Abstract
Horizontal gene transfer commonly occurs from cells to viruses but rarely occurs from viruses to their host cells, with the exception of retroviruses and some DNA viruses. However, extensive sequence similarity searches in public genome databases for various organisms showed that the capsid protein and RNA-dependent RNA polymerase genes from totiviruses and partitiviruses have widespread homologs in the nuclear genomes of eukaryotic organisms, including plants, arthropods, fungi, nematodes, and protozoa. PCR amplification and sequencing as well as comparative evidence of junction coverage between virus and host sequences support the conclusion that these viral homologs are real and occur in eukaryotic genomes. Sequence comparison and phylogenetic analysis suggest that these genes were likely transferred horizontally from viruses to eukaryotic genomes. Furthermore, we present evidence showing that some of the transferred genes are conserved and expressed in eukaryotic organisms and suggesting that these viral genes are also functional in the recipient genomes. Our findings imply that horizontal transfer of double-stranded RNA viral genes is widespread among eukaryotes and may give rise to functionally important new genes, thus entailing that RNA viruses may play significant roles in the evolution of eukaryotes.
Viruses with double-stranded RNA (dsRNA) genomes infect a broad range of hosts, including bacteria, vertebrates, invertebrates, fungi, plants, and protozoa. They are grouped into six families: Birnaviridae, Chrysoviridae, Cystoviridae, Partitiviridae, Reoviridae, and Totiviridae (23). Viruses in the family Totiviridae encompass a broad range of viruses that infect mainly fungi or protozoa and are characterized by a nonsegmented dsRNA genome encoding, in most cases, only a capsid protein (CP) and an RNA-dependent RNA polymerase (RdRp) (23). Partitiviruses, which usually infect fungi or plants, have genomes consisting of two linear monocistronic dsRNA segments; the smaller segment usually encodes the CP, and the larger one typically codes for the RdRp (23). Although these RdRps are classified into the reverse transcriptase (RT)-like superfamily, neither totiviruses nor partitiviruses are known to have RT activity. They are transmitted in nature via cell division and cell fusion or, in the case of plant partitiviruses, by ovule and/or pollen transmission. In addition, they are generally associated with symptomless or persistent infections of their hosts and have no known natural vectors (22, 65).
Phylogenetic analyses of RdRps of dsRNA viruses suggest that they have multiple origins from diverse lineages of positive-strand RNA viruses (1, 20, 35). Recently, based on genomic comparison and phylogenetic analysis, Koonin and coworkers (38) found that the RdRps of viruses in the families Partitiviridae and Totiviridae are related to those of the picorna-like superfamily of eukaryotic positive-strand RNA viruses, and they suggested that the diversification of this superfamily probably evolved in a “Big Bang” that antedated the radiation of the eukaryotic supergroups.
Horizontal gene transfer (HGT)—the transfer of genes between distinct evolutionary lineages—has been recognized as a frequent event occurring from cells to viruses (7, 47, 48, 50) as well as from viruses to viruses (3, 25, 37, 50). Transfer from DNA viruses or retroviruses to eukaryotic cells has also been reported (4, 5, 14, 30, 41, 45). Transfer from nonretroviral RNA viruses to cells is thought to be extremely rare (5, 19). However, several very recent cases of eukaryotes in which nonretroviral integrated RNA viruses (NIRVs) were detected (17, 27, 36, 60, 61) suggest that transfer from nonretroviral RNA viruses to cells might be much more common than previously thought. In the present study, we analyzed eukaryotic genome databases for the presence of dsRNA virus-related sequences, and we present evidence that totiviral and partitiviral genes are transferred frequently into the nuclear genomes of eukaryotes and that some transferred genes even have functions in the recipient genomes.
MATERIALS AND METHODS
Data collection and similarity searches.
All sequences available for genomes and encoded proteins of eukaryotic dsRNA viruses were assembled from viral genome databases at the NCBI website (http://www.ncbi.nlm.nih.gov/genomes/GenomesHome.cgi?taxid=10239). To screen for virus-related sequences in eukaryotic genomes, each protein sequence was searched using the TBLASTn algorithm against the following NCBI databases (http://blast.ncbi.nlm.nih.gov/Blast.cgi): eukaryotic genomic BLAST, nucleotide collection (NT), NCBI genome (chromosome), reference genomic sequence (refseq_genomic), genomic survey sequence (GSS), high-throughput genomic sequence (HTGS), and whole-genome shotgun (WGS) databases. Through an iterative process of screening, all nonredundant hits from these searches with E values of ≤1e−5 were extracted. The relationships between sequences found to be similar in these searches were determined by reverse BLAST comparisons, using each extracted hit as a BLASTx query against the nonredundant (NR) protein database.
To examine the presence of potential transposable elements or repetitive sequences, virus-related sequences in eukaryotic genomes and their flanking sequences were searched against NCBI databases with BLAST or against the Repbase database (28) with Censor (34; http://girinst.org/censor/index.php).
All database searches were performed online and were completed by July 2009.
Phylogenetic analyses.
Multiple alignments of deduced amino acid (aa) sequences of virus-related homologs in eukaryotic genomes and of representative exogenous partitiviruses and totiviruses were constructed using COBALT (52; http://www.ncbi.nlm.nih.gov/tools/cobalt/cobalt.cgi?link_loc=BlastHomeAd) and refined by hand. The in-frame stop codons within sequences of virus-related homologs in eukaryotic genomes were indicated with X's. Protein maximum likelihood (ML) trees were inferred with PhyML mixtures (24, 40), assuming the EX2 mixture model (40) and using the subtree pruning and regrafting (SPR) tree topology search strategy (26). Gaps in alignment were systematically treated as unknown characters. The reliability of internal branches was evaluated based on approximate likelihood ratio test (aLRT) statistics (2).
Tests for selection.
For protein coding sequences, the ratio of nonsynonymous (Ka) to synonymous (Ks) nucleotide substitution rates has been adopted widely as a measure of selective pressure. For example, a ratio of <1 indicates purifying selection to conserve the protein sequence. A Ka/Ks calculator (http://services.cbu.uib.no/tools/kaks) was used to evaluate the Ka/Ks ratio for every branch in the phylogenetic trees for virus-related homologs in eukaryotic genomes. GC contents were estimated from the alignment.
DNA and total RNA extraction.
To obtain genomic DNA and total RNA of Arabidopsis ecotype Col-0, young leaves were collected and ground in liquid nitrogen with a mortar and pestle. DNA was extracted in cetyltrimethylammonium bromide (CTAB) as described by Sambrook et al. (58). Total RNA was prepared with Trizol reagent (MBI Fermentas) according to the manufacturer's instructions. DNA and RNA samples were stored at −80°C before use.
PCR, RT-PCR, and DNA sequencing.
The total volume of each PCR mix was 20 μl and contained 1 μl of DNA template, 2 μl 10× PCR buffer (TaKaRa), 0.4 μl of deoxynucleoside triphosphate (dNTP) mix (10 mM [each dNTP]), 0.2 μl of each primer (20 μM), and 1 unit of Taq DNA polymerase. The PCR temperature profile was 94°C for 4 min; 32 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and final extension at 72°C for 6 min.
RT-PCR detection of transcripts of virus-related homologs in the Arabidopsis genome was performed using RevertAid. A first-strand cDNA synthesis kit (Fermentas) was used with DNase-treated total RNA as described by the manufacturer.
