Abstract
The infectious salmon anemia virus (ISAV), an orthomyxovirus, is the major cause of outbreaks of high mortality rates in salmon in Chile. It has been proposed that the virulence of ISAV isolates lies mainly in hemagglutinin-esterase and fusion glycoproteins. However, based on current information, the contribution of other viral genes cannot be ruled out. To study this, we isolated and determined the complete coding sequence of two high-prevalence Chilean isolates associated with outbreaks of high mortality rates: ISAV752_09 and ISAV901_09. These isolates were compared to 15 Norwegian isolates that exhibit differences in their virulence. For this purpose, we performed bioinformatic analyses of (i) functional domains, (ii) specific mutations, (iii) Bayesian phylogenetics, and (iv) structural comparisons between ISAV and influenza virus glycoproteins by using molecular modeling. Phylogenetic analysis shows two genogroups for each protein, one of them containing the Chilean isolates. The gene sequence of the polymerase complex and nucleoprotein indicated that they are closely related to homologues from highly pathogenic Norwegian viruses. Notably, seven of the eight mutations that are present only in the Chilean isolates are on the polymerase complex and nucleoprotein. Structural modeling of hemagglutinin-esterase shows patches of variable residues on its surface. Fusion protein modeling shows that insertions are flexible regions that could affect proteolytic processing, increasing either the accessibility or the number of recognition sites for specific proteases. We found antigenic drift processes related to insertion into the isolated segment 5 of the ISAV752_09. Our results confirm the European origin of Chilean isolates to be the result of reassortments from Norwegian ancestors.
The infectious salmon anemia virus (ISAV) is a pathogen that principally affects Atlantic salmon, causing multisystemic disorders. It has been associated with high mortality in the aquaculture industry since 1984 (38). The cumulative mortality associated with each outbreak of ISAV in Norway and other countries is very high, reaching 100% in some cases (8, 25, 38, 45, 66). ISAV is a member of the Orthomyxoviridae family, and is the only member of the Isavirus genus (41, 52). ISAV shows a pleomorphic structure, with spiky projections composed of hemagglutinin-esterase protein. It interacts with the sialic acid receptor (28) and the fusion protein that induces the fusion between viral and endosomal membranes (4). Similar to influenza A and B viruses, ISAV displays eight segments of negative single-stranded RNA (10); it has been suggested that ISAV uses its own polymerase to copy and transcribe its genome. The function of most proteins encoded by the segments of ISAV has been assigned according to their similarities to the proteins encoded by the influenza A virus. Thus, polymerase, which in influenza A virus synthesizes both mRNA and vRNA, is constituted by three putative proteins coded by segment 1 (polymerase basic 2, PB2 [72]), segment 2 (polymerase basic 1, PB1 [41]), and segment 4 (polymerase acid, PA [3, 64, 72]). Segment 3 codes for nucleoprotein, NP, which participates in vRNA transport to the nucleus (3).
In the influenza A virus, hemagglutinin (HA) protein is coded by segment 4 and is responsible for sialic acid recognition (28), erythrocyte agglutination (20), and fusion between the viral membrane and the endosome (4). Unlike influenza A viruses, where hemagglutinin and fusion activity are present in the same polypeptide chain, fusion and hemagglutinin activity in ISAV correspond to two independent proteins coded by segments 5 and 6, respectively, making ISAV the first member of the Orthomyxoviridae family having these activities in different proteins (4). Furthermore, for ISAV, hemagglutinin protein displays a receptor-destroying activity in the same protein, like influenza C virus; hence, it is called hemagglutinin-esterase (HE) (40). The fusion protein in ISAV is synthesized as a precursor protein designated as F0. For fusion between viral and cellular membranes, F0 must be cleaved by cellular proteases to generate F1 and F2, which are held together by disulfur bridges. Fusion activity is significantly improved by HA (4). Segment 7 codes for two nonstructural proteins. The NS1 protein is coded by open reading frame 1 (ORF1) and exhibits interferon antagonist activity (24). As with influenza A virus, NS2 is a splicing product of the same transcript, which may correspond to a nuclear export protein since it contains nuclear export signals. A third protein was detected in TO cells infected with the Canadian RPC/NB 980-049-1 isolate that can be recognized by antibodies against NS1. Furthermore, the analysis of North American isolates seems to suggest the theoretical existence of this third protein, while the European isolates are predicted to generate a truncated protein (39, 48).
It has been suggested that segment 8 codes for two proteins from a bicistronic mRNA. The first ORF codes for the matrix protein, which is the major structural protein of the virion (5, 24, 77), and the second ORF codes for an M2 protein that displays two nuclear localization signals. M2 is able to bind RNA and displays anti-interferon activity (24).
Through the sequence comparison of segments 2 and 8, ISAV isolates were classified into two genotypes: European and North American. Both genotypes probably diverged in the year 1900, coinciding with the beginning of European salmon exports to America (6, 31, 42). With the appearance of more isolates, it was established that strains from Norway, Scotland, the Faroe Islands, and Nova Scotia belong to the European genotype. On the other hand, Canadian and North American isolates are considered North American genotypes (13, 36, 63). Based on the similarity to segments 2 and 8 of Canadian virus isolates in 2001, Chilean ISAV isolated from infected Coho salmon was initially classified as the North American genotype (34, 56). However, comparisons made in 2009 of 51 sequences of segment 5 and 78 sequences of segment 6 from Chilean isolates, obtained from Atlantic salmon since 2007, showed that Chilean isolates have a Norwegian origin. Evidence strongly suggests that ISAV was introduced in Chile as an avirulent strain that mutated into a virulent strain (35).
The most variable genomic elements of ISAV are located in segments that code for surface glycoproteins HE and F. The segment that encodes for HE has a mutation rate of 1.13 × 10−3 nucleotides (nt) per site per year and is the major determinant of variability. The variability of HE is located mainly in two regions, one localized in the N-terminal end, in the extracellular region of the protein. This region allows for classifying European isolates into three groups: group 1 contains only a few Norwegian isolates; group 2 contains isolates from Norway, one from the Faroe Islands and one from Scotland; and group 3 contains Scottish and Norwegian isolates (56).
The second region located in the C-terminal end of HE, in the extracellular portion of the protein, close to a transmembrane domain (62), is considered the major determinant of variability. This region is highly variable among genotypes, so it was therefore defined as a highly polymorphic region (HPR) (13). Although it has been proposed that an HPR is the result of recombination (13), the most plausible mechanism is that each HPR comes from partial deletions of precursor HPR0 (12). This is supported by the fact that HPR0 contains every HPR sequences described until now, with HPR0 being the longest sequence. In addition, HPR0 has been reported in healthy fish, suggesting that it is a nonvirulent precursor that could generate the virulent strains described for ISAV (11, 12, 50, 56) through multiple deletions of its sequence. Nearly 30 different HPRs have been identified. It has been suggested that the diversity of HPR sequences provides antigenic variability (56). HPR was proposed as a virulence marker, with the most virulent strains containing HPR4 (36, 37, 50, 51), but some authors do not agree with that conclusion (65). Although every HPR in Chile has been identified, the most abundant is the genotype 7b, which originates from Norway (35).
The other surface protein, the fusion protein, is also used to determine genetic variation in ISAV. The segment 5 sequence has a mutation rate of 0.67 × 10−3 nt per site per year (37), lower than that of HE. Close to the cleavage site, several isolates display an insertion of 8 to 11 amino acids (IN). To date, four insertions called IN1, IN2, IN3, and IN4 have been described. At the genomic level, IN1 is identical to the sequence of segment 3 from positions 1100 to 1123; IN2 is identical to the sequence from segment 5 located between nt 123 to 155, IN3 is identical to nt 93 to 122 of segment 5, and IN4 is identical to nt 399 to 429 of segment 2. This supports the notion of recombination between ISAV segments (14) as a source of variability. Notably, in an analysis of 51 Chilean isolates, 43 showed 11 amino acid insertions identical to IN4. This insertion has only been found in Chilean isolates (35). Recently, the IN region has been associated with virulence (25).
Little is known about the function of the proteins of ISAV; most of the conclusions are based on their homology with their putative counterparts in influenza viruses. We describe here the sequencing of the coding region and the genetic characterization of ISAV752_09 and ISAV901_09, which are the most common Chilean ISAV isolates. ISAV752_09 displays HPR 7b genotype and ISAV901_09 displays HPR 1c genotype. These viruses, which display different HPR and IN sequences, were analyzed for the presence of conserved domains, predicted structural domains, folding homologues, and phylogenetic relationships compared to sequences of coding regions of the European isolates.
MATERIALS AND METHODS
Bioinformatic analysis.
