Abstract
The assembly of retroviruses is driven by oligomerization of the Gag polyprotein. We have used cryo-electron tomography together with subtomogram averaging to describe the three-dimensional structure of in vitro-assembled Gag particles from human immunodeficiency virus, Mason-Pfizer monkey virus, and Rous sarcoma virus. These represent three different retroviral genera: the lentiviruses, betaretroviruses and alpharetroviruses. Comparison of the three structures reveals the features of the supramolecular organization of Gag that are conserved between genera and therefore reflect general principles of Gag-Gag interactions and the features that are specific to certain genera. All three Gag proteins assemble to form approximately spherical hexameric lattices with irregular defects. In all three genera, the N-terminal domain of CA is arranged in hexameric rings around large holes. Where the rings meet, 2-fold densities, assigned to the C-terminal domain of CA, extend between adjacent rings, and link together at the 6-fold symmetry axis with a density, which extends toward the center of the particle into the nucleic acid layer. Although this general arrangement is conserved, differences can be seen throughout the CA and spacer peptide regions. These differences can be related to sequence differences among the genera. We conclude that the arrangement of the structural domains of CA is well conserved across genera, whereas the relationship between CA, the spacer peptide region, and the nucleic acid is more specific to each genus.
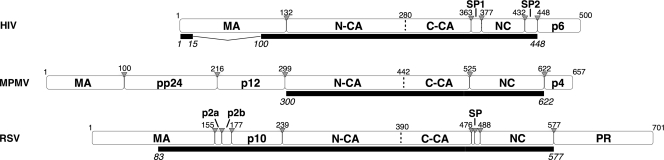
Retrovirus assembly is driven by the oligomerization of Gag, a multidomain protein, including an N-terminal membrane binding domain (MA), a two-domain structural component (CA), and an RNA binding domain (NC). The Gag proteins of all orthoretroviruses, including the alpha-, beta-, and lentiretroviruses discussed here, share this conserved modular architecture (Fig. 1). Despite very weak sequence conservation, the tertiary structures of MA, CA, and NC are conserved among retroviruses. Outside these conserved domains the Gag proteins of different retroviruses exhibit substantial variability. Other domains may be present or absent, and the length and sequence of linker peptides may also vary (12) (Fig. 1).
FIG. 1.
Modular architecture of the full-length Gag proteins of HIV, M-PMV, and RSV. White rectangles illustrate Gag polyprotein cleavage products. The extent of the constructs used in the electron microscopic analysis is specified under each protein as a black rectangle. Gray triangles specify cleavage sites. Residue numbers are counted from the beginning of Gag.
Oligomerization of Gag in an infected cell leads to the formation of roughly spherical immature virus particles, where Gag is arranged in a radial fashion with the N-terminal MA domain associated with a surrounding lipid bilayer, and the more C-terminal NC pointing toward the center of the particle (15, 44, 46). Subsequent multiple cleavages of Gag by the viral protease lead to a rearrangement of the virus. NC and the RNA condense in the center of the particle, CA assembles into a capsid or shell around the nucleoprotein, and MA remains associated with the viral membrane. This proteolytic maturation is required to generate an infectious virion (2). In contrast to the mature CA lattice, which has been extensively studied (11, 16, 36), the Gag lattice in immature particles is incompletely understood.
Gag itself contains all of the necessary determinants for particle assembly. For example, the expression of Gag alone in an insect cell expression system is sufficient to generate viruslike particles (3, 17, 22, 38). Retroviral Gag proteins also can be assembled in vitro in the presence of nucleic acids to form spherical particles (9, 19, 39, 43, 47). The arrangement of Gag within these in vitro-assembled Gag particles is indistinguishable from that found in immature virus particles (6), and the in vitro assembly systems have proved valuable for unraveling the principles of virus assembly (18, 28, 29, 39). Multiple layers of interaction promote the assembly of Gag in vivo, including MA-membrane-MA interactions, CA-CA interactions, and NC-RNA-NC interactions. An extensive body of literature has explored which regions of Gag are required for assembly and which can be replaced or deleted without compromising assembly. MA-membrane-MA interactions contribute but are not essential. NC-RNA-NC interactions appear to function to nonspecifically link Gag molecules together and can be replaced both in vivo and in vitro by other interaction domains such as leucine zippers (4, 13, 20, 32, 48). The C-terminal domain of CA (referred to here as C-CA) and the stretch of amino acids immediately following this domain (termed the spacer peptide [SP] region) are critical for assembly and sensitive to mutation (1, 22, 27, 30).
We set out to understand how the substantial sequence variation among Gag proteins in different retroviruses is manifested in structural differences in the immature Gag lattice. To do this, we studied three retroviruses from different genera: the lentivirus human immunodeficiency virus type 1 (HIV-1), the betaretrovirus Mason-Pfizer monkey virus (M-PMV), and the alpharetrovirus Rous sarcoma virus (RSV). These retroviruses are those for which in vitro assembly was first established and has been most extensively studied (6, 19, 24, 28, 29, 35, 43, 47).
The domain structures of the three retroviruses differ most substantially upstream of CA. Both M-PMV and RSV have domains located between MA and CA that are absent in HIV (Fig. 1). In M-PMV there are 198 residues forming the pp24 and p12 domains; in RSV there are 84 residues forming the p2a, p2b, and p10 domains. The three retroviruses have different requirements for regions upstream of CA during assembly. The C-terminal 25 residues of p10 are essential for proper immature RSV assembly, both in vitro and in vivo, and these residues are inferred to interact directly with N-CA to stabilize the hexamer by forming contacts between adjacent N-CA domains (35). An equivalent assembly domain has not been described for other retroviruses. Within M-PMV p12 is the so-called internal scaffolding domain that is not essential for assembly in vitro (43) but is required for particle assembly when the precursor is expressed under the control of the M-PMV promoter (41). It is a key domain for the membrane-independent assembly of immature capsids (40).