PCR products were fractionated by gel electrophoresis on 1% agarose gels and were stained with ethidium bromide. DNA was sequenced by Sanger methods at the Beijing Genomics Institute (BGI).
Nucleotide sequence accession numbers.
New sequences generated in this study were deposited in GenBank under accession numbers HM068619 and HM068620.
RESULTS
A plant protein homolog of a partitiviral CP.
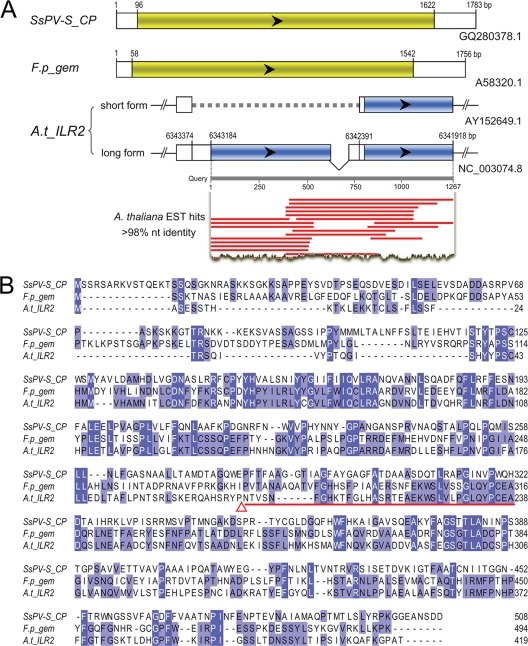
We recently isolated and sequenced the genomic dsRNAs of a partitivirus from Sclerotinia sclerotiorum, an important plant-pathogenic fungus, and designated the virus Sclerotinia sclerotiorum partitivirus S (SsPV-S) (H. Liu et al., unpublished data). BLASTp searches showed that whereas SsPV-S CP has the highest aa sequence similarity to IAA-leucine-resistant protein 2 (ILR2) (42) of Arabidopsis (26% amino acid [aa] identity; E value of <1e−11), its similarity to CPs of other partitiviruses is considerably lower. Furthermore, the proteins encoded by ILR2 and the CP gene of SsPV-S also share significant sequence similarity with the protein encoded by a cDNA clone of a gem gene of meadow fescue (Festuca pratensis) (51% and 25% aa identity, respectively) (62) (Fig. 1). The gem gene is 1,756 bp in length and contains one open reading frame (ORF) followed by an interrupted poly(A) tail at the 3′ end. These properties are very similar to those of partitiviruses. Indeed, we found many cDNA sequences of meadow fescue in the expressed sequence tag (EST) database that share sequence identity with the gem gene, and many others share sequence identity with the RdRp gene of SsPV-S (data not shown). These results suggest the possibility that gem is indeed of partitiviral CP gene origin, even though it was reported as a plant gene.
FIG. 1.
Schematic representation (A) and multiple sequence alignment of predicted amino acid sequences (B) for ILR2 of Arabidopsis thaliana, gem of Festuca pratensis, and the capsid protein gene of SsPV-S. (A) Colored rectangular boxes with arrowheads indicate ORFs. The region indicated by a broken horizontal line is deleted in the Wassilewskija (Ws) and Landsberg erecta (Ler) sequences of Arabidopsis. The matched regions of expressed sequences and their nt identity are indicated. (B) The red triangle indicates predicted frameshift sites in ILR2, and the region of the predicted ILR2 intron is marked with a red line. Identical residues between sequences are shaded. F.p_gem, gem of Festuca pratensis; SsPV-S_CP, capsid protein gene of Sclerotinia sclerotiorum partitivirus S; A.t_ILR2, IAA-leucine-resistant protein 2 of Arabidopsis thaliana; CP, capsid protein; EST, expressed sequence tag.
The ILR2 gene is located on chromosome 3 of Arabidopsis and has two forms, a short form and a long form, among Arabidopsis sequences (42) (Fig. 1A). The long ILR2 form may be the ancestral form of the gene and contains one predicted intron (42). However, the predicted intron of ILR2 is well aligned with coding regions of gem and the CP gene of SsPV-S (Fig. 1B; see Fig. S1 in the supplemental material), suggesting that it is unlikely to be an intron. In fact, many cDNA sequences of ILR2 contain the intron region, but no spliced forms were found in the EST database (Fig. 1A). These results suggest that HGT occurred from a gem-like partitivirus to the genome of an Arabidopsis ancestor.
Partitivirus- and totivirus-related homologs in eukaryotic nuclear genomes.
The finding that the Arabidopsis genome contains a virus-like gene raised our interest in exploring whether other eukaryotic genomes have viral homologs. For this purpose, we used the available protein sequences of various eukaryotic dsRNA viruses to screen publicly accessible genome databases (see Materials and Methods). During this analysis, 22 partitivirus-related and 34 totivirus-related homologs, ranging from short stretches of viral sequences to entire viral genomes (∼123 to 4,473 bp), were found in nuclear genomes of diverse eukaryotic organisms, including plants, arthropods, fungi, nematodes, gastropods, and protozoa (Tables 1 and 2; see Table S1 in the supplemental material). We noted that the presence of NIRVs in Penicillium and three budding yeast species was reported very recently (17, 60). Although partitivirus- and totivirus-like NIRVs occurred widely, NIRVs similar to viruses of other eukaryotic dsRNA families or genera were not detected.
TABLE 1.