Homologues of proteins coded by ISAV genomes were analyzed at three levels: sequence, structure, and folding. Homologous sequences were identified by using BLAST (2) against a nonredundant database of sequence protein deposited in a gene bank. Structural homologues were identified through BLAST against the PDB database. Folding homologues were identified by using PHYRE (http://www.sbg.bio.ic.ac.uk/phyre/) (32) and ROBETTA (http://robetta.bakerlab.org/) servers (9). A cutoff of 60% certainty was used with PHYRE as a limit of folding homology. This means that at least 60% of amino acids are able to fold into this structure. ROBETTA server was used to perform a folding homology search with BLAST, PSIBLAST, HHSEARCH, and PFAM. An E-value cutoff of 0.001 was used for BLAST, PSIBLAST, and PFAM, and a cutoff probe of 85 was used for HHSEARCH. Homology modeling was performed by using Modeler 9v6 Linux 386 (18). Structural homologues were identified by using BLAST against the PDB databank or folding recognition servers. A total of 50 models were generated, and one of them was chosen according to their lowest value of the objective function. The quality of the models was evaluated using PROSA 3.0 (76) (https://prosa.services.came.sbg.ac.at/prosa.php). Structures were visualized with the VMD software (30). Global multialignment was performed with CLUSTAL W (75), using the BLUSOM 62 or BLUSOM 45 matrix. Local multi-alignments were performed by using DIALIGN2 (http://bibiserv.techfak.uni-bielefeld.de/dialign/) (53). RPS-BLAST (46) was used to analyze the conserved domains, with the lowest E-value as a cutoff (0.001). PHYRE server was used to predict secondary protein structures.
Cloning and sequencing genomes from ISAV752_09 and ISAV901_09 isolates.
Viral RNAs were isolated from kidneys of Salmo salar infected with ISAV, and the isolates were named ISAV752_09 and ISAV901_09 (gifts from the GAM Laboratory S.A). Total RNA was extracted by using a total RNA I kit (E.Z.N.A.; Omega Bio-Tek). Reverse transcription-PCR (RT-PCR) for each ISAV segment was carried out separately using the One-Step RT-PCR system with platinum Taq DNA polymerase (Superscript III; Invitrogen), according to the manufacturer's conditions. The following thermal program was used: 30 min at 50°C and 2 min at 94°C, followed by 39 cycles of 15 s at 94°C, 30 s at 55°C, and 30 s at 68°C, with a final extension of 5 min at 68°C. The primers used in each RT-PCR are listed in Table 1.
TABLE 1.
Primers used to amplify the coding region of Chilean isolates ISAV752_09 and ISAV901_09
| Segment | Primer sequence (5′-3′)a |
|
|---|---|---|
| Forward | Reverse | |
| 1 | CACCATGGACTTTATATCAGAAAACAC | TTAAACACCATATTCATCCATCAGGT |
| 2 | CACCATGGAAACTCTAGTAGGTGGG | TCAAACATGTTTTTCTTCTTAATCA |
| 3 | CACCATGGCCGATAAAGGTATG | TCAAATGTCAGTGTCTTCCTCCT |
| 4 | CACCATGGATAACCTCCGTGAA | TTATTGGGTACTGACTGCAATTTTC |
| CATGCCATGGATAACCTCCGTGAATGCATAAACC* | CCGCTCGAGTCATTGGGTACTGACTGCAATTTTC* | |
| 5 | CACCATGGCTTTTCTAACAATTT | TTATCTTCTAATGCATCCCCACAG |
| 6 | TTAAGCAACAGACAGGCTCGATG | CACCATGGCACGATTCATAATTT |
| 7 | CACCATGGATTTCACCAAAGTGTA | TTAATTCTCATTACAAATGTATTTTTCAAC |
| 8 | CACCATGAACGAATCACAATGGA | TTATTGTACAGAGTCTTCCAATTGGTC |
| CATGCCATGGACGAATCACAATGGATAC* | CCGCTCGAGTTATTGTACAGAGTCTTCCAATTG* | |
*, Primers specific for ISAV901_09.
PCR products for each segment were cut from 1% agarose gel, and cDNA was purified by using the E.Z.N.A. (Omega Bio-Tek) gel extraction kit. Purified cDNA was cloned in pGEMT Easy (Promega) according to the manufacturer's instructions and a previously described method (69). The identity of each cloned segment was further confirmed by DNA sequencing of both strands (Macrogen).
Sequences and GenBank access number.
The genomic sequences of ISA viruses chosen for the present study came from isolates with a completely sequenced coding region, which includes the sequences of 15 previously reported Norwegian viruses, plus the complete sequences of two Chilean ISA viruses presented in this study. The GenBank access numbers of the sequences used in the present study are GU830895 to GU830902 for the ISAV752_09 isolate and GU830903 to GU830910 for the ISAV901_09 isolate. The other sequences used here correspond to those previously reported by Markussen et al. (47) (GenBank accession numbers DQ785175 to DQ785285) and the sequences of the SK77-06 isolate (GenBank access numbers EU118815 to EU118822). The sequences used in the present study are summarized in text SA1 in the supplemental material.
Bayesian analysis.
In order to conserve the proper reading frame, each gene sequence was converted into amino acids by using the AlignmentHelper 1.0 program (49). A copy of the original nucleotide sequence was kept. Then each gene was aligned separately using MUSCLE (17), with default parameter settings. The aligned amino acid sequences were back translated into their original nucleotide sequence by using AlignmentHelper. Model selection for each gene was determined by using the Akaike information criterion (1), as implemented in jModelTest (26, 59). The best-fit models for most of the partitions corresponded to general time reversible (GTR) + G. Phylogenies were estimated using Bayesian Inference as implemented in BEAST 1.5.3 (16), with a relaxed uncorrelated lognormal molecular clock (15). Bayesian posterior probabilities were determined by running 200 million generations with a discarded burn-in of 10%, and the trees were sampled every 20,000 generations. Tracer 1.5 (61) was used to check for convergence and mixing. Branch support values are reported as posterior probabilities.
RESULTS
Phylogeny relationship.
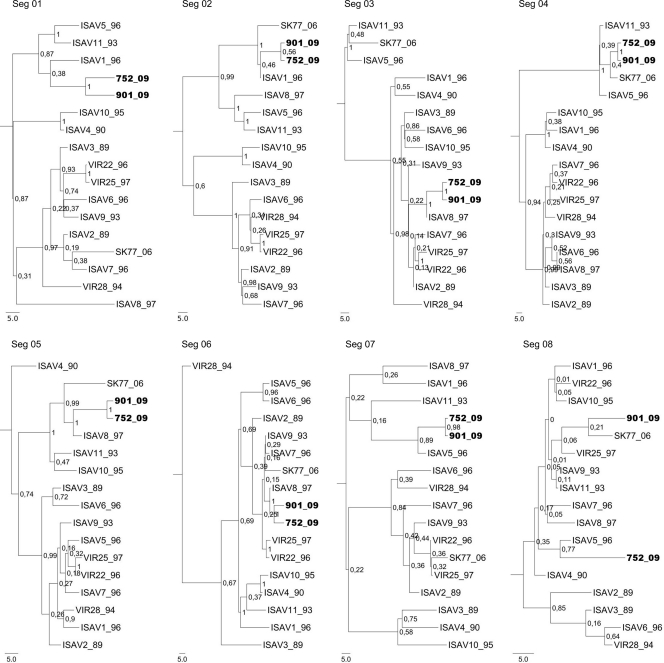
To determine the relation of segments of Chilean ISAV752_09 and ISAV901_09 isolates with 15 Norwegian isolates, a phylogenetic analysis was conducted using the Bayesian method. Global inspection of dendrograms shows that in all segments there are two main genogroups with a posterior probability (PP) of 1. The Chilean isolates in each tree belong to the same genogroup (Fig. 1). Among the Norwegian ISA virus, isolates ISAV1_96, ISAV5_96, ISAV8_97, ISAV11_93, and SK77_06 are associated with Chilean isolates. Within the corresponding genogroup, analysis of segments 1 and 2 shows that Chilean isolates share the same branch as ISAV1_96, with a low PP value (0.38 and 0.46, respectively). However, the interior nodes have a statistically significant PP value (0.87 for segment 1 and 1.0 for segment 2), showing that the Chilean isolates and ISAV1_96 belong to the same clade. Segments 3, 5, and 6 are associated with ISAV8_97, with a PP value of 1.0, 0.99, and 1.0, respectively, whereas segments 4 and 7 are in close association with SK77_06 and ISAV5_96, respectively, with a PP value of 0.4 and 0.89. In the case of segment 4, the main node of its clade has a PP of 1, indicating that both isolates are also in close association with ISAV11_93.
FIG. 1.
Phylogenetic trees of eight segments of the ISA viruses generated by the Bayesian method. The analysis was performed with the coding region for every segment for the 17 viruses used in the present study. The insertion of 33 and 30 nt in segment 5 of isolates ISAV752_09 and ISAV3_89, respectively, was eliminated for the analysis, as was the HPR region from segment 6. Node consistence was determined by the PP value. Its values are indicated in the nodes of the tree. Chilean isolates are shown in boldface type.