In HIV, five residues upstream of CA must be present for assembly of immature virus-like spherical particles in vitro, although larger upstream extensions, including part of MA, are required for efficient assembly of regular particles, both for HIV and RSV. For HIV, if the entire MA domain is included, in vitro assembly requires the presence of inositol penta- or hexakis phosphate (8). If no sequences upstream of CA are present, the in vitro particles in both of these viruses adopt a mature-type tubular morphology (10, 18). It has been hypothesized that cleavage at the N terminus of N-CA during maturation leads to the N-terminal residues of CA folding back into the N-CA structure to form a β-hairpin. The β-hairpin is important for assembly of the mature CA lattice, whereas its absence is important for immature assembly (23, 42). These requirements explain why, in HIV and RSV, immature Gag lattice-like structures are formed only if regions upstream of CA are present (18). In M-PMV, an immature Gag lattice can be produced when the regions upstream of CA are deleted if this is combined with mutations (such as deleting the initial proline of CA), which prevent β-hairpin formation (43).
During maturation, HIV and RSV Gag proteins are cleaved twice between CA and NC to release a small peptide called SP1 or SP. In RSV the most N-terminal of these two cleavages can occur at one of two possible positions such that the released peptide is either 9 or 12 amino acids long (33). In M-PMV only one cleavage occurs between CA and NC, and no short peptide is produced. The region between the final helix of CA and the Zn fingers has been proposed to adopt a helical bundle architecture in HIV and RSV based on bioinformatic prediction, on mutational analysis, and on structural studies (1, 22, 27, 45). In all three viruses, C-CA and the residues immediately downstream are critical for assembly and are sensitive to mutation. C-CA contains the major homology region, a group of residues that are highly conserved across the retroviruses.
Cryo-electron tomography (cET) studies of immature virus particles (6, 45) have resolved the electron density of the HIV Gag lattice in three dimensions at low resolution. Using these methods, we have also described the three-dimensional architecture of in vitro-assembled HIV Gag particles (6). In immature viruses and in vitro-assembled particles, Gag is seen to adopt an 8 nm hexameric lattice, as was predicted from previous Fourier analysis of two-dimensional images (7, 46). The hexameric lattice is interrupted by irregularly shaped holes and cracks in the lattice (6, 45). A similar observation has been made using AFM of in vitro-assembled particles of M-PMV Gag (26). These holes and cracks allow an otherwise planar hexameric lattice to form the surface of an approximately spherical particle.
The radial positions of the MA, CA, and NC domains had been assigned previously from cryo-electron micrographs (44, 46). Based on these assignments and the shape of the density, the position and relative orientations of CA domains can be modeled into the low-resolution structure of the HIV lattice (6, 45). Density ascribed to the N-terminal domain of CA (N-CA) forms rings around large holes at the 6-fold symmetry positions in the lattice. Below this layer, at the expected radius of the C-CA, are 2-fold densities, interpreted as corresponding to dimers of C-CA. These densities are linked by rodlike densities, which descend into the NC-nucleic acid layer.
HIV is the only retrovirus for which the arrangement of Gag in the immature particle has been described in three dimensions. Prior to this work, important open questions were therefore: which features of the arrangement of Gag are conserved between genera and therefore reflect general principles of Gag-Gag interactions, and which features are specific to certain genera? We have applied subtomogram averaging of cryo-electron tomograms to generate reconstructions of in vitro-assembled Gag particles from HIV, M-PMV, and RSV. These allow identification of the general and variable features of the arrangement of Gag and the architecture of immature retroviruses.
MATERIALS AND METHODS
Sample preparation.
Gag proteins were expressed in Escherichia coli and purified. Immature virus-like Gag particles were prepared by in vitro assembly of proteins with nucleic acid as described previously for HIV (19), M-PMV (26), or RSV (35). The constructs used are illustrated in Fig. 1. The nucleic acid was a 50-nucleotide (nt) single-stranded DNA (RSV), a 73-nt single-stranded DNA (HIV), or MS2 phage RNA (M-MPV). The assembled particles were kept at 4°C for at most 6 days until flash frozen in liquid ethane for analysis.
Electron microscopy and image processing.
In vitro-assembled particles of HIV, M-PMV, and RSV Gag were mixed with 10-nm gold beads, deposited on C-flat Holey carbon grids, and vitrified by plunge freezing in liquid ethane. Tilt series were collected on an FEI Tecnai F30 Polara transmission electron microscope with Gatan GIF 2002 post column energy filter and a 2kx2k Multiscan charge-coupled device camera. The data collection was performed at 300 kV using the University of California at San Francisco (UCSF) software package and tomograms reconstructed using the IMOD software package (25) as described previously (6). Tilt series were typically collected between −60° and +60° with an angular increment of 3° and a total electron dose of ∼70 e/Å2. Subtomogram averaging was carried out by using a six-dimensional search (14) in a reference-free manner as described previously (6). Independent reconstructions of half datasets were used to verify the reliability of the structures (see Results) and combined to produce the final reconstruction. Only subtomograms giving a cross correlation with the reference of greater than the mean cross correlation were included in the final reconstruction. The final reconstruction was filtered to the resolution to which the two half datasets are comparable, according to the Fourier shell correlation (see Fig. S3 in the supplemental material). The datasets are summarized in Table 1 .
