Abstract
Retroviral vector-mediated gene therapy has been successfully used to correct genetic diseases. However, a number of studies have shown a subsequent risk of cancer development or aberrant clonal growths due to vector insertion near or within proto-oncogenes. Recent advances in the sequencing technology enable high-throughput clonality analysis via vector integration site (VIS) sequencing, which is particularly useful for studying complex polyclonal hematopoietic progenitor/stem cell (HPSC) repopulation. However, clonal repopulation analysis using the current methods is typically semiquantitative. Here, we present a novel system and standards for accurate clonality analysis using 454 pyrosequencing. We developed a bidirectional VIS PCR method to improve VIS detection by concurrently analyzing both the 5′ and the 3′ vector-host junctions and optimized the conditions for the quantitative VIS sequencing. The assay was validated by quantifying the relative frequencies of hundreds of repopulating HPSC clones in a nonhuman primate. The reliability and sensitivity of the assay were assessed using clone-specific real-time PCR. The majority of tested clones showed a strong correlation between the two methods. This assay permits high-throughput and sensitive assessment of clonal populations and hence will be useful for a broad range of gene therapy, stem cell, and cancer research applications.
Integration of the retroviral DNA provirus into the host genome is an obligatory step in the retroviral life cycle. Because of this unique property, retroviruses have been adapted as vectors (24, 26) and used successfully to correct genetic diseases, such as X-linked severe combined immunodeficiency (SCID), adenosine deaminase (ADA)-deficient SCID, and X-linked adrenoleukodystrophy, by stable genetic modification of hematopoietic progenitor/stem cells (HPSC) (1, 2, 5, 6, 13, 27, 29). However, the risk of insertional mutagenesis from therapeutic vectors has been demonstrated in several cases in which integration events near or within proto-oncogenes triggered leukemia (8, 12, 14, 16, 34). Therefore, it is important to understand the mechanisms for complex hematopoietic repopulation in humans and to study the behaviors of engineered HPSC clones following transplant.
Since retrovirus vectors uniquely “mark” individual HPSC by vector integration sites (VIS), clonal repopulation by HPSC can be analyzed by tracking the VIS. Restriction enzyme-based assays are commonly used for the clonal tracking, where genomic DNA is digested with restriction enzymes to generate VIS DNA fragments of different lengths that can be detected by Southern blotting (9, 17, 18, 22) or nucleotide sequencing via linker-mediated PCR (LM-PCR) (32), inverse PCR (INV-PCR) (33), or linear amplification-mediated PCR (LAM-PCR) (30). These approaches have been widely used in biological and clinical research to study composition of the HPSC pool, stem cell engraftment, regulatory decisions of individual stem cells, and genotoxicity of retroviral vectors (17, 22, 23, 25, 29, 31, 35). While mouse HPSC repopulation is typically mono- or oligoclonal (17), the number of HPSC clones repopulating in humans or nonhuman primates is much larger, manifesting several hundreds to thousands of repopulating clones posttransplant (5, 31, 35). Recent advances in sequencing technology have enabled high-throughput and parallel clonality analysis through large-scale VIS sequencing and enumeration of VIS sequences (5, 15, 35, 36). However, these methods can detect only VIS that are proximal to restriction enzyme sites, and additional experimental limitations may exist (10, 15, 36). As a result, current assays can only roughly estimate clonal frequencies, so the current standard is to perform clone-specific real-time PCR for sensitive and accurate quantification. Recently, novel clonal tracking assays that do not require restriction enzyme usage have been described (10, 11). However, these methods involve experimental steps that are technically challenging, and they require further optimization to achieve reliable, high-throughput quantification.
Here, we present a novel VIS detection and quantification system based on 454 pyrosequencing and accompanying guidelines for high-throughput quantification of multiple clonal populations. We used a novel bidirectional PCR method to concurrently analyze both the 5′ (left) and the 3′ (right) vector-host junctions in peripheral blood repopulating cells (PBC) in a rhesus macaque transplanted with autologous HPSC transduced with lentivirus vectors (3). The reproducibility and conditions for reliable quantification were tested by two independent experiments conducted on the same PBC collected at four posttransplant time points. The lengths of VIS PCR amplicons, the amount of genomic DNA for analysis, and the intensity of sequencing are important factors influencing the reliability and the sensitivity of the assay. Of 964 unique vector integrants analyzed, the relative quantities of a 398-member subset were determined, demonstrating heterogeneous and dynamic clonal frequency changes over time. Clonal frequencies were further confirmed by clone-specific real-time PCR. We show that this assay detects the majority of VIS that are present in a given clonal population and accurately measures their relative frequencies.
MATERIALS AND METHODS
Control assays for quantitative sequencing of different lengths of DNA.