Summary of partitivirus-related sequences in eukaryotic nuclear genomes
| Database | Organism group | Organism | Accession no., position (length [bp]) | Best-matched virusa | Related gene product | % aa identity | E valuea | Junction coverageb | ORF disruption | Adjacent TEsc |
|---|---|---|---|---|---|---|---|---|---|---|
| Refseq_genomic | Plants | Arabidopsis thaliana | NC_003074.8, chr3 positions 6341924 to 6343184 (1,261) | Sclerotinia sclerotiorum partitivirus S | CP | 34 | 1e−14 | Frameshift | DNA/hAT | |
| Plants | Arabidopsis thaliana | NC_003075.7, chr4 positions 8128880 to 8129554 (675) | Raphanus sativus cryptic virus 2 | RNA2 (CP) | 62 | 3e−49 | Deletion | Non-LTR/CR1, DNA | ||
| HTGS | Plants | Brassica rapa subsp. pekinensis (Chinese cabbage) | AC189442.2, positions 81429 to 82709 (1,281) | Raphanus sativus cryptic virus 1 | RasR6-3 (CP) | 56 | 2e−130 | 3× | LTR/Copia | |
| GSS | Plants | Brassica oleracea (wild mustard) | FI711962.1, positions 40 to 594 (555) | Raphanus sativus cryptic virus 1 | RasR6-3 (CP) | 55 | 3e−16 | 3× | ||
| Plants | Nicotiana tabacum (tobacco) | Contig-1, positions 4 to 1406 (1,403)d | Raphanus sativus cryptic virus 1 | RasR6-3 (CP) | 53 | 5e−106 | 8× | Internal stop codon, frameshift | DNA/hAT | |
| Plants | Nicotiana tabacum (tobacco) | Contig-2, positions 237 to 1537 (1,301)d | Raphanus sativus cryptic virus 1 | RasR6-3 (CP) | 43 | 2e−64 | 3× | Internal stop codons, frameshifts | ||
| Plants | Nicotiana tabacum (tobacco) | Contig-3, positions 609 to 1245 (635)d | Fragaria chiloensis cryptic virus | RNA3 (CP) | 43 | 1e−33 | 9× | Frameshift | ||
| Plants | Nicotiana tabacum (tobacco) | Contig-4, positions 91 to 771 (681)d | Raphanus sativus cryptic virus 3 | CP | 40 | 9e−06 | 7× | frameshifts | ||
| Plants | Zea mays (maize) | Contig-1, positions 416 to 905 (490)d | Rose cryptic virus 1 | RNA3 (CP) | 38 | 7e−09 | 2× | Internal stop codon, frameshift | ||
| Plants | Medicago truncatula (barrel medic) | Contig-1, positions 517 to 1145 (629)d | Raphanus sativus cryptic virus 3 | CP | 36 | 2e−17 | 2× | Internal stop codons, frameshift | ||
| Eukaryotic genome database | Arthropods | Acyrthosiphon pisum strain LSR1 (pea aphid) | NW_001916518.1, positions 33930 to 34748 (819) | Vicia faba partitivirus 1 | RdRp | 43 | 9e−42 | 11× | Internal stop codons | DNA/Sola |
| Arthropods | Acyrthosiphon pisum strain LSR1 (pea aphid) | NW_001917032.1, positions 17640 to 18664 (1,025) | Rosellinia necatrix partitivirus 1-W8 | RdRp | 51 | 2e−101 | 12× | Frameshift | Non-LTR/SINE, DNA | |
| Arthropods | Drosophila grimshawi strain TSC 15287-2541.00 (fruit fly) | NW_001961672.1, positions 4532049 to 4533305 (1,257) | Fusarium solani virus 1 | RdRp | 29 | 4e−28 | 4× | DNA/hAT | ||
| Arthropods | Rhodnius prolixus (triatomid bug) | ACPB01059984.1, positions 3884 to 4612 (729) | Penicillium stoloniferum virus F | RdRp | 25 | 7e−07 | 8× | Internal stop codons | ||
| Arthropods | Ixodes scapularis strain Wikel (deer tick) | ABJB010791923.1, positions 7178 to 7417 (240) | Fusarium solani virus 1 | RdRp | 50 | 8e−10 | 10× | Internal stop codon | Non-LTR/L1 | |
| Protozoa | Entamoeba histolytica HM-1:IMSS | NW_001915030.1, positions 31068 to 31789 (721) | Penicillium stoloniferum virus F | RdRp | 29 | 0.05 | 14× | Internal stop codons, frameshift | Non-LTR/SINE, DNA/Polinton |
The best-matched virus and E value were generated by using nuclear virus-related sequences as BLASTx queries against the NR protein database.
The junction coverage indicates the number of trace records/GSS that contain the junctions between viral and host sequences, calculated by using nuclear virus-related sequences and their flanking sequences as megablast queries against the NCBI Trace Archive/GSS database, with a cutoff nt identity of >95%. The two nuclear virus-related sequences in Arabidopsis were verified by PCR and sequencing in this study.
The class or subclass of transposable elements (TEs) or repeats is indicated as follows: DNA, DNA transposon; LTR, long terminal repeat (LTR)-containing retrotransposon; non-LTR, non-LTR-containing retrotransposon. See the genome map of TEs in Fig. S5 in the supplemental material.
Contigs were generated by assembling the GSSs which shared an alignment with at least 97% identity and a 50-bp overlap. See the original GSSs in Table S2 in the supplemental material.
TABLE 2.
Summary of totivirus-related sequences in eukaryotic nuclear genomes
| Database | Organism group | Organism | Accession no., position (length [bp]) | Best-matched virusa | Related gene product | % aa identity | E valuea | Junction coverageb | ORF disruption | Adjacent TEsc |
|---|---|---|---|---|---|---|---|---|---|---|
| Refseq_ genomic | Plants | Populus trichocarpa (black cottonwood) | NC_008473.1, positions 11124146 to 11126110 (1,965) | Southern tomato virus isolate MS-7 | CP-RdRp | 25 | 7e−34 | 7× | Internal stop codons | LTR/Copia, DNA |
| HTGS | Plants | Lotus japonicus (lotus) | AP007812.2, positions 71725 to 72699 (975) | Vicia cryptic virus M | CP | 33 | 1e−46 | |||
| Plants | Lotus japonicus (lotus) | AP007812.2, positions 48764 to 49678 (915) | Vicia cryptic virus M | CP | 36 | 6e−32 | Frameshifts | DNA/Chapaev, LTR/Gypsy | ||
| NR | Plants | Medicago truncatula (barrel medic) | AC196856.3, chr7 positions 68018 to 68767 (750) | Vicia cryptic virus M | CP | 31 | 1e−28 | DNA/Sola | ||
| Plants | Medicago truncatula (barrel medic) | AC175047.3, chr7 positions 74802 to 75279 (478) | Vicia cryptic virus M | CP | 28 | 9e−14 | Non-LTR/R4 | |||
| Plants | Medicago truncatula (barrel medic) | AC175047.3, chr7 positions 90928 to 91353 (426) | Vicia cryptic virus M | CP | 31 | 2e−14 | DNA/MuDR | |||
| Plants | Medicago truncatula (barrel medic) | AC148816.3, chr7 positions 45611 to 46084 (474) | Vicia cryptic virus M | CP | 26 | 1e−04 | Internal stop codon, frameshift | LTR/Copia | ||
| HTGS | Plants | Medicago truncatula (barrel medic) | AC233254.1, chr7 positions 1576 to 2325 (750) | Vicia cryptic virus M | CP | 31 | 1e−28 | DNA/Sola | ||
| Refseq_ genomic | Fungi | Pichia stipitis CBS 6054 (budding yeast) | NC_009047.1, chr7 positions 495820 to 497544 (1,725)e | Saccharomyces cerevisiae virus L-A | CP | 27 | 1e−29 | 14× | DNA/hAT | |
| Fungi | Pichia stipitis CBS 6054 (budding yeast) | NC_009047.1, chr7 positions 498357 to 500279 (1,923)e | Saccharomyces cerevisiae virus L-A | CP | 41 | 3e−117 | 12× | LTR/Tca4 | ||
| Fungi | Pichia stipitis CBS 6054 (budding yeast) | NC_009047.1, chr7 positions 501109 to 505587 (4,479)e | Saccharomyces cerevisiae virus L-A | CP-RdRp | 41 | 0 | 6× | DNA/hAT | ||
| Fungi | Pichia stipitis CBS 6054 (budding yeast) | NC_009047.1, chr7 positions 506568 to 508682 (2,115)e | Saccharomyces cerevisiae virus L-A | CP | 37 | 3e−48 | 11× | DNA/hAT | ||
| Fungi | Debaryomyces hansenii CBS767 (budding yeast) | NC_006044.1, chrB positions 1024483 to 1028772 (4,290)e | Saccharomyces cerevisiae virus L-A | CP-RdRp | 53 | 0 | DNA/hAT | |||
| Fungi | Debaryomyces hansenii CBS767 (budding yeast) | NC_006044.1, chrB positions 1023541 to 1023663 (123) | RdRp | 56 | 1e−06 | Internal stop codons | LTR/Copia | |||
| Fungi | Debaryomyces hansenii CBS767 (budding yeast) | NC_006044.1, chrB positions 1020783 to 1022789 (2,007)e | CP | 42 | 4e−164 | LTR/Gypsy | ||||
| Eukaryotic genome database | Arthropods | Ixodes scapularis strain Wikel (deer tick) | ABJB010328260, positions 9685 to 10343 (659) | Penaeid shrimp infectious myonecrosis virus | RdRp | 44 | 6e−12 | 12× | Frameshifts | |
| Arthropods | Ixodes scapularis strain Wikel (deer tick) | ABJB010888832.1, positions 1 to 278 (278) | Penaeid shrimp infectious myonecrosis virus | RdRp | 39 | 1e−09 | 11× | Internal stop codon | ||
| Arthropods | Ixodes scapularis strain Wikel (deer tick) | ABJB010761541.1, positions 122 to 801 (680) | Penaeid shrimp infectious myonecrosis virus | RdRp | 34 | 9e−15 | 2× | Internal stop codon, frameshift | ||
| Arthropods | Ixodes scapularis strain Wikel (deer tick) | ABJB010453070.1, 2966 to 3470 (505) | Penaeid shrimp infectious myonecrosis virus | CP | 48 | 9e−06 | 4× | Internal stop codon, frameshift | ||
| Arthropods | Aedes aegypti strain Liverpool (yellow fever mosquito) | AAGE02000678.1, positions 54379 to 55086 (708) | Cucurbit yellows-associated virus | RdRp | 38 | 7e−30 | 7× | Non-LTR | ||
| GSS | Nematodes | Strongyloides ratti | Contig-1, positions 506 to 1009 (504)d | Leishmania RNA virus 1-1 | RdRp | 44 | 1e−34 | 4× | Internal stop codons |
The best-matched virus and E value were generated by using totivirus-related sequences as BLASTx queries against the NR protein database.