TABLE 1.
Subtomogram data sets used in this study
| Virus | Defocus (μm) | No. of subtomograms |
Resolution (Å) | |
|---|---|---|---|---|
| Total | In final reconstruction | |||
| HIV | −4.5 | 12,220 | 9,698 | 28 |
| −2.1 | 10,231 | 5,438 | 25.5 | |
| M-PMV | −3.9 | 18,821 | 13,209 | 28 |
| −2.2 | 16,453 | 10,858 | 26.5 | |
| RSV | −3.8 | 9,802 | 4,853 | 28 |
| −2 | 11,001 | 6,732 | 23 | |
Lattice map representations were generated by using Amira (Visage Imaging), together with the EM Package (37). A hexagon is placed at the final aligned position of each tomogram and colored according to the cross-correlation value between the subtomogram and the average. Tomograms aligned to an inappropriate radial position were excluded based on the radii distribution of the set of 20 nearest subtomograms surrounding each subtomogram. If the radius of the selected subtomogram was in the first or in the fourth quartile of the distribution, the subtomogram was excluded. Only hexagons with a cross-correlation over a defined threshold are displayed. Structure interpretation and fitting were carried out using UCSF Chimera (34), which was also used to produce structure figures.
The angles plotted in Fig. 4B for each hexamer are 180° minus the mean angle between the 6-fold axis of a hexamer and the 6-fold axes of its immediate neighbors.
Bioinformatics.
To establish conserved retroviral features present or absent in our proteins of interest, 12 Gag protein sequences of the 27 International Committee on Taxonomy of Viruses (ICTV) reference species from these genera were chosen as follows. From the 27 ICTV reference species, those with incomplete Gag sequences or from defective viruses were not considered. To avoid overweighting the importance of very similar viruses, only one representative sequence was chosen from groups of viruses with sequence similarity in Gag greater than 70%, giving 12 sequences. These sequences were aligned by using MAAFT (21). Conserved features are those identified in >75% of the sequences.
The SP1 region is hypothesized to form an alpha-helix based on bioinformatic prediction, mutational analysis, and structural studies (1, 22, 27). A helical wheel representation of the 18 residues after the CA adjacent cleavage site was created to examine the helical hydrophobic moment of the region for the three viruses of interest. The complete region between the final helix of CA and the first zinc finger of NC was modeled in three dimensions as an alpha-helix using chimera (see Fig. S4 in the supplemental material).
RESULTS AND DISCUSSION
We describe here the local and global architecture of the Gag lattice in in vitro-assembled, immature Gag particles for three retroviral genera, with the aim of identifying general features of Gag assembly and those features which vary among genera. In vitro-assembled Gag particles were produced using expression vectors and assembly conditions that have previously been described (18, 35, 43). Each Gag protein represents approximately the minimal size protein for which in vitro assembly is efficient and correct and can occur without other additions (see the introduction). As a consequence, domains upstream of the conserved CA domains were variably truncated or absent (constructs are illustrated in Fig. 1).
The structure of the Gag lattice in in vitro-assembled immature particles.
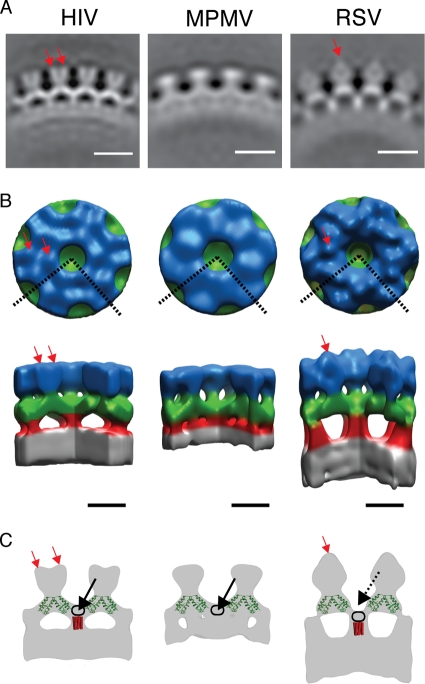
Vitrified, in vitro-assembled Gag particles were imaged by using cryo-electron tomography. Initial data collection was carried out at a defocus of approximately −4 μm. In order to assess the reliability of the reconstructions, for each type of in vitro-assembled Gag particle, the data set was split into two halves and two completely independent reference-free subtomogram averaging reconstructions were carried out. A second round of data collection was carried out with a defocus of approximately −2 μm. Again, the data set for each type of particle was split into two halves, and each half was reconstructed using a different reconstruction from the −4 μm data set as a starting model. Sections through these independent reconstructions are shown in Fig. S1 in the supplemental material. No significant differences were observed between independent reconstructions of particles assembled from the same Gag protein. The independent reconstructions were compared to one another by using Fourier Shell Correlation and were comparable to a resolution of 26.5 Å (see Fig. S2 in the supplemental material). This comparison of two independent reconstructions verified the robustness of the reconstruction process and allowed us to assess the resolution to which the reconstructions could be reliably interpreted. The two independent reconstructions were therefore aligned, averaged, and filtered to 26.5 Å to generate final reconstructions (Fig. 2A). The same process was carried out with in vitro-assembled Gag particles from each of the three different retroviruses. Isosurface representations of the reconstructions were generated (Fig. 2B) and colored for illustrative purposes.
FIG. 2.