Unlike the other segments, sequences of segment 8 of each isolate present low variations with respect to the consensus sequence. Chilean isolate ISAV752_09 has two mutations, and ISAV901_09 has just one, but these isolates do not share any mutations (Fig. SA3 in the supplemental material). Nevertheless, the two Chilean isolates are arranged in the same genogroup, which displays low values of PP in the internodes (<0.35) due to the high identity among the sequences included in this analysis. The exception is the internode that groups ISAV752_09 and ISAV5_96 with a PP value of 0.77. This is because of ISAV752_09 sharing one mutation with ISAV5_96. On the other hand, the similarity between sequences of segment 8 from ISAV901_09 and ISAV77_06 allows placing them together in the same clade, even when they have a low PP value of 0.21 (Fig. 1).
The phylogenetic analysis using the MrBayes and Beast programs with the available sequences of the coding regions of all segments 8 present in the Gene Bank database did not improve the resolution of trees, resulting in low PP values, including ISAV901_09. Nevertheless, there was a clear difference between European and Canadian isolates, which are separated into two clades (data not shown). Given the failure to identify the particular ISA virus isolate containing a segment 8 closely related to the Chilean isolate ISAV901_09, BLAST and FASTA searches were performed to determine the segment with high identity. The results showed that segment 8 of ISAV901_09 is almost identical to VIR22, VIR25, ISAV11, ISAV9, with the same score. On the other hand, the other genogroup displays higher PP values, and includes ISAV2_89, ISAV3_89, ISAV6_96, and VIR28_94 isolated.
Comparisons of amino acid differences between Chilean strains and European isolates.
Alignment of the deduced amino acid sequences of the Chilean ISAV isolates with those of Norwegian isolates shows that each Chilean strain has 22 substitutions. For ISAV752_09, eight are nonconservative substitutions located in proteins PB1, PB2, NP, fusion, and HE, and 14 are conservative substitutions found in proteins PB2, PA, fusion, HE, NS1, NS2, and M1 (Table 2). For ISAV901_09, 14 are nonconservative substitutions located on PB1, NP, PA, fusion, HE, and M2. There are eight conservative substitutions distributed in PB2, PA, fusion, NS1, and NS2 proteins.
TABLE 2.
Comparison of amino acid differences between Chilean strains and other European isolates
| Isolate | Comparison of amino acid differencesa |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PB2 | PB1 | NP | PA | Fusion | HE | NS1 | NS2 | Matrix | M2 | |
| ISAV752_09/others | V/M 146 | F/Y 332 | T/I 67 | L/I 292b | T/A 58c | I/L 20 | E/D 170h | R/K 64i | IPS | A/V 19J |
| A/V 185 | E/D 471b | A/T 71 | L/I 29f | K/R 218 | ||||||
| P/S 229 | R/K 531 | G/N 252d | N/S 154g | |||||||
| N/S 263c | S/P 156 | |||||||||
| Q/L 277e | V/I 158 | |||||||||
| ISAV901_09/others | T/S 85 | R/H 707a | T/I 67 | T/I 204 | T/A 58c | N/S 154g | E/D 170h | R/K 64i | IPS | R/W 116 |
| S/A 105 | L/I 292b | A/T 71 | S/P 156 | |||||||
| S/A 558 | A/T 328 | G/N 252d | D/N 164 | |||||||
| E/D 471b | N/S 263c | G/D 247 | ||||||||
| R/K 531 | A/K 338 | |||||||||
Each value is indicated as first the amino acid difference and then the position number is given. Boldface, nonconservative amino acid substitution; IPS, identical protein sequences. Underlining indicates a novel mutation described for ISAV. Superscript letters: a, amino acid difference also found in virulent isolate ISAV1; b, amino acid difference also found in virulent isolates ISAV1, ISAV4, ISAV5, and low-virulence or avirulent isolates ISAV10, ISAV11, and SK779; c, amino acid difference also found in low-virulence or avirulent isolates ISAV8, ISAV10, ISAV11, and SK779; d, amino acid difference also found in low-virulence or avirulent isolates ISAV8 and SK779; e, amino acid difference also found in virulent isolate ISAV4 and avirulent isolate SK779/06; f, amino acid difference also found in virulent isolate ISAV8; g, amino acid difference also found in avirulent isolate SK779; h, amino acid difference also found in virulent isolates ISAV5, ISAV6, and ISAV7; i, amino acid difference also found in virulent isolates ISAV1, ISAV3, ISAV4, ISAV5, and low-virulence isolates ISAV8, ISAV10, and ISAV11; j, amino acid difference also found in virulent isolate ISAV5.
After several BLAST searches (data not shown), ISAV752_09 presented three new nonconservative substitutions (Table 2, underlined data), P229S in PB2 protein, F332Y in PB1 protein, and T67I in NP protein, which have not been previously reported. ISAV901_09 presented six new nonconservative mutations: T67I, S105A, and S558A in NP protein; T204I and A328T in PA protein; and R116W in M2 protein. Of the new substitutions, only one, at position 67 of the nucleoprotein, is shared by both strains (Table 2, underlined data).
Comparison between the proteins encoded by the isolated Chilean ISAV with other orthomyxoviruses.
In order to look for identity relationships in ISAV proteins, they were compared to other orthomyxoviruses (Table 3) . In general, ISAV proteins show very low identity with proteins from influenza A, B, and C and Thogoto viruses. With PB2, PB1, and PA proteins of ISAV, there was more identity with proteins from influenza virus B, rising to 12, 19, and 18%, respectively, whereas the lowest identity was between nonstructural proteins (NS1 and NS2) and matrix proteins, 8 and 10%, respectively. Analysis of conserved domains using RPSBLAST shows that only PB1 and HE from ISAV have conserved domains (pfam00602 and pfam06215, respectively), which include only members from ISAV proteins. Folding recognition software PHYRE shows that the PB2, PA, and NP proteins have a folding homologue with the bacteriophage N4 virion RNA polymerase (PDB ID 2PO4). In addition, the nucleoprotein of ISAV also shares folding with the nucleoprotein from influenza A virus (PDB ID 2IQH). The fusion protein has a folding homologue with the fusion domain from influenza A virus hemagglutinin (PDB ID 1HA0), and hemagglutinin esterase from ISAV has as a folding homologue the hyluranidase from Streptococcus pyogenes (PDB ID 2YX2) and hemagglutinin esterase fusion (HAF) protein from influenza C virus (PDB ID 1FLC). The ROBETTA server also identifies the nucleoprotein from influenza A virus and hemagglutinin esterase of the HAF protein from influenza C virus as nucleoprotein folding homologues.
TABLE 3.
Comparison of proteins encoded by members of the Orthomyxoviridae family, showing the percent identity, the structural homologue(s), and the folding homologue(s)
| Protein | Size (ISAV901_09) | % Identity with homologues in: |
Conserved domain | Folding homologue(s) |
||||
|---|---|---|---|---|---|---|---|---|
| Influenza A virus | Influenza B virus | Influenza C virus | Thogoto virus | PHYRE | ROBETTA | |||
| PB2 | 722 | 9 | 12 | 9 | 10 | NDb | 2PO4 | 2D59 |
| PB1 | 708 | 18 | 19 | 18 | 16 | pfam00602 | ND | ND |
| PA | 616 | 16 | 18 | 17 | 10 | ND | 2PO4 | ND |
| NP | 616 | 13 | 14 | 12 | 14 | ND | 2PO4, 2IQH | 2IQH |
| F | 444 | NAa | NA | NA | NA | ND | 1HA0, 2F2F | ND |
| HE | 388 | NA | NA | NA | NA | pfam06215 | 2YX2, 1FLC | 1FLC |
| NS1 | 300 | 10 | 8 | 10 | NA | ND | ND | ND |
| NS2 | 159 | 12 | 8 | 14 | NA | ND | ND | ND |
| Matrix | 241 | 8 | 10 | 6 | NA | ND | ND | ND |
| M2 | 196 | 9 | 11 | 9 | NA | ND | ND | ND |
NA, not applicable.
ND, not determined.
Homologues of surface protein in ISAV.
Using PHYRE, we showed that the fusion protein from ISAV can adopt folding similar to that of the hemagglutinin of influenza A virus, with a degree of certainty of 70% (Table 3). A previous report on the sequence of Canadian viruses suggested that the fusion protein of ISAV is a glycoprotein with two putative sites for O glycosylation, one putative site for N-glycosylation, a coiled-coil region, and two putative sites for proteolytic processing by a trypsinlike enzyme (4). In order to determine whether these sites are conserved, we performed a multialignment between every ISAV fusion protein reported in the GenBank database (data not shown). The alignment shows that putative O-glycosylation and N-glycosylation sites (4) are highly conserved; i.e., the T64 O-glycosylation site is 100% conserved. In the case of the T52 O-glycosylation site, even a highly conserved mutation T52M was detected in sequences AAX46235 and AAX46236. On the other hand, the putative N-glycosylation site N370 (numbering according to the fusion protein of ISAV752_09) is 100% conserved and N110 shows a N110D mutation in proteins AAX46238 and AAX46239. This high level of conservation suggests that these sites are indispensable for the function of fusion protein.