A total of 15 DNA clones ranging from 125 bp to 1,680 bp in length were prepared by PCR amplification of the pNL4.3 plasmid (see Table S1 in the supplemental material for the 454 fusion primers). The PCR amplicons were mixed in the same molar ratios and subjected to the amplicon sequencing procedure of 454 pyrosequencing to test the PCR efficiency for different lengths of DNA during the emersion PCR (emPCR) step (Roche). We also examined quantitative VIS sequencing using serially diluted control VIS DNA. Four different lengths of VIS DNA (188, 279, 494, and 1,104 bp) were isolated from the first PCR product of a 5-month PBC sample (see Fig. 2a, experiment 2, and below for details) and cloned into TOPO TA cloning vector (Invitrogen). A mixture of four control DNAs (0.7 ng each) was serially diluted by 2-fold and mixed with 2.5 ng of VIS DNA from an acute infection sample (first PCR product in experiment 2). Serial dilutions were subjected to the first PCR step in experiment 2.
In vitro lentivirus vector transduction.
Peripheral blood CD34+ cells were isolated from the leukapheresis cell product collected after mobilization with granulocyte colony-stimulating factor (G-CSF) and stem cell factor (SCF) as described previously (3, 4). CD34+ cells were transduced with the vesicular stomatitis virus G protein-pseudotyped lentivirus vector (CS-RhMLV-E) on RetroNectin (Takara Bio Inc., Japan)-coated, non-tissue culture-treated six-well plates (Becton Dickinson Labware, Franklin Lake, NJ) as described previously (3, 4). Briefly, 1.6 × 107 CD34+ cells were incubated with lentivirus vectors at a multiplicity of infection (MOI) of 3 in medium supplemented with 100 ng/ml SCF, 50 ng/ml interleukin-6, and 2 μg/ml Polybrene. After 2 h, the culture supernatant was removed and replaced with fresh medium. The genomic DNA of infected cells was isolated 4 days after infection using a DNeasy Blood and Tissue Kit (Qiagen).
Isolation of rhesus peripheral blood cells.
Animal (rhesus macaque 95E132) was transplanted with autologous CD34+ cells transduced with lentivirus vector (CS-RhMLV-E) (3) and maintained in accordance with federal guidelines and the policies of the Veterinary Research Program of the National Institutes of Health. The protocols were approved by the Animal Care and Use Committee of the National Heart, Lung, and Blood Institute. Peripheral blood cells were isolated from EDTA-treated whole blood by ammonium chloride-mediated red blood cell lysis in 2-week to 6-month intervals and cryo-preserved.
Bidirectional VIS PCR amplification.
Two approaches (experiment 1 and experiment 2) were tested for quantitative analysis of both the left and the right vector-host junction DNA (see Fig. 2a for the schematic view of the procedure and Table S2 in the supplemental material for primer sequences).
Experiment 1 (using INV-PCR method).
We modified the inverse-PCR (7) to amplify both the left and the right vector-cellular DNA junctions simultaneously. Two primers specific to vector-host DNA junctions, LgC (0.1 μM) and RgB (0.1 μM), were bound and linearly extended in the 500-μl reaction solution containing 2.5 to 10 μg of genomic DNA, 1× ThermoPol buffer (New England Biolabs), 0.2 mM deoxynucleoside triphosphates (dNTPs), and 25 units of Taq DNA polymerase (New England Biolabs) under the following conditions: 94°C for 3 min, followed by incubation at 55°C for 5 min and at 72°C for 5 min. To remove single-stranded DNA, the reaction mixture was mixed with 10 units of T7 endonuclease I, 60 units of RecJf, 60 units of exonuclease I, and 1× buffer 4 (New England Biolabs) and incubated at 37°C for 1 h. DNA was purified by phenol and chloroform extractions. Double-stranded vector-host junction DNA was digested with TaqαI [(T/C)GA]. TaqαI digestion produces a fixed length of vector DNA (550 bp of vector DNA for the left junction or 812 bp of vector DNA for the right junction) joined by different lengths of cellular DNA for each junction DNA depending on the location of the TaqαI site within the cellular DNA. Digested DNA was purified and ligated under conditions favoring intramolecular ligation. Circularized DNA was amplified by a two-step PCR. The first PCR was carried out using primer Lg1 and primer Rg1 to amplify both the left and the right vector-cellular DNA junctions in 500 μl of PCR mixture with a 0.5 μM concentration of each primer, 0.2 mM dNTPs, and 50 units of Taq DNA polymerase under the following conditions: 2 min of preincubation at 94°C, followed by 20 cycles of 94°C for 25 s, 56°C for 25 s, and 72°C for 2.5 min. Amplified DNA was purified using a QIAquick PCR purification kit (Qiagen). The left and the right DNA junctions were then separately amplified by the junction-specific second PCR. A total of 10 ng of DNA from the first PCR product was used for the second PCR. Primers for the second PCR were designed as described in the amplicon sequencing manual for 454 pyrosequencing (28). For PCR amplification of the left junction, reverse primer B-4-Lg2 and forward primer A-4-LgF were used. The second PCR conditions were the same as those of the first PCR except that 24 cycles were used. For the right vector-cellular DNA junction, forward primer A-4-Rg2 and reverse primer B-4-RgR were used for the second PCR with 28 cycles.