Structure of the in vitro-assembled Gag particles. (A) Radial section through the average structures. The density is white. The scale bar is 10 nm. (B) Surface rendering of the average structures. The color scheme follows an approximate radial division of the structure according to the different domains of Gag as previously assigned (6, 44, 45, 46): gray is the NC-nucleic acid layer, red is the SP1 region, green is the C-CA region, and blue is the N-CA. The dashed lines on the top view represent the section cut out in the side view. Red arrows indicate densities protruding from the N-CA density (see the text). Scale bar, 5 nm. (C) Sections through the isosurface representations (gray), illustrating features referred to in the text. The crystal structure of the CA dimer (PDB: 3ds2) (green) is superimposed on the sections indicated previously (6). Red rods indicate possible positions of a hypothetical 24-Å six-helix bundle. Red arrows indicate densities protruding from the N-CA density. Black arrows indicate density forming the floor of the hole in the C-CA ring. Black rings indicate possible positions of the residues between the final helix in the C-CA and the six-helix bundle. For explanations, see the text.
The three different reconstructions exhibit substantial similarity. An outer density (blue in Fig. 2B) is arranged in hexameric rings around large holes. Where these rings meet, 2-fold densities (green in Fig. 2B) extend between adjacent 6-fold rings. Six of these 2-fold densities converge at each 6-fold axis, beneath the holes in the hexameric rings. Here they connect with a density (red in Fig. 2B), which descends further toward the center of the particle. This density meets the NC-nucleic acid layer (gray in Fig. 2B), which is not ordered in a hexameric manner. Despite these similarities, structural differences are seen at all radii of the Gag layer.
Outermost in the particles (blue in Fig. 2B) are the N termini of the Gag-derived proteins, where the three constructs used differ substantially (Fig. 1). The M-PMV construct begins directly with the N-CA domain, lacking the N-terminal proline residue. Accordingly, the M-PMV reconstruction shows the least featured outer density (blue in Fig. 2B), which can be presumed to represent N-CA.
The HIV construct includes the first 15 and last 33 residues of MA. This protein has been found to efficiently assemble in vitro (18). The reconstruction shows small protrusions around the holes at the 6-fold axis (red arrows in Fig. 2B and C). The extra MA residues may form these densities. Alternatively, the presence of part of MA may lead to a more rigid N-CA lattice than present in M-PMV, allowing individual N-CA domains to be more clearly resolved. In this case, the small protrusions could also represent part of N-CA.
The RSV construct includes p10, p2, and about one-half of MA, which together total 156 amino acid residues upstream of the CA domain. The reconstruction shows large extra densities above the 2-fold axes of N-CA (red arrow in Fig. 2A to C, visible as prominent caplike densities in Fig. 2A). The densities are positioned above the 2-fold axes of the lattice, where they link between adjacent hexamers. These densities could include the N-terminal part of p10 and perhaps sequences further upstream in the p2 and/or MA domains. In this model, each of the caplike densities would be formed from two p10 domains, one from each hexamer, which by linking adjacent hexamers would stabilize the hexameric lattice.
Moving toward the center of the reconstructions, the next layer can be assigned to the C-CA domain as described previously (6, 44-46) (green in Fig. 2B). If the crystal structure of the C-CA dimer (PDB: 3ds2) is superimposed onto the HIV density (as in reference 6 or Fig. 2C), the density occupied by the crystal structure can be assigned to the ordered part of the C-CA domain. The shape of this density is similar in all three reconstructions and, considering the high level of conservation of retroviral CA domains, the same crystal structure has also been superimposed on the RSV and M-PMV reconstructions for illustrative purposes. The density around the 6-fold axis that is not occupied by the crystal structure can be assigned to residues downstream of the final CA helix. In HIV and M-PMV the top of this region forms a density below the hexagonal hole created by the N-CA domains, at the center of the C-CA ring, forming the floor of the hole in the ring (black arrows in Fig. 2C). The position of this floor makes it most likely that it is formed from the residues immediately downstream of the final helix of C-CA. In RSV the floor is farther toward the center of the particle, making the hexagonal hole deeper (compare the positions of the solid and dashed black arrows for HIV and RSV in Fig. 2C).
The lower floor in the center of the C-CA hexamer hole in the RSV reconstruction suggests a difference in the arrangement of the residues immediately downstream of C-CA and in their interactions with the structured domain of C-CA. This suggests that the arrangements of C-CA, and the dimensions of the hexamer, which are well conserved across the retroviruses, do not require a specific conserved arrangement of the amino acid sequences that comprise the floor or a specific conserved interaction with the downstream structure present on the 6-fold axis. Instead, the conserved arrangement of C-CA is presumably predominantly mediated by interactions between CA dimers around the 6-fold axis.
The region between C-CA and the NC-nucleic acid layer is strikingly different in the three viruses. In RSV a strong rodlike feature descends from below C-CA to the NC-nucleic acid layer along the 6-fold axis (red in Fig. 2B). In HIV a similar feature is seen, but as described above the density begins within the C-CA layer, where it forms the floor of the hole at the 6-fold axis in the C-CA lattice. In M-PMV the floor is present, but there is no clear rodlike density between C-CA and the NC-nucleic acid layer.
Comparing structural differences with sequence differences.