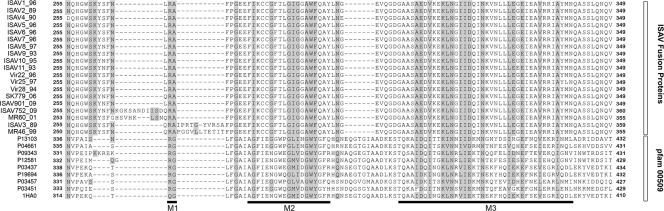
In order to identify functional relationships between the fusion protein of ISA and influenza viruses, a multialignment was performed by using CLUSTAL W with Blusom45. To compensate for the low identity between ISAV fusion and influenza virus hemagglutinin proteins, this alignment was constructed using the most diverse members of pfam00509 (conserved fusion domain present in influenza A virus HA protein), ISAV fusion proteins of all previous sequences, plus the fusion protein representatives of inserts containing IN1 (MR60_01) and IN2 (MR46_49), and the sequence of HA whose three-dimensional structure has been solved by X-ray diffraction (1HA0, Table 3) (Fig. 2). ISAV fusion proteins share an average of 11% identity with the most diverse members of the hemagglutinin family. This alignment shows at least three short motifs conserved between the ISAV fusion protein and influenza virus hemagglutinin proteins. The first (M1) region is located in the proteolytic processing site of the fusion peptide, indicating that an arginine is the residue of proteolytic processing in these two orthomyxoviruses, as has been reported by others (25). It is notable that in the influenza virus, arginine is followed by a glycine residue, but in ISAV arginine is followed by alanine, with both residues being nonpolar. The second (M2) region is a transmembrane region close to the catalytic processing site. This GIGGAWF sequence is totally conserved in ISAV, while GxxxxWx is conserved in the influenza virus. The third motif (M3) is located in the predicted coiled-coil region (4). In ISAV, the M3 motif containing the sequence AEDVKEKLNGIIDQINKVNLLLEGEIEAVRRIA is completely conserved, with the IDxIxxK(I/V/L)Nx(I/L/V)(I/V)(D/E)xxNxxxxx(I/V)(E/D/R/G/Q)xxExx(V/I/L)xxx(I/V/L) pattern of the function of the amino acid properties conserved in human influenza A virus, showing that the nature and position of the amino acid are conserved between the two orthomyxoviruses. The amino acids highlighted above are highly conserved between both orthomyxovirus members (Fig. 2).
FIG. 2.
Conserved regions between the fusion protein from influenza and ISA viruses. The figure shows a multialignment between most diverse members of the pfam00509 family and fusion proteins from ISAV. The shaded background indicates the conserved residues. The horizontal black bars show the putative proteolytic site (M1), the transmembrane of ISAV fusion protein (M2), and the coiled-coil (M3) regions.
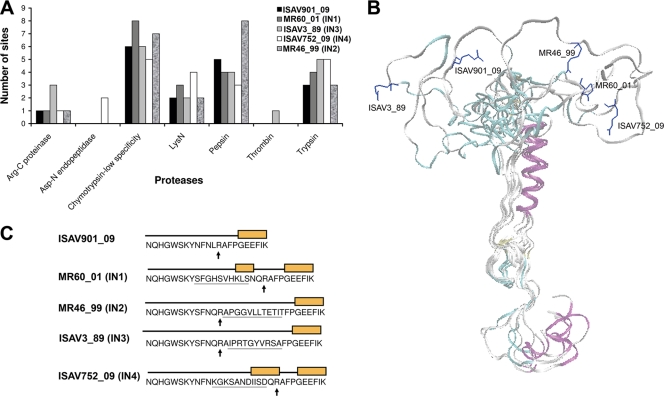
The fusion protein of ISAV from different isolates has high identity along the protein, with one exception: the region close to the predicted proteolytic processing site (Fig. 3). In some viral isolates an insertion (IN) has been determined in this region. Specifically, MR60_01 displays IN1; MR46_99, IN2; ISAV3_89, IN3 (14); and ISAV752_09, IN4. Of them, ISAV3_89 is experimentally determined as virulent (47), and ISAV752_09 has almost 80% prevalence in Chile (HPR 7b) (35). This insertion is inside the putative protease-processing site, which could improve the processing of the fusion protein either by increasing the number of recognition sites for trypsinlike protease processing or by increasing accessibility for cellular proteases. To explore both possibilities, the putative site for several proteases was predicted using the “peptide cutter” server. Interestingly, the variable region of the fusion protein from ISAV752_09 shows more sites for trypsin, Asp-N endopeptidase and N-lysin endopeptidases, and a lower number of sites for the digestive enzymes pepsin or chymotrypsin. The variable region of the peptide fusion protein from ISAV752_09 is unique in showing a cleavage site for the metalloprotease Asp-N endopeptidase. In the case of ISAV752_09 and ISAV3_89, the number of sites of cleavage for trypsin enzyme increases from three to five with respect to the fusion protein without insertion. On the other hand, the number of sites of cleavage for pepsin and chemotrypsin enzymes in the ISAV752_09 (IN4) isolate decreases by two sites and one site, respectively, compared to the fusion protein of ISAV901_09 without insertion (Fig. 3A).
FIG. 3.
Homology modeling of fusion proteins of ISAV. (A) Number of sites for several proteases present in IN of fusion proteins. (B) Superposition of models of the ISAV fusion protein with different insertions, and putative site of the proteolytic processing site (residue R) (the amino acid residue is shown in a stick representation). The models were generated by homology modeling, using the fusion domain from the HA protein of influenza A virus as a template. (C) Putative secondary structure of IN. The boxes show the helix structure.
A three-dimensional model of the ISAV fusion protein was constructed with a homology model using the fusion domain of influenza A virus HA protein (PDB ID 1HA0, Fig. 3B) as a template. Compared to ISAV901_09, which has no insertion, secondary structure prediction of the insertion region in the fusion protein shows that this region can adopt a helix structure in ISAV752_09 (IN4) and ISAVMR60_01 (IN1) in which the insertions are located in the amino-terminal position of the putative site of cleavage-arginine. Moreover, this region adopts a random coil structure in ISAV3_89 (IN3) and MR46/99 (IN2) in which the insertions are located on the carboxy-terminal position of arginine. The homology model shows that the flexibility of this region increases with the size of the insertion (Fig. 3B and C).
Esterase protein.
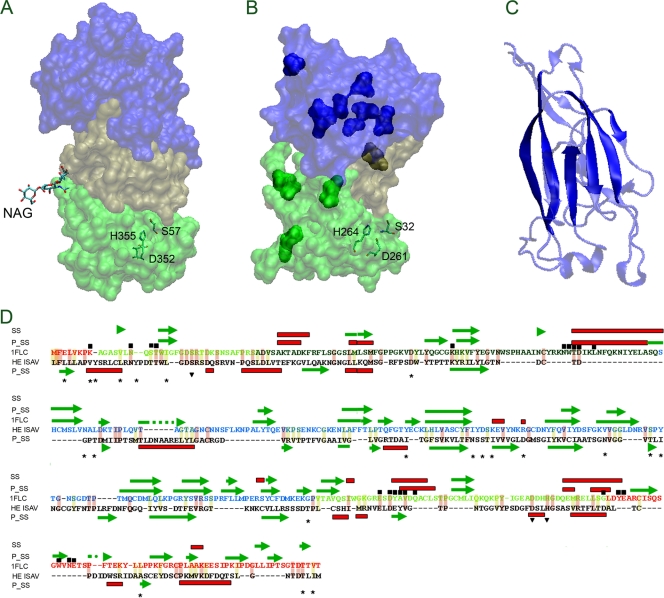
The protein encoded by segment 6 of the ISAV genome has hemagglutinin and receptor-destroying activities. Fold recognition using PHYRE and ROBETTA shows that HE of ISAV shares folding homology with the hyluronidase (PDB ID 2YX2) and HEF of influenza C virus (PDB ID 1FLC; Table 3). Homology modeling using the receptor and esterase regions of HEF from influenza C virus as a template (chain E, PDB ID 1FLC) (67) Fig. 4 A) shows that the esterase from ISAV (Fig. 4B) shares the E1 and E2 regions and partially the E′ region with HEF. This domain forms the esterase enzyme, suggesting a similar catalytic mechanism, in agreement with the conservation of the residues (indicated in Fig. 4A and B, inverted triangles in Fig. 4D) that constitute the catalytic site in influenza C virus esterase. Multialignment among proteins encoded by segment 6 from ISAV isolates shows several changes distributed along the protein. In addition to the HPR region, the location of variable positions distributed along the protein in the model shows that most of these positions are in the region involved in the recognition of the receptor (solid blue domain in Fig. 4B and asterisk in Fig. 4D) in influenza C virus esterase. A few variable positions are located in the region corresponding to the esterase activity (E1, E2, and E′, light and dark solid green in Fig. 4B). Analysis of the predicted conserved secondary structure (Fig. 4D) and crystallographic results between the HEF R domain and HE shows that the conserved beta strand is located in the main beta sheet, suggesting that these structures are conserved between both proteins (Fig. 4C). The location of the residues close to N-acetylglucosamine (NAG, the receptor of HEF) (dark boxes in Fig. 4D) shows that most of them are conserved in HE, especially residues located in the E1, E2 and E′ domains, suggesting that the binding mechanisms to sugar and further enzymatic processes are shared between both viruses (Fig. 4D).