Experiment 2 (using both LM-PCR and INV-PCR methods).
The left and the right vector-cellular DNA junctions were processed using INV-PCR and LM-PCR, respectively, with modifications. The initial steps for the junction-specific linear extension and single-stranded DNA digestion were performed as described above, except that 5′ end-biotinylated primer, B_RgC (5′-biotin-GGTACCTTTAAGACCAATGAC) was used instead of RgB. The right-junction DNAs, which were biotinylated at the 5′ end of the vector side DNA, were bound to streptavidin-agarose Dynabeads (Dynal magnetic beads; Invitrogen) and separated from the left-junction DNAs (in the supernatant) using a magnetic separator, according to the manufacturer's instructions. After separation, the right-junction DNAs were processed following the conventional LM-PCR method as described previously (19) with modifications. The bead-bound DNA was digested with TaqαI. Since the vector DNA does not harbor a TaqαI site, only cellular DNA was cleaved. Then the TaqαI cleavage site in cellular DNA was ligated to a linker DNA containing a CG nucleotide 5′ overhang complementary with the TaqαI-digested DNA. The linker-ligated DNA was subjected to the two-step PCR. The first PCR was carried out using primers Rg1 and Link1 (5′-TAACTGTCACACCTGGAGATA-3′) in a final volume of 300 μl with a 0.5 μM concentration of each primer, 0.2 mM dNTPs, and 12 U of Taq DNA polymerase under the following conditions: 2 min of preincubation at 94°C, followed by 24 cycles at 94°C for 25 s, 58°C for 25 s, and 72°C for 2.5 min. The PCR product was purified using a PCR purification kit (Qiagen) and was used as the template for the second PCR. Ten nanograms of DNA from the first PCR product was used for the second PCR. The conditions were identical to those for the first PCR, except that the second PCR was conducted with 18 cycles using 454 fusion primers, A-5-Rg2 and B-4-Link2. After bead separation, left-junction DNA in the supernatant was cleaned by phenol and chloroform extractions and digested with TaqαI. Digested DNA was purified and ligated under conditions favoring intramolecular ligation. Circularized VIS DNA was amplified by a two-step PCR. The first PCR and the second PCR were carried out under the identical conditions described above for the left-junction amplification in the experiment 1.
454 Pyrosequencing.
The VIS PCR products were purified and subjected to 454 pyrosequencing (Roche) using Genome Sequencer FLX and GS FLX standard series kits for amplicon sequencing according to the company manual (28), except that we modified emPCR conditions as follows: 4 min at 94°C followed by 40 cycles of 94°C for 30s, 58°C for 3 min, and 68°C for 2 min; this was followed by an additional 13 cycles of 94°C for 30 s and 58°C for 6 min. Different samples were run and analyzed in parallel using an eight-region gasket and the 4-bp tag (barcode) sequences within the fusion primers.
Analysis of VIS sequences.
VIS sequences were collected and analyzed after two runs of 454 pyrosequencing. Initially, VIS DNA sequences from the left and the right junctions were separately processed (see Fig. S1 in the supplemental material). Sequences were analyzed according to the following program.
(i) VIS authentication.
VIS sequences were authenticated by the existence of a vector-cellular DNA junction. The cellular DNA sequences were confirmed by alignment to the rhesus macaque genome (January 2006 rheMac2 assembly) and the human genome (March 2006 hg18 assembly; NCBI build 36.1) using BLAT (www.genome.ucsc.edu) and GMAP (www.gene.com/share/gmap/).
(ii) Sequence enumeration of unique VIS.
The numbers of individual VIS sequences were counted based on the results of sequence comparison as well as sequence alignment onto the genome. Initially, individual sequences were clustered into groups of identical sequences based on sequence similarity (>95% homology) between sequences and their genomic alignment results, and the total number of sequences for each group and its representative nucleotide sequence were determined. Of total 73,470 sequences analyzed, we found that about 26% contain sequencing errors, mostly homopolymer errors (21), and failed to have a good match (>95% homology) to a correct VIS sequence (a sequence without errors). Other errors including nucleotide substitution, insertions, and deletions were relatively rare and were allowed by the >95% homology criterion. Homopolymer errors were apparent when an error-containing sequence was compared to the correct VIS sequence. Hence, we identified and added those sequences containing homopolymer errors into the counts of the correct VIS sequences by a three-step search/match process. In step 1, the intermediate sequence data from the previous process were clustered into different groups based on sequence similarity at a lower stringency condition (>90% homology). In step 2, we assigned a different confidence level to individual sequences within a group and chose the highest ranking one to use as a standard sequence, representing the correct VIS sequence, for comparison with other error-containing sequences. The ranking criteria determining the confidence levels are as follows: first, a sequence whose authenticity is confirmed by the presence of the other side-junction sequence in our data set (see “Matching the left and the right junctions” below); second, a sequence that aligns best, with at least 95% homology, onto the genome; third, a sequence with higher sequence counts; fourth, a sequence with a TaqαI site at the 3′ end; and fifth, a sequence with longer sequence read. In step 3, any sequence within a group showing >95% sequence homology, excluding mismatches at homopolymer stretches, to the standard sequence in the same group was added into the count of the standard sequence. We repeated one more cycle of the three-step process with the matching stringency of 80% at step 1 to cover more error-containing sequences. Lastly, for some sequences that were apparently identical to one of the VIS sequences but remained uncombined, we manually added those sequences into the appropriate VIS sequences.