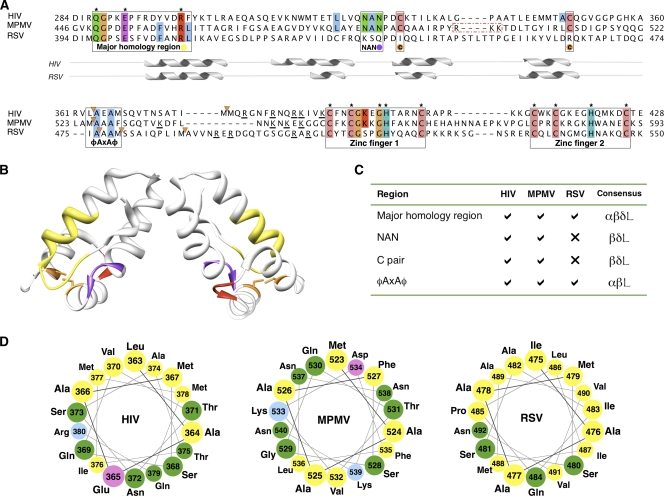
A sequence alignment of the C-CA-NC region highlights features conserved across multiple orthoretroviruses (see Materials and Methods and see Fig. S3 in the supplemental material). The alignment for the three viruses of interest is shown in Fig. 3A. This alignment identifies a number of previously described conserved features, the major homology region (MHR), nucleotide-binding Zn finger motifs, a CA-adjacent cleavage site, and a conserved cysteine pair. Most strikingly conserved is the MHR, which is known to be present in all orthoretroviruses. A pair of well-conserved cysteine residues, which approach close to one another between helices 3 and 4 of C-CA in the HIV CA crystal structure, is present in betaretroviruses, lentiviruses, and deltaretroviruses but absent in the alpharetroviruses. Not previously described is a conserved NAN motif in the turn between helices 2 and 3, which is also absent in the alpharetroviruses. These regions are highlighted on the structure of the C-CA dimer from HIV-1 in Fig. 3B, and summarized in Fig. 3C.
FIG. 3.
Comparisons of the Gag sequences from HIV, M-PMV, and RSV. (A) Multiple alignment of Gag C-CA-NC for RSV, M-PMV, and HIV. Regions of interest are boxed, and colored circles indicate the coloring of these regions in panels B and C. Residues conserved across >75% of viruses in an extended alignment, including 12 viral species (see Fig. S3 in the supplemental material), are colored using ClustalX coloring; residues conserved across all species are specified by an asterisk. Orange triangles denote cleavage sites. Basic residues adjacent to the NC are underlined. A dashed red box surrounds an RKK motif unique to M-PMV C-CA. The position of helices from the solved structures for RSV (PDB: 1d1d) and HIV (PDB: 3ds2) are shown under the alignment for illustrative purposes. Residues are numbered from the beginning of Gag. (B) HIV C-CA (PDB: 3ds2) with residues colored to indicate the positions of regions of interest: yellow, MHR; purple, NAN; orange, paired cysteine residues; and red, RKK motif (see the text and panels A and C). (C) Table of regions of interest defined in panel A; the colors correspond to the coloring in the multiple alignment (A) and HIV structure (A). Consensus signifies genera in which the feature is found (α signifies alpha, β signifies beta, δ signifies delta, and L signifies lentiretroviruses). There is currently no solved structure for the CA region in M-PMV, the final helix of M-PMV is defined assuming structural homology to HIV (PDB: 3ds2). (D) Helical wheel representation of the 18 residues after the CA adjacent cleavage site for the proteins of interest.
The conserved cysteine pair and NAN turn motif within C-CA, which are absent in RSV but present in HIV and M-PMV, are located away from the dimer axis, in a position that would lie close to the hole at the 6-fold axis (Fig. 3B). This is the region where the position of the floor differs between the retroviruses. We speculate that the cysteine pair and NAN sequence motifs may contribute to the organization of this region in HIV and M-PMV.
A stretch of amino acids roughly corresponding to the HIV-1 SP1 has been suggested to have helical propensity based upon computational prediction, mutational analysis, and nuclear magnetic resonance (NMR) (1, 31). It has been proposed that this region assembles into a six-helix bundle in the immature HIV-1 Gag lattice (45). We examined this proposal by considering the helical hydrophobic moment of this region (Fig. 3D and see Fig. S4 in the supplemental material). Both HIV and RSV show a 16-residue stretch, starting from the residue immediately upstream of the cleavage site and running into SP1 or SP, with a strong helical hydrophobic moment, a finding consistent with the assembly of this region into a six-helix bundle held together by hydrophobic side chains in its center. M-PMV does not show a strong helical hydrophobic moment. The calculated hydrophobic face in RSV is slightly larger than that in HIV. In both HIV and RSV, the 16-residue alpha-helical bundle would be a maximum of ∼24 Å long depending on the angle between the helices and the axis of the bundle. A 24-Å bundle is superimposed onto the structures in Fig. 2C for illustrative purposes.
The resolution of the reconstructions is not sufficient to resolve the secondary structural details, which would be necessary to describe the structure of the CA-NC linker region unambiguously, and multiple models are therefore consistent with the observed structures. In one such model, the region between the final helix of C-CA and the beginning of the proposed helical bundle is arranged to form the floor density in HIV (outlined by black ring in Fig. 2C), while the rodlike structure is formed by the proposed six-helix bundle. Below this region, basic residues close to the Zn fingers are involved in binding nucleic acid. In RSV, the region between the final helix of C-CA and the start of the helical bundle is not held in the floor position. It instead forms the top part of the rodlike structure (outlined by black ring in Fig. 2C). In the model, the lower part of the rod would then be formed by the six-helix bundle. In M-PMV the residues after the final helix of CA also form a floorlike structure as for HIV. The rodlike structure is not visible in M-PMV, and the nucleic acid directly abuts C-CA. This observation can be interpreted in two ways. Either the helical bundle is absent or the helical bundle is present despite the absence of a strong helical hydrophobic moment, but the rodlike density is obscured because it is buried in the nucleic acid layer. Presently available data do not allow these two possibilities to be distinguished.