The junction coverage indicates the number of trace records/GSS that contain the junctions between viral and host sequences, calculated by using nuclear virus-related sequences and their flanking sequences as megablast queries against the NCBI Trace Archive/GSS database, with a cutoff nt identity of >95%. The totivirus-related sequences found in lotus and barrel medic were confirmed by genomic comparison (see details in the text).
The class or subclass of transposable elements (TEs) or repeats is indicated as follows: DNA, DNA transposon; LTR, LTR-containing retrotransposon; non-LTR, non-LTR-containing retrotransposon. See the genome map of TEs in Fig. S5 in the supplemental material.
The contig was generated by assembling the GSSs which shared an alignment with at least 97% identity and a 50-bp overlap. See the original GSSs in Table S2 in the supplemental material.
Excluding the possibility that NIRVs are contaminant sequences.
Because routine DNA sequencing does not involve the use of reverse transcriptase, there is little chance for contamination of eukaryotic genomic DNA with RNA viral sequences. To examine any possible artifacts, we performed the following tasks. (i) We amplified and sequenced the proposed junctions of host-virus sequences for two NIRVs in Arabidopsis (Fig. 2A; see Fig. S2A and B in the supplemental material). The results revealed that the PCR products were of the expected sizes and that the experimental sequences had 100% identity with sequences of the Arabidopsis genome containing both the expected host sequences and the virus-like sequences. (ii) We examined all available trace data in the NCBI Trace Archive for the genomes containing NIRVs and found that in the genomes whose trace records were available, each junction between NIRVs and host sequences was covered by at least two trace records (>95% nucleotide [nt] identity) (Tables 1 and 2; see examples in Fig. S2C and E in the supplemental material). This supports the contention that these are not artifacts. (iii) We searched various NCBI databases (NT, EST, GSS, HTGS, and WGS) for sequences that included at least portions of the NIRV loci whose trace records were not available. We found some sequences that also showed the junctions between viral and host sequences (see examples in Fig. S2B and D to G in the supplemental material). (iv) We carefully examined the NIRVs in contigs which we assembled from GSSs (see Table S2 in the supplemental material). For each contig, at least two GSS records containing the junctions between viral and host sequences were found (Tables 1 and 2; see examples in Fig. S2H and I in the supplemental material). In addition, these NIRVs had apparently degenerated, since they contained frameshifts, internal stop codons, or deletions compared to the viral genes (Tables 1 and 2). Since at least two GSSs covered these regions, this indicated that mutations should not be due merely to sequencing errors. (v) We also compared the NIRVs and their flanking cellular sequences. For example, the two NIRVs as well as their flanking sequences in one clone each of Medicago truncatula (barrel medic) chromosome 7 and Lotus japonicus chromosome 1 are homologous (see Fig. S2J in the supplemental material). Likewise, the flanking sequences of NIRVs from two other clones of barrel medic (GenBank accession no. AC196856.3 and AC233254.1) are also homologous. These results suggest that genomic segments were probably duplicated after viral integration.
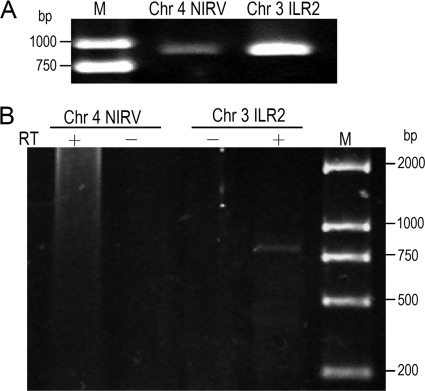
FIG. 2.
PCR amplification of total DNA of Arabidopsis ecotype Col-0 (A) and RT-PCR with DNase-treated total RNA (B). (A) The sequences of the PCR products were deposited under GenBank accession numbers HM068619 and HM068620. (B) Reverse transcriptase is marked as either present (+) or absent (−). PCR products were fractionated by gel electrophoresis on 1% agarose gels and were stained with ethidium bromide. M, DNA marker DL 2000; NIRV, nonretroviral integrated RNA virus.
From these analyses, we believe that these viral integrations are real rather than being chimeric clones or misassembled from contaminated sequences of exogenous incidental viral sequences. A few putative NIRVs have not been confirmed due to a lack of sufficient sequence information. They are regarded as candidates for viral integration (listed in Table S1 in the supplemental material), and experimental verification is pending.
Phylogenetic analysis of NIRVs and exogenous viruses.