(iii) Removal of sequence count data for VIS DNA of >500 bp.
Since quantitative sequencing of the PCR amplicons of >500 bp was not efficient under current experimental conditions, sequence count data for VIS DNA of >500 bp were removed from quantitative analysis. The VIS DNA lengths were determined based on the nearest restriction enzyme site (TaqαI).
(iv) Matching the left and the right junctions.
When a VIS sequence from one of two vector-host junctions was aligned onto the genome in opposite orientation of another VIS sequence from the other junction with a 5-bp overlap, which is a typical phenomenon of genomic DNA duplication following HIV integration, we considered these pairs to have originated from the same vector integrant. Sequence counts from these pairs were averaged (see Fig. 5a).
(v) Compensation factor.
To adjust for overestimation of clonal frequencies, we applied a compensation factor (0.39) (see Fig. 5b) to the relative sequence frequencies of individual vector integrants.
(vi) Statistical analysis.
To test the correlation of the clonal frequencies between two experiments, we used the Pearson product-moment correlation coefficient (r). We used r × c contingency table analysis (chi-square approximation) and a Student's t test to test the null hypothesis of no correlation between the two experiments and equal numbers of unique VIS, respectively. Continuous variables are summarized by the mean ± standard deviation (SD).
Clone-specific real-time PCR.
Primer sequences and their annealing locations are listed in Table S2 in the supplemental material. The clone-specific primers (LcA, LcB, Lc1, Lc2, RcA, RcB, Rc1, and Rc2) were designed based on the rhesus genome sequence at the vicinity of the VIS, and the general primers specific to vector DNA (LgA, LgB, LgC, Lg1, Lg2, Lv1, Lv2, LgF, RgR, RgA, RgB, RgC, Rg1, Rg2, Rv1, and Rv2) were designed based on the vector DNA sequences. To obtain accurate quantitative measurements, the clone-specific primers (Lc1, Lc2, Rc1, and Rc2) were chosen after testing six to seven different primers for each junction. Primers were synthesized by Integrated DNA Technologies, Inc. Copy numbers were determined by (i) standard assays and (ii) clonal real-time PCR.
(i) Standard assays.
The vector-host DNA junctions of seven chosen clones were cloned to use as copy number standards for individual clone-specific real-time PCRs. The junction DNA was PCR amplified using primers LcA, LgA, LcB, and LgB for the left junctions and primers RcA, RgA, RcB, and RgB for the right junctions and cloned using TOPO-TA cloning kit (Invitrogen). The copy number of standard plasmid DNA was calculated based on the DNA concentration measured by a NanoDrop 1000 instrument (Thermo Scientific) and molecular weight of the plasmid. The plasmid DNAs were serially diluted into 20 ng/μl rhesus genomic DNA in the range of 100 to 106 plasmid copies.
(ii) Clonal real-time PCR.
To quantify the total copy number of the clone of interest, a real-time nested-PCR procedure was performed. In the first round of PCR, the target DNA was amplified with 0.5 μM LgC and Lc1 primers for the left junctions or RgC and Rc1 primers for the right junctions, respectively, using 500 ng of genomic DNA or 100 to 106 copies of clone-specific plasmids for standards under the following conditions: 2 min of preincubation at 94°C, followed by 16 cycles of 94°C for 25 s, 58°C for 25 s, and 72°C 1.5 min. After purification of the first PCR product using a QIAquick PCR Purification Kit (Qiagen), 1/10 of the eluted DNA was subjected to the second round of PCR using Lg1 and Lc2 primers for the left junctions or Rg1 and Rc2 for the right junctions, respectively, in a 25-μl mixture containing iTaq SYBR Green Supermix with ROX (6-carboxy-X-rhodamine; Bio-Rad). All reactions were performed on an IQ-5 real-time PCR detection system (Bio-Rad) under the following conditions: 1.5 min of preincubation at 94°C, followed by 40 cycles of 95°C for 10 s, 58°C for 20 s, and 72°C for 30 s. Total vector copies were also measured by the identical real-time PCR procedure described above, except that internal primer sets, Lv1 and Lv2 for the left junctions and Rv1 and Rv2 for the right junctions, respectively, were used instead of clone-specific primers. All the real-time PCRs were conducted in parallel with standard assays using serially diluted clone-specific standard plasmids. The total copy number of the target junction DNA was determined in reference to the clone-specific standard curve.