FIG. 4.
Structural characterization of the HE protein from ISAV by homology modeling. (A) Structure of HEF from influenza C virus (PDB ID 1FLC); (B) structural model of HE from ISAV. Their characteristic subdomains E1 and E2, R, and E′ are indicated in light green, blue, and dark green, respectively. The catalytic residues of HAF from influenza C virus (S57, D352, and H355) and putative catalytic residues from ISAV (H264, D261, and S32) are indicated. (C) Structure of the R domain from influenza C virus. The solid colors indicate conserved structural elements with the predicted secondary structure of HE from ISAV. (D) Alignment between influenza C virus HEF and ISAV HE using secondary structural prediction (using Phyre). The figure shows the predicted secondary structure (P_SS), experimental secondary structure (SS), the catalytic residues (residues over the inverted triangle), residues close to (6 A) NAG (residues under the black box), the variable position among HE proteins coming from ISA virus with its entire genome sequenced (residues over the stars). The helixes are shows in red boxes and β-strands in green arrows. The conserved changes are indicated with light yellow background and the identical residues with light red background. The secondary element b-strand (green arrow) and helix (red box) are also indicated.
DISCUSSION
The phylogenetic analysis of the coding region reported here for Chilean ISAV isolates and their comparison to Norwegian sequences allows for classifying every segment analyzed into two genogroups. These results are in agreement with a previous report, in which just segments 5 and 6 were analyzed, that also shows two genogroups (35). Thus, our report is an expansion of this observation for every segment of ISAV.
It is noteworthy that Chilean isolates always match in the same genogroup, indicating a common ancestor. Furthermore, Chilean isolates matched a subgroup of Norwegian viruses that includes mainly ISAV1-96, ISAV5-96, ISAV8-97, ISAV11-93, and SK77-06, a group of viruses with considerable differences in their virulence. ISAV1-96 and ISAV 5-96 are highly virulent; ISAV8 and ISAV11-93 are slightly virulent; and SK77-06 is a nonculturable virus that has been associated with avirulent behavior (47). The fact that virulent, slightly virulent, and avirulent viruses are associated with the same genogroup suggests that virulence may be related to specific changes in the sequence, such as point mutation, short sequence inserts, or deletion, rather than major changes in the sequence.
In this context, it has been widely accepted that there is a relationship between the HPR type and the level of virulence, with HPR0 being avirulent. In this respect, ISAV4_90 and ISAV7_96 share the same HPR4; however, in the dendrogram of segment 6, the two isolates belong to different genogroups. This is explained by the excision of the HPR in the analysis and the presence of 12 mutations between the two sequences. If the HPR is the only variable involved in virulence, both isolates should exhibit the same virulence; however, there is a controversy about the virulence of the two isolates. Mjaaland et al. in 2005 (51) showed that the viruses cause different levels of mortality of Salmo salar, with ISAV4_90 causing seven times as many deaths as ISAV7_96 and an increased immune response. In contrast, in 2009 Ritchie et al. (65) reported that two different ISAV isolates with common HPR4 and different farm virulence showed the same mortality under controlled conditions. The authors explained that these differences may be caused by changes in temperature, feeding, stress, and husbandry procedures. We hypothesize that HPR is not by itself a virulence factor.
The IN region of segment 5 has been associated with virulence in ISAV752_09, which displays an IN4, showing more prevalence than ISAV901_09, which has no insertion. This is very noteworthy, because the fifth segments of these viruses have almost identical sequences, differing just in this insertion by one nucleotide that generates one amino acid substitution. Furthermore, their HPR differs only in two nucleotides. This implies that the differences between the two isolates are mainly located on the sequence of segment 5 because of the IN4 insertion. This argues for discarding the HPR region as the only virulence marker, supporting the idea that virulence is the consequence of multiple factors rather than a specific change in only one segment.
In agreement with IN4 being a virulence factor is the fact that IN4 of ISAV752_09 is a unique insertion with a site for cleavage for the metalloprotease Asp-N endopeptidase (Fig. 3A), originally described in Pseudomonas fragi, but also detected in the skin mucosa of salmonids (21, 68), suggesting that it could increase the activation of ISAV752_09 in comparison to ISAV901_09. In addition, IN4 shows an increase from three to five sites for the trypsin enzyme compared to isolated ISAV901_09 without insertion. This increase in processing sites is also seen in IN3 from ISAV3-89, which is a highly virulent isolate (47). It suggests that additional proteolytic processing sites in IN4 or IN3 favor viral infection, probably by increasing the fusion of membranes in early stages of the viral cycle. Indeed, it has been reported that the increased number of sites for proteolytic processing in the fusion protein is a virulence factor in influenza A (29, 33, 57, 58). Interestingly, the number of cleavage sites for pepsin and chemotrypsin is lower in the ISAV752_09 and ISAV3_89 isolates, perhaps favoring a specific activation of the virus on endosomes or an alternative route of activation of membrane fusion. There are precedents that support this idea: in the case of influenza A virus it has been shown that low pathogenic avian influenza (LPAI) virus shifts to highly pathogenic avian influenza (HPAI) virus by insertion on the region of cleavage sites of a multibasic cleavage site (MBCS) present on the H5 genotype of HPAI virus. The insertion of an MBCS into HA0 of the LPAI virus, which displays an H6N1 genotype, yields H6N1MBCS. The increase in the number of cleavage sites for proteases other than trypsin increases the amount of tissue susceptible to infection, enhancing the virus titer similar to H5N1 titer grown in MDCK cells, indicating that MBCS insertion increases the virulence of the virus. Moreover, chicken infected with the H6N1 influenza virus do not present clinical symptoms, but the pathogenesis increases in H6N1MBCS virus due to the presence of the insertion, resulting in a shift from an LPAI to an HPAI phenotype. From all the virus samples that we studied in the case of segment 5, only 1 of the 50 Chilean ISA isolates does not contain IN4 in the fusion protein. Indeed, Kibenge et al. (35) reported that 80% of Chilean isolates contain IN4. This is in agreement with the proposal that part of the pathogenicity of the virus is given by the cleavage sites in the fusion protein.
However, there are more factors affecting the pathogenicity because the introduction of LPAI MBCS in H0 is not achieved a phenotype of the same magnitude as HPAI, so that the pathogenicity is the result of a multifactorial process, like a polymerase single amino acid mutations (23).
Alignment analysis of ISAV F protein suggests that N-glycosylation sites are indispensable to function as fusion proteins. It has been shown that in other myxoviruses, N glycosylation of the F domain in Sendai and influenza C viruses is important for protein fusion activity. The absence of N glycosylation decreases the transport from the endoplasmic reticulum to the cytoplasmic membrane and alters protein trimerization (70, 71, 74). A glycosylated form of fusion protein has been reported (50 kDa) (4, 19) with an approximate mass difference of 2 kDa compared to its immature isoform. This difference is equivalent to that observed in HE from influenza A viruses (7).
HE, the other glycoprotein of ISAV, was analyzed by folding homology compared to HEF of influenza C virus. It is noteworthy that several mutations among different ISAV isolates are located in the putative receptor domain (Fig. 4). This can imply that each mutation can confer a diverse capacity to recognize the receptor or provide an antigenic variation to evade the immune system. This is partially supported by the fact that these mutations are clustered. In contrast, the esterase activity domain shows lower sequence variation, indicating lower selection pressure. This may serve to conserve catalytic activity, with as few changes as possible. In addition, the structural analysis of esterase based on the homology model shows that the putative catalytic residues H264, D261 and S32 of HE are not only conserved in HEF (influenza C virus), as has been reported elsewhere (67), but they also display a spatial distribution that agrees with catalytic activity, sustaining their participation in this process. HEF residues arranged to bind to sialic acid NAG (dark boxes in Fig. 4D), are conserved in ISAV HE. This suggests that there is a conserved mechanism of recognition of target cells and is consistent with the fact that esterase activity of HE cleaves neuroaminic acid. Furthermore, after this paper was submitted, a report was published online in which a structural analysis of HE of ISAV was performed using GeneSilico and BioInfo metaserver (54). The research in this report used ISAV HE modeling with HE from porcine torovirus as a template and revealed a structure similar to the HEF protein of influenza C virus. Briefly, in agreement with our results, these researchers have demonstrated a highly conserved esterase motif and less conserved receptor domain.
Interestingly, of the eight novel substitutions described for both Chilean isolates, seven correspond to changes located in the proteins that form the polymerase complex and nucleoprotein (3, 41, 64, 72), which may have a major role in the virus replication machinery. The analysis of all individual amino acid mutations for 15 Norwegian ISA viruses shows that 6 of 8 correspond to PB2 protein, 12 of 15 correspond to PB1, 8 of 10 correspond to NP, and 6 of 11 correspond to PA of the virulent strain. In contrast, only one of three of the individual single mutations present on the NS2 protein of Norwegian viruses corresponds to virulent strains (data not show). This finding indicates that single amino acid changes in polymerase could be related to increases in virulence and suggests that this phenomenon occurs in Chilean isolates, increasing their replicative efficiency by changing these residues to adapt to host and environment. It is very probable that these changes accelerate the virus spreading.