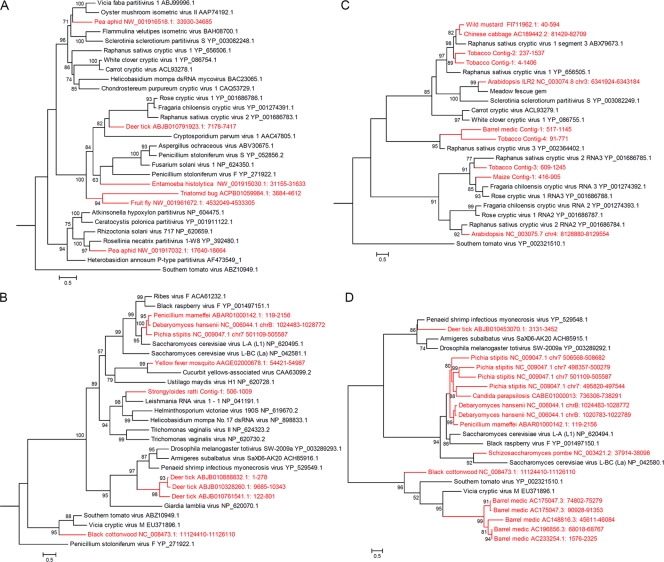
To evaluate the genetic relationships of NIRVs and exogenous viruses, we constructed maximum likelihood phylogenetic trees for deduced aa sequences of NIRVs, using the RdRp or CP protein sequences of representative partitiviruses and totiviruses. As shown in Fig. 3, the NIRVs were clearly located within the partiti- or totiviral phylogenetic trees, suggesting that they originated from these exogenous viruses. NIRVs from the same genome clustered within different clades (such as those with Arabidopsis, tobacco, and pea aphid), implying that integration of different viruses in one genome could have occurred. The NIRVs from Chinese cabbage and wild mustard clustered together, and their flanking sequences also shared sequence identity (see Fig. S2D in the supplemental material), indicating that NIRV integration preceded the separation of these two species. In contrast, the Saccharomyces cerevisiae virus L-A (ScV L-A)-like NIRVs from different fungal genomes clustered together (Fig. 3B and D), but their phylogenetic topologies did not follow species evolution. In addition, NIRVs were not detected in closely related fungal species. These results suggested that multiple independent integration events rather than a single insertion from a common ancestor had taken place. Alternatively, HGTs might have occurred among these species. Indeed, we found one sequence (GenBank accession no. AM669000) of the protozoan Entamoeba terrapinae M that shared sequence identity not only with the NIRV but also with its flanking sequence (80% nt identity) in the yeast Debaryomyces hansenii (see Fig. S2F in the supplemental material). This supports the idea that HGT occurred between these two species.
FIG. 3.
Phylogenetic analysis of NIRVs and representative partitiviruses and totiviruses. Maximum likelihood trees were estimated for the aligned amino acid sequences by using PhyML mixtures (24, 40). Gaps in alignment were systematically treated as unknown characters. Because many of the NIRVs were short, corresponding to different parts of the viral gene, there were only short or no common sites for all sequences aligned. Phylogenetic trees for the individual NIRVs were also constructed (data not shown), and their topologies were consistent with the trees presented in this figure. (A and C) RdRp (A) and CP (C) trees for partitiviruses and their related NIRVs. The trees were rooted with the totivirus-like southern tomato virus. (B and D) RdRp (B) and CP (D) trees for totiviruses and their related NIRVs. The RdRp tree was rooted with corresponding sequences from the partitivirus Penicillium stoloniferum virus F. The CP tree was not rooted because outgroup sequences were too divergent. Only P values for the approximate likelihood ratios (SH test) of >0.5 (50%) are indicated. All scale bars correspond to 0.5 amino acid substitution per site. The NIRV branches and taxa are indicated in red.
Expression and function of transferred genes in recipient genomes.
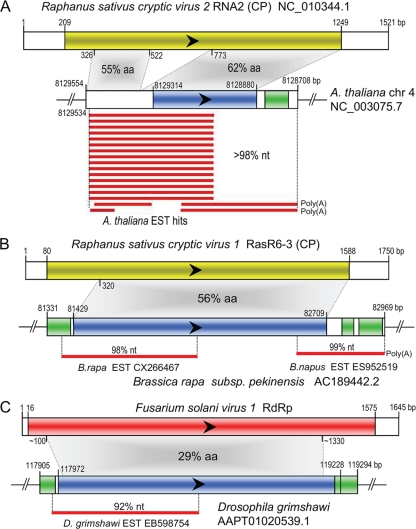
Comparison of NIRVs and their related viruses suggested that most were 5′- and/or 3′-truncated sequences and even contained frameshifts, internal stop codons, and/or deletions (Tables 1 and 2; see Table S1 in the supplemental material). However, some of the NIRVs retained intact or nearly intact reading frames of viral genes (Fig. 4 and 5; see Fig. S3 in the supplemental material). This suggested that they could have functions when expressed. We performed RT-PCR to examine the transcripts of NIRVs in the Arabidopsis genome. We detected transcripts for ILR2 but were unable to detect those for the chromosome 4 NIRV (Fig. 2B). However, by searching the EST database with the BLASTn program, it was clearly shown that the chromosome 4 NIRV was also expressed in Arabidopsis (Fig. 4A). The expressed sequences for chromosome 4 NIRV came from pooled cDNA populations from Arabidopsis, which were reconstructed from roots, inflorescence, calluses, young seedlings, and Arabidopsis plants subjected to various abiotic and biotic stresses (Y. Xiao et al., unpublished data). Since we did not detect expression from Arabidopsis young leaves, this suggests that the expression of chromosome 4 NIRV may occur in specialized cells or may be activated by stresses. By searching the translated EST database, we also found that the NIRV in Chinese cabbage was expressed (Fig. 4B). It is worth noting that the EST from rapeseed (Brassica napus) corresponded to the 3′ part of the NIRV and the adjacent genomic sequence of Chinese cabbage (Fig. 4B), indicating that there was a similarly expressed insertion in rapeseed. ESTs of NIRVs were also found in insects and yeasts (Fig. 4C; see Fig. S2G in the supplemental material). Interestingly, the southern tomato virus-like NIRV in the Populus genome is a pseudogene, but its 3′ partial cDNA sequence was also found (see Fig. S2E in the supplemental material). Transcriptions of other NIRVs were not detected, but the possibility of low levels of expression could not be ruled out. Alternatively, the currently available EST data are nevertheless limited.
FIG. 4.
Schematic representation of some NIRVs, their expressed sequences, and their most related viruses. Colored rectangular boxes with arrowheads indicate ORFs. Green rectangular boxes indicate repeated sequences in eukaryotic genomes detected by BLASTn searches. Gray sectors connect corresponding homologous regions, and the % aa identity is indicated. Similar regions of expressed sequences are identified, and the % nt identity with NIRVs is indicated. CP, capsid protein; RdRp, RNA-dependent RNA polymerase; EST, expressed sequence tag; aa, amino acid; nt, nucleotide.
FIG. 5.
Alignment of putative amino acid sequences of fruit fly NIRV and its best-matched virus. The alignment was obtained using the COBALT program. Identical residues between two sequences are shaded. The regions of RdRp conserved motifs of partitiviruses are boxed. The invariant amino acid residues in partitiviruses are highlighted in red. FsV-1, Fusarium solani virus 1 (GenBank accession no. NP_624350.1).
Partitiviruses in the genera Alphacryptovirus and Betacryptovirus are common in plants, and thus plant EST sequences might be contaminated with such cryptoviral sequences. For example, the CP sequence of White clover cryptic virus 1 has been discovered in an EST library of Trifolium repens (51). We indeed detected numerous viral sequences in various plant EST data. However, we were convinced that the expressed sequences from the NIRVs mentioned above were not contaminating viral sequences because high levels of nt sequence identity were detected between the expressed NIRV sequences and their corresponding genomic NIRVs rather than between them and partitiviruses. Furthermore, such sequences also included flanking nuclear genomic sequences characteristics of repeated elements (Fig. 4). In contrast, no significant nt sequence identity was found between the viral sequences we detected in the EST database and any NIRVs.