RESULTS
Quantitative sequencing of different lengths of DNA.
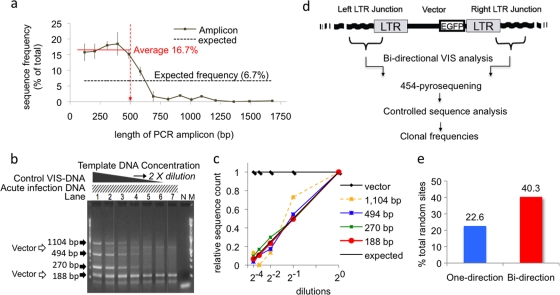
Popular VIS sequencing methods, such as LM-PCR, INV-PCR, and LAM-PCR, use restriction enzymes to generate different lengths of vector-host junction DNA (VIS DNA). The recovery of integration sites relies on the genomic distribution of restriction enzyme sites as only a fraction of VIS that generate short VIS DNA may be efficiently amplified by PCR (10). To test the efficiency of 454 pyrosequencing for VIS sequences of various lengths, two control experiments were performed (Fig. 1 a to c). First, 15 DNA clones ranging in size from 125 bp to 1,680 bp were mixed in the same molar ratios and subjected to 454 pyrosequencing. Although all DNA clones were detectable, sequence frequencies were severely biased by the length of DNA. Sequence frequencies assayed for DNA clones of ≤488 bp were relatively even, but the efficiency dropped for longer DNA clones under the current experimental conditions (Fig. 1a). Second, to test the accuracy of our quantification method, four different VIS DNA clones (188, 279, 494, and 1,104 bp) were serially diluted and mixed with a pool of VIS DNA from CD34+ HPSC that were acutely infected with lentivirus vectors (Fig. 1b). Diluted samples were subjected to the first PCR step of the LM-PCR procedure (see Fig. 2a) and sequenced in parallel. The sequencing results showed that the relative frequencies of three control VIS DNAs of <500 bp in different dilutions showed a linear correspondence to the dilution factor (correlation γ ranging from 0.989 to 0.999) (Fig. 1c). The longer DNA clone (1,104 bp control VIS DNA) showed a weak positive correlation to the dilution factor, and its sequence frequency was 10 to 100 times lower than that of the other controls. These experiments demonstrate that 454 pyrosequencing accurately enumerates DNA of <500 bp but is unsuitable for quantifying larger sequences.
FIG. 1.
Quantitative sequencing via pyrosequencing-based, bidirectional vector integration site analysis. (a) Sequencing efficiency for different lengths of DNA. The sequencing efficiency dropped for DNA of >500 bp but was relatively stable for DNA of ≤500 bp when 15 control DNAs (lengths ranging from 125 to 1,680 bp) were tested by the 454 pyrosequencing. (b and c) Analysis of the serially diluted control VIS DNA showed linear correspondence of the relative sequence frequencies to the dilution factor for DNA of <500 bp. A mixture of four control VIS DNAs was serially diluted and mixed with the VIS DNA from acutely infected cells (b). Panel B shows a 2% agarose gel run after the second PCR. Lanes 1 to 7, 2-fold serial dilutions of the control DNAs; lane N, water; lane M, 2-log DNA marker; dark arrows, four control VIS DNA; empty arrows, internal control (vector, LTR circles from acute infection). The relative sequence counts of the control VIS DNAs of <500 bp are in linear correlation with the expected values after 2-fold dilution (c). (d) The schematic view of the assay. The bidirectional VIS analysis enabled concurrent analysis of both the left and the right vector-host DNA junctions. Amplified junction DNA was then subjected to 454 pyrosequencing, followed by controlled sequence analysis. (e) Percentage of random vector integrants that can generate VIS DNA of <500 bp. The length of VIS DNA was determined based on the location of the nearest TaqαI site. With the one-directional approach, about 22.6% of random VIS generated VIS DNA of <500 bp. With the bidirectional approach, 40.3% could generate VIS DNA of <500 bp in either one of two directions (upstream or downstream from the vector).
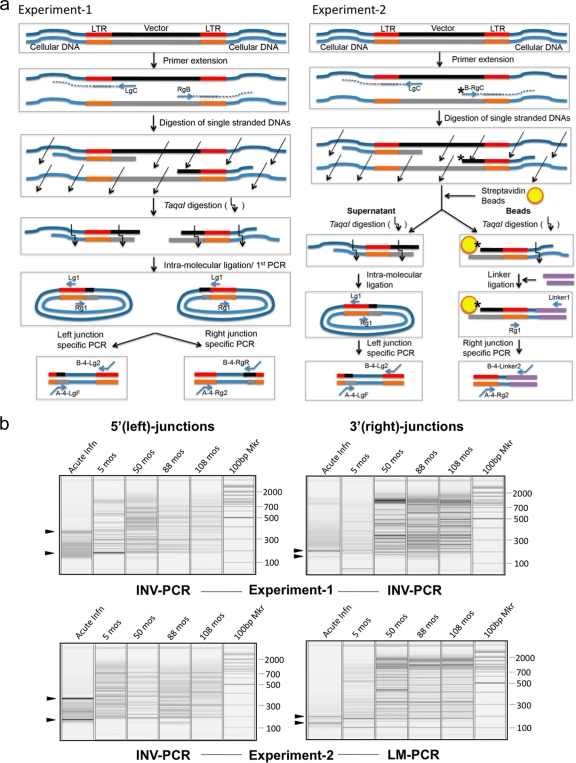
FIG. 2.