This idea is supported by studies in avian influenza viruses, in which a single mutation on position 627 of PB2 allows the virus to replicate in mammals with high pathogenicity (27, 73). In addition, duck influenza virus, containing a single mutation on position 701 of PB2, infects mice with high lethality (43). In the case of other polymerase mutations, it has been reported that seven mutations in polymerase proteins—three in PB2, two in PB1, one in PA, and NP, respectively—allow the infection of mammalian species with very high virulence, in which polymerase activity increases by between 200 and 1,000% (23). These observations support the concept that mutations of the polymerase complex are critical for adaptation to the new environment (i.e., temperature or host) once the virus has been transmitted to a new host (22, 44). The seven novel single amino acid mutations displayed by Chilean strains suggest that these could be an adaptation to a new environment and allow the wide distribution that both strains have displayed along the Chilean coast, mainly the ISAV 752_09 isolate, with 80% prevalence.
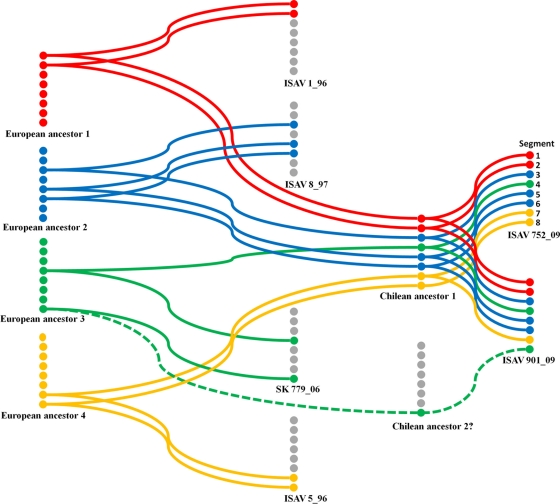
The high value of PP seen in the phylogenetic tree allowed for the arrangement of each genomic segment of the Chilean and Norwegian ISA viruses (Fig. 1). This makes it possible to generate a hypothetical model for the reassortment of genes (antigenic shift) that could have given rise to the two Chilean isolates analyzed in the present study (Fig. 5). The hypothetical reassortment indicates that segments 1 to 7 from isolates ISAV752_09 and ISAV901_09 share a close ancestor (Fig. 1), allowing us to suggest that this is a Chilean ancestor I (Fig. 5). In this sense, segments 1 and 2 are strongly related to segments 1 and 2 of the virulent isolate ISAV1_96. The two isolates possibly share what we have termed the first European ancestor. Similarly, segments 3, 5, and 6 of the two Chilean isolates came from the same Chilean ancestor, which would be related to at least the second European ancestor that gave rise to the same segments also shared with the European low-virulent strain ISAV8_97. Furthermore, segments 4 of the Chilean isolates might come from the first Chilean ancestor. This first ancestor could be related to a third European ancestor that gave rise to segment 4 of the avirulent strain SK77_06. An analogous situation was observed in segment 7 of the Chilean isolates that might have come from the first Chilean ancestor. It comes from a fourth European ancestor that probably gave rise to segment 7 of the virulent isolate ISAV5_96. Finally, the phylogenetic analysis of segment 8, with high PP value support, indicates that it is possible that segment 8 of the Chilean isolate ISAV752_09 comes from a fourth European ancestor that might also be related to the virulent European strain ISAV5_96. However, the common node for segment 8 from isolates ISAV901_09 and SK77_06 has a low PP value, which is not supported statistically (0.21), which suggests a low degree of certainty that this segment comes from of a putative second Chilean ancestor that in turn comes from the third European ancestor. These results suggest that both sequenced Chilean isolates possess a common ancestor, although they have different HPRs in segment 6 and ISAV752_09 has an insert of 33 nucleotides in segment 5. There is a high degree of uncertainty in the genealogy of ISAV901_09 segment 8, although a new phylogenetic analysis was performed. The high identity displayed by segment 8 in every isolate (shown in BLAST and FASTA) makes it difficult to resolve the relationship between ISAV901_9 segment 8 and the other segments 8 reported in GenBank.
FIG. 5.
Hypothetical model of segment reassortment of Chilean isolates. The Chilean isolates come from at least four European isolates that give rise to two possible Chilean ancestral originators of ISAV752_09 and ISAV901_09.
The Chilean isolates come from four European ancestors (Fig. 5). It is interesting that there are common ancestors for segments that code for proteins with related functions. Indeed, segments 1 and 2 from two Chilean isolates that encode the putative polymerase subunits PB2 and PB1, respectively, and segments 5 and 6, which encode the superficial HE and fusion proteins, evolved from a common ancestor. Thus, we can find clues on the replicative activity-virulent behavior relationships of these two viruses in farms located in southern Chile.
Reassortment is one of the mechanisms used by members of the Orthomyxoviridae family to generate new variants of the virus. However, as can be seen from the example of influenza virus, there are two important considerations: first, the process does not involve change in all segments of the viral genome, indicating that during a particular period of time only one variant is active, causing outbreaks of disease and using drift to evade the immune system. At some point, the virus can be coinfected with a second variant, generating a more pathogenic virus. Thus, the appearance of a reassortment or interspecies transfer could produce a pandemic (55, 60). In the case of Chilean ISAV isolates, their origin is rearranged. Unlike influenza virus, this derives from perhaps four different viruses, suggesting that there not numerous variants of the virus circulating among the fish. Although in ISAV in general the emergence of highly pathogenic reassortments is more common than in influenza virus, it has been shown that nonlethal isolates are in circulation only during the winter, when influenza A outbreaks occur (60). In contrast, in a given period of time, ISAV can generate two or more viral variants capable of causing infections with high mortality, as in the case of the two Chilean isolates.
To our knowledge, this is the first report of the complete coding region of Chilean ISAV isolates, with comparisons to their Norwegian counterparts. Using phylogenetic analysis and the predicted three-dimensional structure, we have adequately assessed the evolution of this virus using information contained in all segments, with a broad consideration of the molecular epidemiology of ISAV in Chile.
Supplementary Material
Acknowledgments
This study was supported by DICYT grant 020943CSM from the Universidad de Santiago de Chile, by PBCT-CONICYT grant PDA-20 (Government of Chile), and by Innova-Corto grants 09MCSS-6691, 09MCSS-6698, and 07CN13PBT-90. L.C. was supported by a fellowship from CONICYT (Government of Chile) and belongs to the Doctorate in Biotechnology Program, USACH.
We thank Siri Mjaaland for providing the primer set used for amplification and for sequence coding the genome region of ISAV. We thank Eduardo Castro Nallar, Brigham Young University, Provo, UT, for bioinformatic support in phylogeny analysis and Maria Teresa Castillo for assistance in cell culture.