Although we attribute the differences in tomographic density features in the NC region to differences in amino acid sequences among the three viruses studied, we cannot entirely exclude a role for the type of nucleic acid used to promote assembly. Whereas M-PMV particles were formed with RNA, the HIV and RSV particles were formed with DNA oligonucleotides. These respective nucleic acids were found previously to promote assembly in the most robust manner. We think it is unlikely that the HIV and RSV lattices are affected by the use of DNA since reconstructions of immature HIV particles (6, 45), which contain RNA, show the same arrangement of the NC layer.
The observation that the density interpreted to be the NC-nucleic acid layer directly abuts the base of the CA dimer in M-PMV suggests a possible direct interaction between the two. If the sequence of the C-CA domain of M-PMV is mapped onto the crystal structure of the HIV dimer, a basic RKK motif present in M-PMV but absent in RSV and HIV (see the dashed red boxes in Fig. 3A and the red area in Fig. 3B) is exposed on the underside of each monomer. These basic residues provide a potentially potent binding site for nucleic acid, and may explain the proximity of the nucleic acid layer to C-CA in the M-PMV structure. We hypothesize that such an interaction may be sufficient to pull the nucleic acid layer toward the CA layer, even if the Zn fingers are held at a distance from the CA layer by a rodlike structure, leading to the rod becoming buried within the nucleic acid layer.
Global arrangement of Gag in the in vitro-assembled Gag particles.
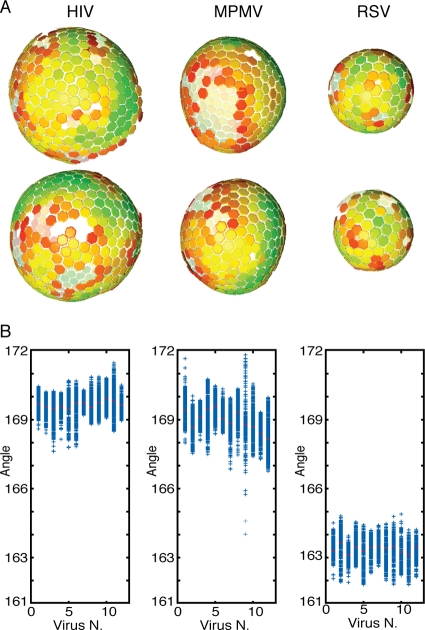
The global arrangement of the Gag shell within an individual in vitro-assembled particle can be visualized by placing a hexamer at the position and orientation to which each subtomogram has converged during the alignment and reconstruction procedure (6). Two example particles for each genus are illustrated in Fig. 4A. The relative positions and orientations of the placed hexamers are not random but instead are consistent with a hexameric lattice. All three types of particles are assembled from a single assembled hexameric lattice with irregularly shaped defects, indicating that this pattern is a conserved outcome of retroviral Gag protein assembly.
FIG. 4.
Global arrangement of Gag lattice in the particles. (A) Global lattice maps of in vitro-assembled Gag particles. The centers of each hexameric unit cell are marked with hexamers, which are colored according to cross correlation on a scale from low (red) to high (green). Higher cross-correlation values indicate that the subtomogram is more similar to the average structure. The cross-correlation range in the each map has been set between the minimum and the maximum cross-correlation value present in the map. Maps are shown in perspective, such that hexamers on the rear surface of the particle appear smaller and fainter. Two representative particles are shown for each genus. (B) Scatter plot showing the distribution of the angles between each hexamer and its neighbors (see Materials and Methods). The mean value for each particle is marked in red. For each construct, 12 particles have been analyzed.
The mean curvature of the particles is not the same in all cases. Similar to previous observations using conventional cryo-EM (5, 47) in vitro-assembled HIV Gag particles had a diameter of (94 ± 4 nm, n = 12), measured to the C-CA domain, which is similar to the diameter of M-PMV particles (90 ± 7 nm, n = 12) and significantly larger than RSV particles (60 ± 3 nm, n = 12). The differing diameters are reflected in the different curvatures of the lattice visible in Fig. 4A. This change in size reflects changes in the angle between adjacent hexamers, which in HIV and M-PMV are approximately 170° and 169°, compared to 163° in RSV (Fig. 4B). It is tempting to speculate that the tighter curvature in RSV may result from a difference in the relative orientations of the C-CA domains around the 6-fold axis, facilitated by the structural differences observed in the CA-SP1 region. This speculation is supported by the observation that, upon in vitro assembly of chimeric HIV-RSV Gag-derived proteins, the main determinant for particle size was CA (3). RSV CA was found to induce the formation of smaller, more tightly curved particles than HIV-CA in these chimeric proteins.
Although less dramatic than the differences between the genera, there is also variability in the angle between hexamers within each genus, when different particles are compared and also within individual particles. This variability is manifested in the formation of particles with slightly varying diameters and with deviations from a perfect spherical shape. Flexibility in the angle between the hexamers is a necessary basis for the formation of heterogeneous particles, a known feature of all retroviruses.
Summary.