Bidirectional VIS analysis. (a) Schematic diagram outlining the major steps of the two approaches (experiment 1 and experiment 2) for the bidirectional VIS amplification. See Materials and Methods for a detailed description of the experimental procedures. Oblique arrows indicate nuclease digestion of single-stranded DNA. Bent arrows indicate TaqαI digestion. Ovals and stars (*) represent streptavidin beads and 5′ end biotins, respectively. DNA linkers for experiment 2 are denoted by double purple lines. (b) QIAEXEL capillary electrophoresis analysis. Bidirectional PCR analysis of acutely infected CD34+ cells (Acute Infn) and peripheral blood repopulating cells collected at 5, 50, 88, and 108 months (mos) posttransplant. Left junctions and right junctions were concurrently analyzed by either INV-PCR or LM-PCR during experiment 1 or experiment 2. A 100-bp marker (Mkr) is included in each panel. Arrowheads indicate DNA bands resulting from LTR circles after acute infection.
Vector integration site sequencing for repopulating nonhuman primate HPSC clones.
Current VIS sequencing methods analyze only one of two vector-host DNA junctions at a time. To improve the chance to detect vector integrants, we developed a novel bidirectional method with which both the left and the right junctions of the same vector integrant can be concurrently analyzed in a single experiment (Fig. 1d and Fig. 2 a). The two junctions were concurrently processed and amplified by either INV-PCR or LM-PCR with modifications. We used TaqαI (TCGA) enzyme to fragmentize genomic DNA. When random VIS were modeled for the conventional one-directional VIS analysis using TaqαI, only about 22% of random vector integrants generated VIS DNA of <500 bp. However, when random integrants were analyzed with the bidirectional approach, about 40.3% generated VIS DNA of <500 bp at either one of two junctions (Fig. 1e).
We tested the technique by analyzing repopulation of HPSC following transplant in a nonhuman primate. A rhesus macaque (95E132) was transplanted 10 years ago with autologous CD34+ cells following ex vivo transduction with self-inactivating lentiviral vectors (3). The animal has maintained stable marking in all hematopoietic lineages to date without adverse effects. Peripheral blood cells (PBC) collected from the animal at four time points over 9 years as well as from acutely infected CD34+ HPSC, as a control sample, were subjected to the assay. Two independent experiments were conducted on the same sample to test assay conditions and reproducibility. During the analyses, the left junctions were amplified by the modified INV-PCR method (Fig. 2a, both experiments), and the right junctions were amplified by the INV-PCR (experiment 1) or by the modified LM-PCR (experiment 2). When PCR-amplified VIS DNA clones from PBC samples were visualized by QIAEXEL capillary electrophoresis analysis (Fig. 2b), various intensities of distinctive bands in the range of 150 to 2,000 bp were observed, reflecting differential clonal expansions. The band patterns of experiments 1 and 2 were similar for the left and right junctions, demonstrating the consistency of VIS amplification by either INV-PCR or LM-PCR. In contrast, VIS DNA from acute infection appeared more uniformly distributed, except DNA bands resulting from long terminal repeat (LTR) circles.
As observed by VIS DNA band patterns, sequence counts for individual unique VIS showed dissimilar patterns between PBC and acutely infected cells. After pyrosequencing of VIS DNA from PBC at four time points, 28,118 and 26,151 VIS sequences were obtained from the left and the right vector-host junctions, respectively, and a total of 562 and 616 unique VIS, respectively, were identified (Table 1; see also data files in the supplemental material). Sequence counts for individual VIS were highly variable, ranging from 1 to 674 sequences per unique VIS (with 11.2 ± 27.1 [SD] to 27.8 ± 46.8 sequences on average, depending on the junctions and time points), indicating differential clonal expansion of certain repopulating clones. In contrast, the acute infection samples had 2,996 unique VIS with an average of 1.4 ± 1.1 sequences per unique VIS, indicating no major clonal expansions of transduced cells after random vector integration.
TABLE 1.