Footnotes
Published ahead of print on 1 September 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Akaike, H. 1974. A new look at the statistical model identification. IEEE Trans. Automatic Control 16:716-723. [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Aspehaug, V., K. Falk, B. Krossoy, J. Thevarajan, L. Sanders, L. Moore, C. Endresen, and E. Biering. 2004. Infectious salmon anemia virus (ISAV) genomic segment 3 encodes the viral nucleoprotein (NP), an RNA-binding protein with two monopartite nuclear localization signals (NLS). Virus Res. 106:51-60. [DOI] [PubMed] [Google Scholar]
- 4.Aspehaug, V., A. B. Mikalsen, M. Snow, E. Biering, and S. Villoing. 2005. Characterization of the infectious salmon anemia virus fusion protein. J. Virol. 79:12544-12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bierin, E., K. Falk, E. Hoel, J. Thevarajan, M. Joerink, A. Nylund, C. Endresen, and B. Krossoyl. 2002. Segment 8 encodes a structural protein of infectious salmon anaemia virus (ISAV); the co-linear transcript from segment 7 probably encodes a nonstructural or minor structural protein. Dis. Aquat. Organ. 49:117-122. [DOI] [PubMed] [Google Scholar]
- 6.Blake, S., D. Bouchard, W. Keleher, M. Opitz, and B. L. Nicholson. 1999. Genomic relationships of the North American isolate of infectious salmon anemia virus (ISAV) to the Norwegian strain of ISAV. Dis. Aquat. Organ 35:139-144. [DOI] [PubMed] [Google Scholar]
- 7.Blake, T. A., T. L. Williams, J. L. Pirkle, and J. R. Barr. 2009. Targeted N-linked glycosylation analysis of H5N1 influenza hemagglutinin by selective sample preparation and liquid chromatography/tandem mass spectrometry. Anal. Chem. 81:3109-3118. [DOI] [PubMed] [Google Scholar]
- 8.Bouchard, D., W. Keleher, H. M. Opitz, S. Blake, K. C. Edwards, and B. L. Nicholson. 1999. Isolation of infectious salmon anemia virus (ISAV) from Atlantic salmon in New Brunswick, Canada. Dis. Aquat. Organ. 35:131-137. [DOI] [PubMed] [Google Scholar]
- 9.Chivian, D., D. E. Kim, L. Malmstrom, P. Bradley, T. Robertson, P. Murphy, C. E. Strauss, R. Bonneau, C. A. Rohl, and D. Baker. 2003. Automated prediction of CASP-5 structures using the Robetta server. Proteins 53(Suppl. 6):524-533. [DOI] [PubMed] [Google Scholar]
- 10.Clouthier, S. C., T. Rector, N. E. Brown, and E. D. Anderson. 2002. Genomic organization of infectious salmon anaemia virus. J. Gen. Virol. 83:421-428. [DOI] [PubMed] [Google Scholar]
- 11.Cook-Versloot, M., S. Griffiths, R. Cusack, S. McGeachy, and R. Ritchie. 2004. Identification and characterisation of infectious salmon anaemia virus (ISAV) haemagglutinin gene highly polymorphic region (HPR) type 0 in North America. Bull. Eur. Assoc. Fish Pathol. 24:203-208. [Google Scholar]
- 12.Cunningham, C. O., A. Gregory, J. Black, I. Simpson, and R. S. Raynard. 2002. A novel variant of the infectious salmon anaemia virus (ISAV) haemagglutinin gene suggests mechanisms for virus diversity. Bull. Eur. Assoc. Fish Pathol. 22:366-374. [Google Scholar]
- 13.Devold, M., K. Falk, B. Dale, B. Krossoy, E. Biering, V. Aspehaug, F. Nilsen, and A. Nylund. 2001. Strain variation, based on the hemagglutinin gene, in Norwegian ISA virus isolates collected from 1987 to 2001: indications of recombination. Dis. Aquat. Organ. 47:119-128. [DOI] [PubMed] [Google Scholar]
- 14.Devold, M., M. Karlsen, and A. Nylund. 2006. Sequence analysis of the fusion protein gene from infectious salmon anemia virus isolates: evidence of recombination and reassortment. J. Gen. Virol. 87:2031-2040. [DOI] [PubMed] [Google Scholar]
- 15.Drummond, A. J., S. Y. Ho, M. J. Phillips, and A. Rambaut. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drummond, A. J., and A. Rambaut. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edgar, R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eswar, N., B. Webb, M. A. Marti-Renom, M. S. Madhusudhan, D. Eramian, M. Y. Shen, U. Pieper, and A. Sali. 2007. Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. Chapter 2:Unit 2-9. [DOI] [PubMed] [Google Scholar]
- 19.Falk, K., V. Aspehaug, R. Vlasak, and C. Endresen. 2004. Identification and characterization of viral structural proteins of infectious salmon anemia virus. J. Virol. 78:3063-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falk, K., E. Namork, E. Rimstad, S. Mjaaland, and B. H. Dannevig. 1997. Characterization of infectious salmon anemia virus, an orthomyxo-like virus isolated from Atlantic salmon (Salmo salar L.). J. Virol. 71:9016-9023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fast, M. D., D. E. Sims, J. F. Burka, A. Mustafa, and N. W. Ross. 2002. Skin morphology and humoral nonspecific defense parameters of mucus and plasma in rainbow trout, coho and Atlantic salmon. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 132:645-657. [DOI] [PubMed] [Google Scholar]
- 22.Gabriel, G., M. Abram, B. Keiner, R. Wagner, H. D. Klenk, and J. Stech. 2007. Differential polymerase activity in avian and mammalian cells determines host range of influenza virus. J. Virol. 81:9601-9604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gabriel, G., B. Dauber, T. Wolff, O. Planz, H. D. Klenk, and J. Stech. 2005. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc. Natl. Acad. Sci. U. S. A. 102:18590-18595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Rosado, E., T. Markussen, O. Kileng, E. S. Baekkevold, B. Robertsen, S. Mjaaland, and E. Rimstad. 2008. Molecular and functional characterization of two infectious salmon anaemia virus (ISAV) proteins with type I interferon antagonizing activity. Virus Res. 133:228-238. [DOI] [PubMed] [Google Scholar]
- 25.Godoy, M. G., A. Aedo, M. J. Kibenge, D. B. Groman, C. V. Yason, H. Grothusen, A. Lisperguer, M. Calbucura, F. Avendano, M. Imilan, M. Jarpa, and F. S. Kibenge. 2008. First detection, isolation and molecular characterization of infectious salmon anaemia virus associated with clinical disease in farmed Atlantic salmon (Salmo salar) in Chile. BMC Vet. Res. 4:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 27.Hatta, M., P. Gao, P. Halfmann, and Y. Kawaoka. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840-1842. [DOI] [PubMed] [Google Scholar]
- 28.Hellebo, A., U. Vilas, K. Falk, and R. Vlasak. 2004. Infectious salmon anemia virus specifically binds to and hydrolyzes 4-O-acetylated sialic acids. J. Virol. 78:3055-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirst, M., C. R. Astell, M. Griffith, S. M. Coughlin, M. Moksa, T. Zeng, D. E. Smailus, R. A. Holt, S. Jones, M. A. Marra, M. Petric, M. Krajden, D. Lawrence, A. Mak, R. Chow, D. M. Skowronski, S. A. Tweed, S. Goh, R. C. Brunham, J. Robinson, V. Bowes, K. Sojonky, S. K. Byrne, Y. Li, D. Kobasa, T. Booth, and M. Paetzel. 2004. Novel avian influenza H7N3 strain outbreak, British Columbia. Emerg. Infect. Dis. 10:2192-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph 8:27-28. [DOI] [PubMed] [Google Scholar]
- 31.Inglis, J. A., J. Bruce, and C. O. Cunningham. 2000. Nucleotide sequence variation in isolates of infectious salmon anaemia virus (ISAV) from Atlantic salmon Salmo salar in Scotland and Norway. Dis. Aquat. Organ. 43:71-76. [DOI] [PubMed] [Google Scholar]
- 32.Kelley, L. A., and M. J. Sternberg. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363-371. [DOI] [PubMed] [Google Scholar]
- 33.Khatchikian, D., M. Orlich, and R. Rott. 1989. Increased viral pathogenicity after insertion of a 28S rRNA sequence into the haemagglutinin gene of an influenza virus. Nature 340:156-157. [DOI] [PubMed] [Google Scholar]
- 34.Kibenge, F. S., O. N. Garate, G. Johnson, R. Arriagada, M. J. Kibenge, and D. Wadowska. 2001. Isolation and identification of infectious salmon anaemia virus (ISAV) from Coho salmon in Chile. Dis. Aquat. Organ. 45:9-18. [DOI] [PubMed] [Google Scholar]
- 35.Kibenge, F. S., M. G. Godoy, Y. Wang, M. J. Kibenge, V. Gherardelli, S. Mansilla, A. Lisperger, M. Jarpa, G. Larroquete, F. Avendano, M. Lara, and A. Gallardo. 2009. Infectious salmon anaemia virus (ISAV) isolated from the ISA disease outbreaks in Chile diverged from ISAV isolates from Norway around 1996 and was disseminated around 2005, based on surface glycoprotein gene sequences. Virol. J. 6:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kibenge, F. S., M. J. Kibenge, P. K. McKenna, P. Stothard, R. Marshall, R. R. Cusack, and S. McGeachy. 2001. Antigenic variation among isolates of infectious salmon anaemia virus correlates with genetic variation of the viral haemagglutinin gene. J. Gen. Virol. 82:2869-2879. [DOI] [PubMed] [Google Scholar]
- 37.Kibenge, F. S., M. J. Kibenge, Y. Wang, B. Qian, S. Hariharan, and S. McGeachy. 2007. Mapping of putative virulence motifs on infectious salmon anemia virus surface glycoprotein genes. J. Gen. Virol. 88:3100-3111. [DOI] [PubMed] [Google Scholar]
- 38.Kibenge, F. S., K. Munir, M. J. Kibenge, T. Joseph, and E. Moneke. 2004. Infectious salmon anemia virus: causative agent, pathogenesis and immunity. Anim. Health Res. Rev. 5:65-78. [DOI] [PubMed] [Google Scholar]
- 39.Kibenge, F. S., H. Xu, M. J. Kibenge, B. Qian, and T. Joseph. 2007. Characterization of gene expression on genomic segment 7 of infectious salmon anaemia virus. Virol. J. 4:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kristiansen, M., M. K. Froystad, A. L. Rishovd, and T. Gjoen. 2002. Characterization of the receptor-destroying enzyme activity from infectious salmon anaemia virus. J. Gen. Virol. 83:2693-2697. [DOI] [PubMed] [Google Scholar]
- 41.Krossoy, B., I. Hordvik, F. Nilsen, A. Nylund, and C. Endresen. 1999. The putative polymerase sequence of infectious salmon anemia virus suggests a new genus within the Orthomyxoviridae. J. Virol. 73:2136-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krossoy, B., F. Nilsen, K. Falk, C. Endresen, and A. Nylund. 2001. Phylogenetic analysis of infectious salmon anaemia virus isolates from Norway, Canada, and Scotland. Dis. Aquat. Organ. 44:1-6. [DOI] [PubMed] [Google Scholar]
- 43.Li, Z., H. Chen, P. Jiao, G. Deng, G. Tian, Y. Li, E. Hoffmann, R. G. Webster, Y. Matsuoka, and K. Yu. 2005. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J. Virol. 79:12058-12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Londt, B. Z., J. Banks, R. Gardner, W. J. Cox, and I. H. Brown. 2007. Induced increase in virulence of low virulence highly [corrected] pathogenic avian influenza by serial intracerebral passage in chickens. Avian Dis. 51:396-400. [DOI] [PubMed] [Google Scholar]
- 45.Lovely, J. E., B. H. Dannevig, K. Falk, L. Hutchin, A. M. MacKinnon, K. J. Melville, E. Rimstad, and S. G. Griffiths. 1999. First identification of infectious salmon anaemia virus in North America with haemorrhagic kidney syndrome. Dis. Aquat. Organ. 35:145-148. [DOI] [PubMed] [Google Scholar]
- 46.Marchler-Bauer, A., J. B. Anderson, F. Chitsaz, M. K. Derbyshire, C. DeWeese-Scott, J. H. Fong, L. Y. Geer, R. C. Geer, N. R. Gonzales, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, S. Lu, G. H. Marchler, M. Mullokandov, J. S. Song, A. Tasneem, N. Thanki, R. A. Yamashita, D. Zhang, N. Zhang, and S. H. Bryant. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205-D210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Markussen, T., C. M. Jonassen, S. Numanovic, S. Braaen, M. Hjortaas, H. Nilsen, and S. Mjaaland. 2008. Evolutionary mechanisms involved in the virulence of infectious salmon anaemia virus (ISAV), a piscine orthomyxovirus. Virology 374:515-527. [DOI] [PubMed] [Google Scholar]
- 48.McBeath, A. J., M. Snow, C. J. Secombes, A. E. Ellis, and B. Collet. 2007. Expression kinetics of interferon and interferon-induced genes in Atlantic salmon (Salmo salar) following infection with infectious pancreatic necrosis virus and infectious salmon anaemia virus. Fish Shellfish Immunol. 22:230-241. [DOI] [PubMed] [Google Scholar]
- 49.McClellan, D. A., and S. Woolley. 2004. AlignmentHelper, ver. 1.0. Brigham Young University, Department of Integrative Biology, Provo, UT.