To test whether NIRVs are evolving as functional genes, we calculated the Ka/Ks ratio for each branch in the phylogenetic trees of some NIRVs (see Fig. S4 in the supplemental material). The ratios for all branches were <1, indicating that evolution of NIRVs was dominated by purifying selection. However, these Ka/Ks values for NIRVs and viruses were all larger than is typical for strong purifying selection. This may be due to the fact that the sequences used for the Ka/Ks test were highly divergent, and as a consequence, saturation of the number of Ks may occur. The NIRVs not only were expressed but also evolved under pressure of purifying selection, suggesting that they carry functional genes. However, we cannot differentiate between past and present functions or function at the protein or RNA level. The RdRps from fruit fly and yeast species retained the conserved residues of these viruses (Fig. 5; see Fig. S3 in the supplemental material), suggesting that they may have enzymatic activity when expressed. However, we cannot rule out the possibility that the NIRVs evolved from the precise mechanistic function of the viral homolog in subtle ways by positive selection while still maintaining the broader function.
DISCUSSION
Direction of HGTs.
In this study, we provide evidence that genes of totiviruses and partitiviruses have widespread homologs in the nuclear genomes of eukaryotic organisms. Traditionally, viruses are viewed as pickpockets of cellular genes, with the direction of gene transfer occurring predominantly from cell to virus rather than in the opposite direction (50). With this in mind, it is easier to envision viruses that are derived from their nuclear homologs as escaped entities from early eukaryotic or prokaryotic cell environments (16) or as capturing the relevant genes from the host during infection. For instance, the protein synthesis-related genes of mimiviruses were acquired from the host by HGT (48, 49). Other examples include the Hsp70 gene, acquired from host plants by a common ancestor of closteroviruses (11, 29), and the host 28S rRNA gene sequence, acquired by an influenza virus (32). Our data, however, support Taylor and Bruenn's proposal (60) that the direction of transfer was from virus to cell. First, if toti- and partitiviruses represented escaped fragments of their host NIRVs from early cellular chromosomes, one would expect that the NIRVs would cluster together and that their phylogenetic topologies would follow that of species evolution. Our data showed the phylogenetic distribution of the NIRVs to be patchy, suggesting that the transfer of relevant genes between cells and viruses occurred recently. Second, the fact that the RdRps of many RNA viruses belonging to the picorna-like superfamily of eukaryotic positive-strand RNA viruses are phylogenetically related to those of toti- and partitiviruses (38) suggests an ancient origin for these viruses. Third, the CP trees of toti- and partitiviruses have similar topologies to their RdRp trees (6, 21), implying parallel evolution of CPs and RdRps. It is unlikely that the respective genes were acquired independently from the host by HGTs in recent times. In addition, most of the nuclear viral homologs are pseudogenes. Hence, the most likely explanation is that these genes were transferred horizontally from viruses to eukaryotic genomes.
Integration mechanism and integration preference.
Both totiviruses and partitiviruses have no recognized RT and integrase functions. How, then, do viral sequences become integrated? Integration of viral RNA sequences into the nuclear genome must involve reverse transcription and illegitimate recombination events. Recently, Geuking et al. (19) demonstrated that an endogenous retrotransposon can recombine with lymphocytic choriomeningitis virus, resulting in its integration into the mouse genome. We found that the flanking sequences of many viral insertions within eukaryotic genomes had the typical traits of transposable elements or multiple repeat sequences (see Fig. S5 in the supplemental material). It seems likely that these elements are involved in the integration of viral genes into eukaryotic genomes (43, 59). However, integration near an endogenous retrotransposon can also be explained by a common preference for integrating viral sequences into retrotransposable element-rich regions. Given that these regions are particularly unstable and easily form double-strand breaks, such integration events also could occur by double-strand-break repair (55). Indeed, some integrations (such as those in Ixodes scapularis) (see Fig. S5 in the supplemental material) most likely occurred via double-strand-break repair rather than by a retrotransposon-mediated mechanism. Hence, it seems likely that there have been different mechanisms of integration.
Frank and Wolfe (17) proposed that double-strand-break repair is responsible for the capture of sequences of viral or plasmid origin by the yeast nuclear genome. However, the use of viral sequences to repair breaks has not been detected in laboratory experiments associated with double-strand-break repair in S. cerevisiae (46, 55, 67), even though most laboratory strains do contain the ScV L-A virus at high copy numbers (66). In our study, although the ScV L-A-infected S. cerevisiae genome lacked any related homologs, L-A-related homologs were found in other eukaryotic species that are not known to be infected with ScV L-A. In fact, to date, no eukaryotic species with integrated viral genes have been reported to be infected by the respective viruses. Likewise, virus-infected eukaryotic host species are not subject to integration by the invading viral sequences. Furthermore, we found by cloning in silico that at least 40 eukaryotic species contained potential toti- and partitivirus sequences (Liu et al., unpublished data). However, none of these eukaryotic species contained integrated viral genes, except for Chinese cabbage, which harbored viruses distinct from its nuclear viral homologs. Otherwise, we did not find potential toti- and partitiviruses in the eukaryotic species with integrated viral genes, except for the expressed sequences by nuclear homologs discussed above. Considering that toti- and partitiviruses are intimately associated with their host cells over long periods of evolutionary time (22), this persistent association is expected to enhance the likelihood of their integration into the infected host genome. However, integration events most likely occurred when the viruses were first introduced into noninfected species. Hence, it seems feasible that there was a triggering factor for occurrence of integration.
Potential triggering factors for prevalence of virus-to-eukaryote genome HGTs.
Toti- and partitiviruses do not have extracellular routes for infection and are transmitted intracellularly during cell division and sporogenesis (vertical transmission) or, in the case of plant partitiviruses, via ovule and/or pollen transmission. Horizontal or lateral transmission of fungal partitiviruses and totiviruses occurs during cell fusion as a result of hyphal anastomosis. Such horizontal transmission occurs only between individuals within the same species belonging to the same or closely related vegetative compatibility groups. The natural host ranges of these viruses are therefore limited to individuals within the same or closely related vegetative compatibility groups (20, 65). However, physical contacts of unicellular eukaryotes that share the same habitats (such as yeasts and protozoa) can increase the likelihood of interspecies virus transmission. The pathogenic or endosymbiotic relationship between plants and fungi, insects, protozoa, nematodes, etc., can also provide an opportunity for interkingdom transmission of viruses. Toti- and partitiviruses are generally associated with symptomless or persistent infections of their hosts, suggesting coevolutionary interactions between viruses and hosts (20, 64). Hence, they may be recognized as self elements by the infected host's pattern recognition receptors (31, 44, 68) and therefore do not trigger the host's innate immune system. However, when these viruses are transferred to noninfected hosts, or occasionally to nonhost species, the antiviral innate immune response of the cell is induced (31, 44, 68). Virus infection can also activate transposable elements (8, 10, 53) and systemically enhance the frequency of DNA recombination (12, 39). The cDNA fragments of viruses can be synthesized randomly by RTs encoded by these transposable elements (18, 33) and subsequently transported to the nuclear genome. Viral cDNA integration could occur either by hitchhiking on retrotransposons (19, 43, 59) or by double-strand-break repair (17, 54). The integrated viral genes could provide the species with a new phenotype of an immune system in order to recognize and prevent similar infections.
Role of NIRVs in antiviral immunity.