Summary of VIS sequence analysis
| Sample and expt. | Time (mos.) | Amt of genomic DNA (μg) | Junctiond | No. of sequences |
No. of sequence counts/per unique VIS |
No. of unique VIS ≤450 bp | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Totala | Uncertainb | VISc | Unique VIS | Maxf | Median | AvG | SD | |||||
| In vivo repopulating PBC (95E132) | ||||||||||||
| Expt. 1 | 5 | 5.3 | L | 3,873 | 543 | 3,332 | 192 | 346 | 6 | 17.35 | 41.35 | 109 |
| R | 2,649 | 524 | 2,124 | 174 | 179 | 5 | 12.28 | 19.63 | 99 | |||
| 50 | 2.8 | L | 4,165 | 358 | 3,814 | 187 | 300 | 5 | 20.40 | 41.34 | 85 | |
| R | 3,802 | 461 | 3,341 | 147 | 367 | 7 | 22.73 | 47.65 | 76 | |||
| 88 | 2.5 | L | 3,747 | 211 | 3,537 | 192 | 222 | 8 | 18.42 | 27.77 | 84 | |
| R | 3,157 | 104 | 3,203 | 196 | 322 | 8 | 16.34 | 31.97 | 77 | |||
| 108 | 10.7 | L | 4,692 | 228 | 4,475 | 225 | 371 | 5 | 19.89 | 43.00 | 125 | |
| R | 6,756 | 365 | 6,241 | 258 | 674 | 5 | 24.19 | 63.27 | 106 | |||
| Expt. 2 | 5 | 5.3 | L | 3,669 | 648 | 3,025 | 178 | 183 | 7 | 16.99 | 28.44 | 110 |
| R | 2,828 | 1,192 | 1,636 | 170 | 179 | 5 | 9.62 | 17.29 | 89 | |||
| 50 | 2.8 | L | 3,193 | 147 | 3,049 | 121 | 287 | 7 | 25.20 | 41.99 | 67 | |
| R | 3,003 | 497 | 2,506 | 224 | 214 | 3 | 11.19 | 27.13 | 112 | |||
| 88 | 2.5 | L | 3,936 | 216 | 3,725 | 134 | 265 | 8 | 27.80 | 46.79 | 71 | |
| R | 2,558 | 293 | 2,265 | 178 | 258 | 4 | 12.72 | 27.94 | 90 | |||
| 108 | 10.7 | L | 3,372 | 220 | 3,161 | 233 | 169 | 4 | 13.57 | 24.60 | 110 | |
| R | 5,947 | 1,112 | 4,835 | 306 | 523 | 3.5 | 15.80 | 41.52 | 124 | |||
| Total | L | 30,647 | 2,571 | 28,118 | 559 | 216 | ||||||
| R | 30,700 | 4,548 | 26,151 | 615 | 205 | |||||||
| Combined (L-R match)e | 61,347 | 7,119 | 54,269 | 1,174 (212) | 422 (24) | |||||||
| In vitro acutely infected CD34+ cells | ||||||||||||
| Expt. 1 | 10 | L | 2,670 | 1,288 | 1,330 | 861 | 22 | 1 | 1.54 | 1.39 | 739 | |
| R | 1,541 | 995 | 493 | 370 | 7 | 1 | 1.33 | 0.84 | 299 | |||
| Expt. 2 | 10 | L | 2,478 | 1,252 | 1,165 | 752 | 15 | 1 | 1.55 | 1.39 | 642 | |
| R | 5,434 | 4,175 | 1,196 | 1,039 | 8 | 1 | 1.15 | 0.55 | 871 | |||
| Total | L | 5,148 | 2,540 | 2,495 | 1,591 | 1,362 | ||||||
| R | 6,975 | 5,170 | 1,689 | 1,405 | 1,167 | |||||||
| Combined (L-R match)e | 12,123 | 7,710 | 4,184 | 2,996 (3) | 2,529 (2) | |||||||
Total number of sequences that passed filtering after pyrosequencing.
Uncertain sequences include low-quality sequences and vector sequences.
Authenticated VIS sequences (vector DNA joined to cellular DNA).
Analyzed vector-cellular DNA junctions L, left; R, right.
Number of VIS pairs from the left and the right junctions originating from the same vector integrant. The number of matches is shown in parentheses.
Max, maximum.
Controlled sequence frequency analysis to calculate the quantities of individual clones.
Selective amplification and sequencing efficiency for DNA of <500 bp was observed in our control analyses, and rhesus PBC samples showed similar results. A total of 39.3 to 61.8% of unique VIS DNA clones were <500 bp, accounting for 54.1 to 86.2% of total VIS sequences (Fig. 3 and Table 1). There was no notable sequence frequency bias among VIS DNA of <500 bp; VIS DNA clones of >500 bp had lower frequencies, consistent with our reconstitution studies using cloned DNA (Fig. 1a to c). Hence, we used VIS DNA of <500 bp to estimate the relative frequencies of individual vector integrants. The number of unique VIS selected for quantitative analysis ranged from 67 to 125 depending on the left/right junction type and time points analyzed, but we did not see significant differences between results from experiments 1 and 2 (95.4 ± 17.7 versus 96.6 ± 20.6, respectively) or between the left and the right junctions (95.4 ± 21.3 versus 96.6 ± 16.9, respectively).