The comparison of the immature Gag lattice from three retroviral genera highlights the general features of particle architecture. All three of the particles assemble from a single hexameric lattice with irregular defects. The arrangement of the two CA domains is strikingly conserved between different genera, whereas the angles between adjacent hexamers appear to vary, changing the degree of curvature and thus the particle size. Outside of the CA domains, sequence differences among the three genera are manifested as structural differences. Based on mutational data and chimeric Gag proteins containing foreign domains, the C-CA-SP region seems to contain the critical assembly determinants for the immature Gag shell. Nevertheless, the inferred structure of this region, and its relationship to CA and the NC-nucleic acid layer, appear to be specific to each genus.
Supplementary Material
Acknowledgments
We thank Pasi Laurinmäki and Sarah Butcher for sharing unpublished data.
This study was supported by the grants from the Deutsche Forschungsgemeinschaft within Schwerpunktprogramm 1175 to J.A.G.B. and H.-G.K., from the European Union (FP6 LSHP-CT-2007-036793; HIV PI resistance) to J.A.G.B. and H.-G.K., from the Czech Ministry of Education (1M6837805002 and MSM 6046137305) to T.R., and from the U.S. Public Health Service (CA20081) to V.M.V.
P.U., J.M.P., and V.L. prepared in vitro-assembled Gag particles, with contributions from T.F. A.D.M. and J.D.R. collected electron microscopy data. A.D.M. and J.A.G.B. analyzed data. N.E.D. contributed bioinformatics analyses. J.A.G.B., V.M.V., H.-G.K., and P.U. conceived the experiments. A.D.M. and J.A.G.B. wrote the paper with help from N.E.D., P.U., T.R., H.-G.K., and V.M.V. N.E.D. and A.D.M. prepared figures.
Footnotes
Published ahead of print on 1 September 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Accola, M. A., S. Hoglund, and H. G. Gottlinger. 1998. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J. Virol. 72:2072-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamson, C. S., and E. O. Freed. 2007. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv. Pharmacol. 55:347-387. [DOI] [PubMed] [Google Scholar]
- 3.Ako-Adjei, D., M. C. Johnson, and V. M. Vogt. 2005. The retroviral capsid domain dictates virion size, morphology, and coassembly of gag into virus-like particles. J. Virol. 79:13463-13472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borsetti, A., A. Ohagen, and H. G. Gottlinger. 1998. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J. Virol. 72:9313-9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briggs, J. A., M. C. Johnson, M. N. Simon, S. D. Fuller, and V. M. Vogt. 2006. Cryo-electron microscopy reveals conserved and divergent features of gag packing in immature particles of Rous sarcoma virus and human immunodeficiency virus. J. Mol. Biol. 355:157-168. [DOI] [PubMed] [Google Scholar]
- 6.Briggs, J. A., J. D. Riches, B. Glass, V. Bartonova, G. Zanetti, and H. G. Krausslich. 2009. Structure and assembly of immature HIV. Proc. Natl. Acad. Sci. U. S. A. 106:11090-11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briggs, J. A., M. N. Simon, I. Gross, H. G. Krausslich, S. D. Fuller, V. M. Vogt, and M. C. Johnson. 2004. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 11:672-675. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, S., R. J. Fisher, E. M. Towler, S. Fox, H. J. Issaq, T. Wolfe, L. R. Phillips, and A. Rein. 2001. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc. Natl. Acad. Sci. U. S. A. 98:10875-10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell, S., and V. M. Vogt. 1997. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J. Virol. 71:4425-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell, S., and V. M. Vogt. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardone, G., J. G. Purdy, N. Cheng, R. C. Craven, and A. C. Steven. 2009. Visualization of a missing link in retrovirus capsid assembly. Nature 457:694-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coffin, J. M., S. H. Huges, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 13.Crist, R. M., S. A. Datta, A. G. Stephen, F. Soheilian, J. Mirro, R. J. Fisher, K. Nagashima, and A. Rein. 2009. Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: implications for retrovirus assembly. J. Virol. 83:2216-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forster, F., O. Medalia, N. Zauberman, W. Baumeister, and D. Fass. 2005. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc. Natl. Acad. Sci. U. S. A. 102:4729-4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuller, S. D., T. Wilk, B. E. Gowen, H. G. Krausslich, and V. M. Vogt. 1997. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr. Biol. 7:729-738. [DOI] [PubMed] [Google Scholar]
- 16.Ganser-Pornillos, B. K., A. Cheng, and M. Yeager. 2007. Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell 131:70-79. [DOI] [PubMed] [Google Scholar]
- 17.Gheysen, D., E. Jacobs, F. de Foresta, C. Thiriart, M. Francotte, D. Thines, and M. De Wilde. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103-112. [DOI] [PubMed] [Google Scholar]
- 18.Gross, I., H. Hohenberg, C. Huckhagel, and H. G. Krausslich. 1998. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J. Virol. 72:4798-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross, I., H. Hohenberg, and H. G. Krausslich. 1997. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur. J. Biochem. 249:592-600. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, M. C., H. M. Scobie, Y. M. Ma, and V. M. Vogt. 2002. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 76:11177-11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katoh, K., K. Misawa, K. Kuma, and T. Miyata. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keller, P. W., M. C. Johnson, and V. M. Vogt. 2008. Mutations in the spacer peptide and adjoining sequences in Rous sarcoma virus Gag lead to tubular budding. J. Virol. 82:6788-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kingston, R. L., T. Fitzon-Ostendorp, E. Z. Eisenmesser, G. W. Schatz, V. M. Vogt, C. B. Post, and M. G. Rossmann. 2000. Structure and self-association of the Rous sarcoma virus capsid protein. Structure 8:617-628. [DOI] [PubMed] [Google Scholar]