There is growing evidence that retroviruses and endogenous retroviruses have played a major role in the evolution of animal and, most particularly, human adaptive immunity (56, 57, 63, 65). Recently, nonretroviral integration-based immunity was proposed (5, 15, 36). Our data support the hypothesis that integration of virus-specific sequences into eukaryotic genomes results in acquired immunity. The integrated NIRVs could provide the basic elements to recognize and prevent cell infection with related viruses and therefore might affect host immunity. A new phenotype of the host immune system may thus have evolved. This phenomenon might be widespread in diverse organisms with various viruses. To address this question, worldwide surveys of other viral gene integration events are necessary. In fact, we also detected plant positive-strand RNA virus-like sequences in genomes of insects (Liu et al., unpublished data). Hence, with ongoing eukaryotic genome sequencing projects and the availability of sequence information for more viruses, more viral gene integration events will be brought to light.
In conclusion, our study provides substantial confirmation that RNA viruses can donate genetic material horizontally to wide-ranging eukaryotic species and give rise to functionally significant new genes. Given that RNA viruses are widespread (13, 38), they not only may have played important roles in early cellular evolution (9, 16, 37, 64) but also may continue to do so in the subsequent evolution of eukaryotes.
Supplementary Material
Acknowledgments
This research was supported in part by the National Basic Research Program (2006CB101901), the Special Fund for Agro-Scientific Research in the Public Interest (3-21), and the Program for New Century Excellent Talents in University (NCET-06-0665).
We thank the anonymous reviewers for their constructive and helpful comments.
Footnotes
Published ahead of print on 1 September 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ahlquist, P. 2006. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double stranded RNA viruses. Nat. Rev. Microbiol. 4:371-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anisimova, M., and O. Gascuel. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55:539-552. [DOI] [PubMed] [Google Scholar]
- 3.Awadalla, P. 2003. The evolutionary genomics of pathogen recombination. Nat. Rev. Genet. 4:50-60. [DOI] [PubMed] [Google Scholar]
- 4.Bejarano, E. R., A. Khashoggi, M. Witty, and C. Lichtenstein. 1996. Integration of multiple repeats of geminiviral DNA into the nuclear genome of tobacco during evolution. Proc. Natl. Acad. Sci. U. S. A. 93:759-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertsch, C., M. Beuve, V. V. Dolja, M. Wirth, F. Pelsy, E. Herrbach, and O. Lemaire. 2009. Retention of the virus-derived sequences in the nuclear genome of grapevine as a potential pathway to virus resistance. Biol. Direct 4:21. doi: 10.1186/1745-6150-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blawid, R., D. Stephan, and E. Maiss. 2007. Molecular characterization and detection of Vicia cryptic virus in different Vicia faba cultivars. Arch. Virol. 152:1477-1488. [DOI] [PubMed] [Google Scholar]
- 7.Bratke, K. A., and A. McLysaght. 2008. Identification of multiple independent horizontal gene transfers into poxviruses using a comparative genomics approach. BMC Evol. Biol. 8:67. doi: 10.1186/1471-2148-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capy, P., G. Gasperi, C. Biemont, and C. Bazin. 2000. Stress and transposable elements: co-evolution or useful parasites? Heredity 85:101-106. [DOI] [PubMed] [Google Scholar]
- 9.Claverie, J. M. 2006. Viruses take center stage in cellular evolution. Genome Biol. 7:110. doi: 10.1186/gb-2006-7-6-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dellaporta, S. L., P. S. Chomet, J. P. Mottinger, J. A. Wood, S. M. Yu, and J. B. Hicks. 1984. Endogenous transposable elements associated with virus infection in maize. Cold Spring Harbor Symp. Quant. Biol. 49:321-328. [DOI] [PubMed] [Google Scholar]
- 11.Dolja, V. V., J. F. Kreuze, and J. P. Valkonen. 2006. Comparative and functional genomics of closteroviruses. Virus Res. 117:38-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong, X. 2004. Pathogen-induced systemic DNA rearrangement in plants. Trends Plant Sci. 9:60-61. [DOI] [PubMed] [Google Scholar]
- 13.Edwards, R. A., and F. Rohwer. 2005. Viral metagenomics. Nat. Rev. Microbiol. 3:504-510. [DOI] [PubMed] [Google Scholar]
- 14.Filée, J., P. Forterre, and J. Laurent. 2003. The role played by viruses in the evolution of their hosts: a view based on informational protein phylogenies. Res. Microbiol. 154:237-243. [DOI] [PubMed] [Google Scholar]
- 15.Flegel, T. W. 2009. Hypothesis for heritable, anti-viral immunity in crustaceans and insects. Biol. Direct 4:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forterre, P. 2006. The origin of viruses and their possible roles in major evolutionary transitions. Virus Res. 117:5-16. [DOI] [PubMed] [Google Scholar]
- 17.Frank, A. C., and K. H. Wolfe. 2009. Evolutionary capture of viral and plasmid DNA by yeast nuclear chromosomes. Eukaryot. Cell 8:1521-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabriel, A., and S. C. Teng. 1998. LCMV cDNA formation: which reverse transcriptase is responsible? Trends Genet. 14:220-221. [DOI] [PubMed] [Google Scholar]
- 19.Geuking, M. B., J. Weber, M. Dewannieux, E. Gorelik, T. Heidmann, H. Hengartner, R. M. Zinkernagel, and L. Hangartner. 2009. Recombination of retrotransposon and exogenous RNA virus results in nonretroviral cDNA integration. Science 323:393-396. [DOI] [PubMed] [Google Scholar]
- 20.Ghabrial, S. A. 1998. Origin, adaptation and evolutionary pathways of fungal viruses. Virus Genes 16:119-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghabrial, S. A., and M. L. Nibert. 2009. Victorivirus, a new genus of fungal viruses in the family Totiviridae. Arch. Virol. 154:373-379. [DOI] [PubMed] [Google Scholar]
- 22.Ghabrial, S. A., and N. Suzuki. 2008. Fungal viruses, p. 284-291. In B. W. J. Mahy and M. H. V. Van Regenmortel (ed.), Encyclopedia of virology, 3rd ed., vol. 2. Elsevier, Oxford, United Kingdom. [Google Scholar]
- 23.Ghabrial, S. A., and N. Suzuki. 2009. Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 47:353-384. [DOI] [PubMed] [Google Scholar]
- 24.Guindon, S., and O. Gascuel. 2003. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 25.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. U. S. A. 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hordijk, W., and O. Gascuel. 2005. Improving the efficiency of SPR moves in phylogenetic tree search methods based on maximum likelihood. Bioinformatics 21:4338-4347. [DOI] [PubMed] [Google Scholar]
- 27.Horie, M., T. Honda, Y. Suzuki, Y. Kobayashi, T. Daito, T. Oshida, K. Ikuta, O. Jern, T. Gojobori, J. M. Coffin, and K. Tomonaga1. 2010. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature 463:84-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kapitonov, V. V., and J. Jurka. 2008. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat. Rev. Genet. 9:411-412. [DOI] [PubMed] [Google Scholar]
- 29.Karasev, A. V. 2000. Genetic diversity and evolution of closteroviruses. Annu. Rev. Phytopathol. 38:293-324. [DOI] [PubMed] [Google Scholar]
- 30.Katzourakis, A., M. Tristem, O. G. Pybus, and R. J. Gifford. 2007. Discovery and analysis of the first endogenous lentivirus. Proc. Natl. Acad. Sci. U. S. A. 104:6261-6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawai, T., and S. Akira. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131-137. [DOI] [PubMed] [Google Scholar]
- 32.Khatchikian, D., M. Orlich, and R. Rott. 1989. Increased viral pathogenicity after insertion of a 28S ribosomal RNA sequence into the haemagglutinin gene of an influenza virus. Nature 340:156-157. [DOI] [PubMed] [Google Scholar]
- 33.Klenerman, P., H. Hengartner, and R. M. Zinkernagel. 1997. A non-retroviral RNA virus persists in DNA form. Nature 390:298-301. [DOI] [PubMed] [Google Scholar]
- 34.Kohany, O., A. J. Gentles, L. Hankus, and J. Jurka. 2006. Annotation, submission and screening of repetitive elements in Repbase: Repbase Submitter and Censor. BMC Bioinformatics 7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koonin, E. V. 1992. Evolution of double-stranded RNA viruses: a case for polyphyletic origin from different groups of positive-stranded RNA viruses. Semin. Virol. 3:327-339. [Google Scholar]
- 36.Koonin, E. V. 2010. Taming of the shrewd: novel eukaryotic genes from RNA viruses. BMC Biol. 8:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koonin, E. V., and V. V. Dolja. 2006. Evolution of complexity in the viral world: the dawn of a new vision. Virus Res. 117:1-4. [DOI] [PubMed] [Google Scholar]
- 38.Koonin, E. V., Y. I. Wolf, K. Nagasaki, and V. V. Dolja. 2008. The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat. Rev. Microbiol. 6:926-939. [DOI] [PubMed] [Google Scholar]
- 39.Kovalchuk, I., O. Kovalchuk, V. Kalck, V. Boyko, J. Filkowski, M. Heinlein, and B. Hohn. 2003. Pathogen-induced systemic plant signal triggers DNA rearrangements. Nature 423:760-762. [DOI] [PubMed] [Google Scholar]
- 40.Le, S. Q., N. Lartillot, and O. Gascuel. 2008. Phylogenetic mixture models for proteins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363:3965-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Linial, M. L., H. Fan, B. Hahn, R. Lwer, J. Neil, S. Quackenbush, A. Rethwilm, P. Sonigo, J. Stoye, and M. Tristem. 2005. Retroviridae, p. 421-440. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, London, United Kingdom.