- 50.Mjaaland, S., O. Hungnes, A. Teig, B. H. Dannevig, K. Thorud, and E. Rimstad. 2002. Polymorphism in the infectious salmon anemia virus hemagglutinin gene: importance and possible implications for evolution and ecology of infectious salmon anemia disease. Virology 304:379-391. [DOI] [PubMed] [Google Scholar]
- 51.Mjaaland, S., T. Markussen, H. Sindre, S. Kjoglum, B. H. Dannevig, S. Larsen, and U. Grimholt. 2005. Susceptibility and immune responses following experimental infection of MHC compatible Atlantic salmon (Salmo salar L.) with different infectious salmon anaemia virus isolates. Arch. Virol. 150:2195-2216. [DOI] [PubMed] [Google Scholar]
- 52.Mjaaland, S., E. Rimstad, K. Falk, and B. H. Dannevig. 1997. Genomic characterization of the virus causing infectious salmon anemia in Atlantic salmon (Salmo salar L.): an orthomyxo-like virus in a teleost. J. Virol. 71:7681-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morgenstern, B. 2004. DIALIGN: multiple DNA and protein sequence alignment at BiBiServ. Nucleic Acids Res. 32:W33-W36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muller, A., T. Markussen, F. Drablos, T. Gjoen, T. O. Jorgensen, S. T. Solem, and S. Mjaaland. 2010. Structural and functional analysis of the hemagglutinin-esterase of infectious salmon anaemia virus. Virus Res. 151:131-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neumann, G., T. Noda, and Y. Kawaoka. 2009. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 459:931-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nylund, A., M. Devold, H. Plarre, E. Isdal, and M. Aarseth. 2003. Emergence and maintenance of infectious salmon anaemia virus (ISAV) in Europe: a new hypothesis. Dis. Aquat. Organ. 56:11-24. [DOI] [PubMed] [Google Scholar]
- 57.Orlich, M., H. Gottwald, and R. Rott. 1994. Nonhomologous recombination between the hemagglutinin gene and the nucleoprotein gene of an influenza virus. Virology 204:462-465. [DOI] [PubMed] [Google Scholar]
- 58.Orlich, M., and R. Rott. 1994. Thermolysin activation mutants with changes in the fusogenic region of an influenza virus hemagglutinin. J. Virol. 68:7537-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Posada, D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253-1256. [DOI] [PubMed] [Google Scholar]
- 60.Potter, C. W. 2001. A history of influenza. J. Appl. Microbiol. 91:572-579. [DOI] [PubMed] [Google Scholar]
- 61.Rambaut, A., and A. J. Drummond. 2007. Tracer v1.4. http://beast.bio.ed.ac.uk/Tracer.
- 62.Rimstad, E., S. Mjaaland, M. Snow, A. B. Mikalsen, and C. O. Cunningham. 2001. Characterization of the infectious salmon anemia virus genomic segment that encodes the putative hemagglutinin. J. Virol. 75:5352-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ritchie, R. J., M. Cook, K. Melville, N. Simard, R. Cusack, and S. Griffith. 2001. Identification of infectious salmon anaemia virus in Atlantic salmon from Nova Scotia (Canada): evidence for functional strain differences. Dis. Aquat. Organ. 44:171-178. [DOI] [PubMed] [Google Scholar]
- 64.Ritchie, R. J., J. Heppell, M. B. Cook, S. Jones, and S. G. Griffiths. 2001. Identification and characterization of segments 3 and 4 of the ISAV genome. Virus Genes 22:289-297. [DOI] [PubMed] [Google Scholar]
- 65.Ritchie, R. J., J. T. McDonald, B. Glebe, W. Young-Lai, E. Johnsen, and N. Gagne. 2009. Comparative virulence of Infectious salmon anaemia virus isolates in Atlantic salmon, Salmo salar L. J. Fish Dis. 32:157-171. [DOI] [PubMed] [Google Scholar]
- 66.Rodger, H. D., T. Turnbull, F. Muir, S. Millar, and R. H. Richards. 1998. Infectious salmon anaemia (ISA) in the United Kingdom. Bull. Eur. Assoc. Fish Pathol. 18:115-116. [Google Scholar]
- 67.Rosenthal, P. B., X. Zhang, F. Formanowski, W. Fitz, C. H. Wong, H. Meier-Ewert, J. J. Skehel, and D. C. Wiley. 1998. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature 396:92-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salles, C. M. C., P. Gagliano, S. A. T. Leita, J. B. Salles, H. L. M. Guedes, V. P. F. Cassano, and S. Giovanni De-Simone. 2007. Identification and characterization of proteases from skin mucus of tambacu, a neotropical hybrid fish. Fish Physiol. Biochem. 33:173-179. [Google Scholar]
- 69.Sambrook, J. G., and R. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 70.Segawa, H., A. Inakawa, T. Yamashita, and H. Taira. 2003. Functional analysis of individual oligosaccharide chains of Sendai virus hemagglutinin-neuraminidase protein. Biosci. Biotechnol. Biochem. 67:592-598. [DOI] [PubMed] [Google Scholar]
- 71.Segawa, H., T. Yamashita, M. Kawakita, and H. Taira. 2000. Functional analysis of the individual oligosaccharide chains of Sendai virus fusion protein. J. Biochem. 128:65-72. [DOI] [PubMed] [Google Scholar]
- 72.Snow, M., R. Ritchie, O. Arnaud, S. Villoing, V. Aspehaug, and C. O. Cunningham. 2003. Isolation and characterisation of segment 1 of the infectious salmon anaemia virus genome. Virus Res. 92:99-105. [DOI] [PubMed] [Google Scholar]
- 73.Subbarao, E. K., W. London, and B. R. Murphy. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 67:1761-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sugahara, K., S. Hongo, K. Sugawara, Z. N. Li, E. Tsuchiya, Y. Muraki, Y. Matsuzaki, and K. Nakamura. 2001. Role of individual oligosaccharide chains in antigenic properties, intracellular transport, and biological activities of influenza C virus hemagglutinin-esterase protein. Virology 285:153-164. [DOI] [PubMed] [Google Scholar]
- 75.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wiederstein, M., and M. J. Sippl. 2007. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 35:W407-W410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Workenhe, S. T., M. J. Kibenge, G. M. Wright, D. W. Wadowska, D. B. Groman, and F. S. Kibenge. 2008. Infectious salmon anaemia virus replication and induction of alpha interferon in Atlantic salmon erythrocytes. Virol. J. 5:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.