- 24.Klikova, M., S. S. Rhee, E. Hunter, and T. Ruml. 1995. Efficient in vivo and in vitro assembly of retroviral capsids from Gag precursor proteins expressed in bacteria. J. Virol. 69:1093-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kremer, J. R., D. N. Mastronarde, and J. R. McIntosh. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71-76. [DOI] [PubMed] [Google Scholar]
- 26.Kuznetsov, Y. G., P. Ulbrich, S. Haubova, T. Ruml, and A. McPherson. 2007. Atomic force microscopy investigation of Mason-Pfizer monkey virus and human immunodeficiency virus type 1 reassembled particles. Virology 360:434-446. [DOI] [PubMed] [Google Scholar]
- 27.Liang, C., J. Hu, R. S. Russell, A. Roldan, L. Kleiman, and M. A. Wainberg. 2002. Characterization of a putative alpha-helix across the capsid-SP1 boundary that is critical for the multimerization of human immunodeficiency virus type 1 gag. J. Virol. 76:11729-11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma, Y. M., and V. M. Vogt. 2004. Nucleic acid binding-induced Gag dimerization in the assembly of Rous sarcoma virus particles in vitro. J. Virol. 78:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma, Y. M., and V. M. Vogt. 2002. Rous sarcoma virus Gag protein-oligonucleotide interaction suggests a critical role for protein dimer formation in assembly. J. Virol. 76:5452-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melamed, D., M. Mark-Danieli, M. Kenan-Eichler, O. Kraus, A. Castiel, N. Laham, T. Pupko, F. Glaser, N. Ben-Tal, and E. Bacharach. 2004. The conserved carboxy terminus of the capsid domain of human immunodeficiency virus type 1 Gag protein is important for virion assembly and release. J. Virol. 78:9675-9688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morellet, N., S. Druillennec, C. Lenoir, S. Bouaziz, and B. P. Roques. 2005. Helical structure determined by NMR of the HIV-1 (345-392) Gag sequence, surrounding p2: implications for particle assembly and RNA packaging. Protein Sci. 14:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muriaux, D., S. Costes, K. Nagashima, J. Mirro, E. Cho, S. Lockett, and A. Rein. 2004. Role of murine leukemia virus nucleocapsid protein in virus assembly. J. Virol. 78:12378-12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pepinsky, R. B., R. J. Mattaliano, and V. M. Vogt. 1986. Structure and processing of the p2 region of avian sarcoma and leukemia virus gag precursor polyproteins. J. Virol. 58:50-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 35.Phillips, J. M., P. S. Murray, D. Murray, and V. M. Vogt. 2008. A molecular switch required for retrovirus assembly participates in the hexagonal immature lattice. EMBO J. 27:1411-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pornillos, O., B. K. Ganser-Pornillos, B. N. Kelly, Y. Hua, F. G. Whitby, C. D. Stout, W. I. Sundquist, C. P. Hill, and M. Yeager. 2009. X-ray structures of the hexameric building block of the HIV capsid. Cell 137:1282-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pruggnaller, S., M. Mayr, and A. S. Frangakis. 2008. A visualization and segmentation toolbox for electron microscopy. J. Struct. Biol. 164:161-165. [DOI] [PubMed] [Google Scholar]
- 38.Royer, M., S. S. Hong, B. Gay, M. Cerutti, and P. Boulanger. 1992. Expression and extracellular release of human immunodeficiency virus type 1 Gag precursors by recombinant baculovirus-infected cells. J. Virol. 66:3230-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rumlova-Klikova, M., E. Hunter, M. V. Nermut, I. Pichova, and T. Ruml. 2000. Analysis of Mason-Pfizer monkey virus Gag domains required for capsid assembly in bacteria: role of the N-terminal proline residue of CA in directing particle shape. J. Virol. 74:8452-8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakalian, M., and E. Hunter. 1999. Separate assembly and transport domains within the Gag precursor of Mason-Pfizer monkey virus. J. Virol. 73:8073-8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sommerfelt, M. A., S. S. Rhee, and E. Hunter. 1992. Importance of p12 protein in Mason-Pfizer monkey virus assembly and infectivity. J. Virol. 66:7005-7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang, C., Y. Ndassa, and M. F. Summers. 2002. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Struct. Biol. 9:537-543. [DOI] [PubMed] [Google Scholar]
- 43.Ulbrich, P., S. Haubova, M. V. Nermut, E. Hunter, M. Rumlova, and T. Ruml. 2006. Distinct roles for nucleic acid in in vitro assembly of purified Mason-Pfizer monkey virus CANC proteins. J. Virol. 80:7089-7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilk, T., I. Gross, B. E. Gowen, T. Rutten, F. de Haas, R. Welker, H. G. Krausslich, P. Boulanger, and S. D. Fuller. 2001. Organization of immature human immunodeficiency virus type 1. J. Virol. 75:759-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wright, E. R., J. B. Schooler, H. J. Ding, C. Kieffer, C. Fillmore, W. I. Sundquist, and G. J. Jensen. 2007. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 26:2218-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeager, M., E. M. Wilson-Kubalek, S. G. Weiner, P. O. Brown, and A. Rein. 1998. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc. Natl. Acad. Sci. U. S. A. 95:7299-7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu, F., S. M. Joshi, Y. M. Ma, R. L. Kingston, M. N. Simon, and V. M. Vogt. 2001. Characterization of Rous sarcoma virus Gag particles assembled in vitro. J. Virol. 75:2753-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, Y., and E. Barklis. 1997. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J. Virol. 71:6765-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.