- 42.Magidin, M., J. K. Pittman, K. D. Hirschi, and B. Bartel. 2003. ILR2, a novel gene regulating IAA conjugate sensitivity and metal transport in Arabidopsis thaliana. Plant J. 35:523-534. [DOI] [PubMed] [Google Scholar]
- 43.Maori, E., E. Tanne, and I. Sela. 2007. Reciprocal sequence exchange between non-retro viruses and hosts leading to the appearance of new host phenotypes. Virology 362:342-349. [DOI] [PubMed] [Google Scholar]
- 44.Mathieu, P., and S. J. Saupe. 2009. Fungal incompatibility: evolutionary origin in pathogen defense? BioEssays 31:1201-1210. [DOI] [PubMed] [Google Scholar]
- 45.Monier, A., A. Pagarete, C. de Vargas, M. J. Allen, B. Read, J. M. Claverie, and H. Ogata. 2009. Horizontal gene transfer of an entire metabolic pathway between a eukaryotic alga and its DNA virus. Genome Res. 19:1441-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore, J. K., and J. E. Haber. 1996. Capture of retrotransposon DNA at the sites of chromosomal double-strand breaks. Nature 383:644-646. [DOI] [PubMed] [Google Scholar]
- 47.Moreira, D. 2000. Multiple independent horizontal transfers of informational genes from bacteria to plasmids and phages: implications for the origin of bacterial replication machinery. Mol. Microbiol. 35:1-5. [DOI] [PubMed] [Google Scholar]
- 48.Moreira, D., and C. Brochier-Armanet. 2008. Giant viruses, giant chimeras: the multiple evolutionary histories of mimivirus genes. BMC Evol. Biol. 8:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moreira, D., and P. López García. 2005. Comment on “The 1.2 megabase genome sequence of mimivirus.” Science 308:1114. [DOI] [PubMed] [Google Scholar]
- 50.Moreira, D., and P. López-García. 2009. Ten reasons to exclude viruses from the tree of life. Nat. Rev. Microbiol. 7:306-311. [DOI] [PubMed] [Google Scholar]
- 51.Nakatsukasa-Akune, M., K. Yamashita, Y. Shimoda, T. Uchiumi, M. Abe, T. Aoki, A. Kamizawa, S. Ayabe, S. Higashi, and A. Suzuki. 2005. Suppression of root nodule formation by artificial expression of the TrEnodDR1 (coat protein of white clover cryptic virus 1) gene in Lotus japonicus. Mol. Plant Microbe Interact. 18:1069-1080. [DOI] [PubMed] [Google Scholar]
- 52.Papadopoulos, J. S., and R. Agarwala. 2007. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23:1073-1079. [DOI] [PubMed] [Google Scholar]
- 53.Pouteau, S., M. A. Grandbastien, and M. Boccara. 1994. Microbial elicitors of plant defense responses activate transcription of a retrotransposon. Plant J. 5:535-542. [Google Scholar]
- 54.Puchta, H. 2005. The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J. Exp. Bot. 56:1-14. [DOI] [PubMed] [Google Scholar]
- 55.Ricchetti, M., C. Fairhead, and B. Dujon. 1999. Mitochondrial DNA repairs double-strand breaks in yeast chromosomes. Nature 402:96-100. [DOI] [PubMed] [Google Scholar]
- 56.Ryan, F. P. 2004. Human endogenous retroviruses in health and disease: a symbiotic perspective. J. R. Soc. Med. 97:560-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ryan, F. P. 2009. An alternative approach to medical genetics based on modern evolutionary biology. 2. Retroviral symbiosis. J. R. Soc. Med. 102:324-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sambrook, J., E. F. Frisch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 59.Tanne, E., and I. Sela. 2005. Occurrence of a DNA sequence of a non-retro RNA virus in a host plant genome and its expression: evidence for recombination between viral and host RNAs. Virology 332:614-622. [DOI] [PubMed] [Google Scholar]
- 60.Taylor, D. J., and J. Bruenn. 2009. The evolution of novel fungal genes from non-retroviral RNA viruses. BMC Biol. 7:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor, D. J., R. W. Leach, and J. Bruenn. 2010. Filoviruses are ancient and integrated into mammalian genomes. BMC Evol. Biol. 10:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas, H., C. M. Griffiths, B. Hauck, Dorothea, and P. H. Schuenmann. 28 November 1996. Gene controlling senescence in plants. WO patent 9637613-A1.
- 63.Villarreal, L. P. 2009. The source of self: genetic parasites and the origin of adaptive immunity. Ann. N. Y. Acad. Sci. 1178:194-232. [DOI] [PubMed] [Google Scholar]
- 64.Villarreal, L. P. 2005. Viruses and the evolution of life. American Society for Microbiology, Washington, DC.
- 65.Villarreal, L. P. 2009. Origin of group identity: viruses, addiction and cooperation. Springer Press, New York, NY.
- 66.Wickner, R. B. 1996. Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol. Rev. 60:250-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu, X., and A. Gabriel. 1999. Patching broken chromosomes with extranuclear cellular DNA. Mol. Cell 4:873-881. [DOI] [PubMed] [Google Scholar]
- 68.Zipfel, C. 2008. Pattern-recognition receptors in plant innate immunity. Curr. Opin. Immunol. 20:10-16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.