-anchored envelope glycoproteins of herpes simplex virus type 1. Virology 324:213-228. [DOI] [PubMed] [Google Scholar]
- 35.Kadlec, J., S. Loureiro, N. G. Abrescia, D. I. Stuart, and I. M. Jones. 2008. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat. Struct. Mol. Biol. 15:1024-1030. [DOI] [PubMed] [Google Scholar]
- 36.Kemble, G. W., T. Danieli, and J. M. White. 1994. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell 76:383-391. [DOI] [PubMed] [Google Scholar]
- 37.Kinzler, E. R., and T. Compton. 2005. Characterization of human cytomegalovirus glycoprotein-induced cell-cell fusion. J. Virol. 79:7827-7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozlovsky, Y., L. V. Chernomordik, and M. M. Kozlov. 2002. Lipid intermediates in membrane fusion: formation, structure, and decay of hemifusion diaphragm. Biophys. J. 83:2634-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krummenacher, C., V. M. Supekar, J. C. Whitbeck, E. Lazear, S. A. Connolly, R. J. Eisenberg, G. H. Cohen, D. C. Wiley, and A. Carfi. 2005. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J. 24:4144-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin, E., and P. G. Spear. 2007. Random linker-insertion mutagenesis to identify functional domains of herpes simplex virus type 1 glycoprotein B. Proc. Natl. Acad. Sci. U. S. A. 104:13140-13145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Markosyan, R. M., F. S. Cohen, and G. B. Melikyan. 2000. The lipid-anchored ectodomain of influenza virus hemagglutinin (GPI-HA) is capable of inducing nonenlarging fusion pores. Mol. Biol. Cell 11:1143-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milne, R. S., A. V. Nicola, J. C. Whitbeck, R. J. Eisenberg, and G. H. Cohen. 2005. Glycoprotein D receptor-dependent, low-pH-independent endocytic entry of herpes simplex virus type 1. J. Virol. 79:6655-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 44.Muggeridge, M. I. 2000. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. J. Gen. Virol. 81:2017-2027. [DOI] [PubMed] [Google Scholar]
- 45.Nicola, A. V., A. M. McEvoy, and S. E. Straus. 2003. Roles for endocytosis and low pH in herpes simplex virus entry into HeLa and Chinese hamster ovary cells. J. Virol. 77:5324-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parry, C., S. Bell, T. Minson, and H. Browne. 2005. Herpes simplex virus type 1 glycoprotein H binds to alphavbeta3 integrins. J. Gen. Virol. 86:7-10. [DOI] [PubMed] [Google Scholar]
- 47.Peng, T., M. Ponce-de-Leon, H. Jiang, G. Dubin, J. M. Lubinski, R. J. Eisenberg, and G. H. Cohen. 1998. The gH-gL complex of herpes simplex virus (HSV) stimulates neutralizing antibody and protects mice against HSV type 1 challenge. J. Virol. 72:65-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pereira, L., M. Ali, K. Kousoulas, B. Huo, and T. Banks. 1989. Domain structure of herpes simplex virus 1 glycoprotein B: neutralizing epitopes map in regions of continuous and discontinuous residues. Virology 172:11-24. [DOI] [PubMed] [Google Scholar]
- 49.Perez-Romero, P., A. Perez, A. Capul, R. Montgomery, and A. O. Fuller. 2005. Herpes simplex virus entry mediator associates in infected cells in a complex with viral proteins gD and at least gH. J. Virol. 79:4540-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pertel, P. E., A. Fridberg, M. L. Parish, and P. G. Spear. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313-324. [DOI] [PubMed] [Google Scholar]
- 51.Roche, S., S. Bressanelli, F. A. Rey, and Y. Gaudin. 2006. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 313:187-191. [DOI] [PubMed] [Google Scholar]
- 52.Roop, C., L. Hutchinson, and D. C. Johnson. 1993. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J. Virol. 67:2285-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Satoh, T., J. Arii, T. Suenaga, J. Wang, A. Kogure, J. Uehori, N. Arase, I. Shiratori, S. Tanaka, Y. Kawaguchi, P. G. Spear, L. L. Lanier, and H. Arase. 2008. PILRalpha is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell 132:935-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shukla, D., and P. G. Spear. 2001. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J. Clin. Invest. 108:503-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spear, P. G. 2004. Herpes simplex virus: receptors and ligands for cell entry. Cell. Microbiol. 6:401-410. [DOI] [PubMed] [Google Scholar]
- 56.Spear, P. G., R. J. Eisenberg, and G. H. Cohen. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1-8. [DOI] [PubMed] [Google Scholar]
- 57.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. 77:10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Subramanian, R. P., J. E. Dunn, and R. J. Geraghty. 2005. The nectin-1alpha transmembrane domain, but not the cytoplasmic tail, influences cell fusion induced by HSV-1 glycoproteins. Virology 339:176-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Subramanian, R. P., and R. J. Geraghty. 2007. Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc. Natl. Acad. Sci. U. S. A. 104:2903-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turner, A., B. Bruun, T. Minson, and H. Browne. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Warner, M. S., R. J. Geraghty, W. M. Martinez, R. I. Montgomery, J. C. Whitbeck, R. Xu, R. J. Eisenberg, G. H. Cohen, and P. G. Spear. 1998. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology 246:179-189. [DOI] [PubMed] [Google Scholar]
- 62.Wilson, D. W., N. Davis-Poynter, and A. C. Minson. 1994. Mutations in the cytoplasmic tail of herpes simplex virus glycoprotein H suppress cell fusion by a syncytial strain. J. Virol. 68:6985-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]