FIG. 3.
Sequence frequency distribution for different lengths of VIS DNAs. The length of VIS DNA was calculated based on the distance of the nearest TaqαI site from the junction added to 50 bp of vector DNA. Individual VIS were distributed primarily from 25 to 450 bp.
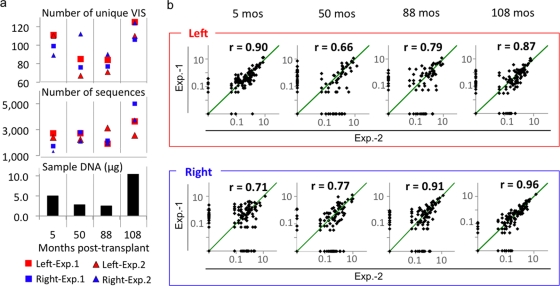
We tested the reproducibility of this analysis by comparing VIS frequencies between experiments 1 and 2. Pearson's product-moment coefficient (r) was calculated for the two experiments at each vector-host junction for samples obtained at four different posttransplant time points. The correlation between experiments ranged from 0.66 to 0.96, depending on the time points and junctions, and may be attributed to differences in the amount of genomic DNA and sequencing intensity (Fig. 4). The experimental correlation declined when fewer than 2,000 VIS sequences were present or when less than 5 μg of genomic DNA was used. The highest correlation (r = 0.96) was observed for the right junction of PBC of 108 months, for which 6,241 (experiment 1) and 4,835 (experiment 2) sequences were generated from 10.7 μg of genomic DNA. Since the overall marking efficiency was maintained at a relatively constant level (data not shown), the amount of genomic DNA is a reflection of the absolute number of vector integrants in the sample (3.8 × 104 to 4.8 × 104 vector copies per μg of DNA). Hence, these results indicate that both the amount of sample (vector copies) and the sequencing intensity are important factors for the sensitivity as well as the reproducibility of the assay.
FIG. 4.
Sequence frequency analysis for unique VIS of <450 bp. (a) Both the amount of sample DNA and the number of available VIS sequences influence detection of unique VIS. The number of unique VIS and the number of VIS sequences from the left (red) and the right (blue) junctions generated by experiment 1 or experiment 2 as well as the amount of genomic DNA used in the analysis were displayed at each time point. (b) Reproducibility of the assay. The relative frequencies (percentage of total sequences for VIS DNA of <500 bp) for individual sites from the two experiments were plotted on logarithmic scales. Reproducibility was tested using a Pearson product-moment correlation coefficient (r). Green diagonal lines indicate complete frequency match (r = 1). Higher correlation was observed when >5 μg of genomic DNA and >2,000 VIS sequences were used for the analysis (PBC from 5 months and 108 months, respectively).
To achieve reliability in clonal frequency analysis, sequence data from experiments 1 and 2 were combined so that the analysis could be based on a sufficient DNA sample and VIS sequence count (total of >2 × 105 vector copies and >3,000 VIS sequences). Individual vector integrants were quantified based on the relative sequence frequencies available at either one of the two junctions (Fig. 5 a). At this step, the frequency for each vector integrant was calculated as a fraction of only the quantifiable rather than total vector integrants. Hence, assuming that 40% of total vector integrants were quantifiable, we estimated a 2.56-fold overestimation for the calculated frequencies (Fig. 5b). Hence, we adjusted them with this compensation factor (1/2.56 = 0.39) and present these adjusted frequencies as the percentage of total vectors or total marked cells (Fig. 5c and d).
FIG. 5.
Determination of relative frequencies for individual clones. (a) Calculation of clonal frequencies. The relative frequencies of VIS DNA of <500 bp from the left (x) and the right (y) junction were combined as described and represented as quantifiable vector integrants (QVIs). (b) The relation between the fraction of QVI in total vector (vertical axis) and expected overestimation (n-fold) of individual QVI frequency (horizontal axis). When 40.3% of vector integrants are QVIs, individual QVI frequencies are 2.56-fold overestimated. (c) Clonal frequency changes at four time points. The adjusted frequencies for individual QVIs were displayed according to the following color scheme: white to black to red, representing 0% to 0.1% to 3.1%. The frequency change of the top 15 highest-frequency QVIs was magnified on the right side. Ten of them were unambiguously mapped onto the rhesus genome (genomic locations are indicated on the right). Among those, seven were further tested with clone-specific real-time PCR. ND, nondetermined. (d) Clone-specific real-time PCR. The relative frequencies of seven QVIs were confirmed by clone-specific real-time PCR. Dark bars denote the adjusted relative frequencies determined by quantitative VIS sequencing. The values obtained by clone-specific real-time PCR (percentage of total vector copies) at the right junction and at the left junction of the vector integrants are shown with yellow and green bars, respectively.
Differential and polyclonal repopulation in a rhesus macaque.
At any given time point, the frequencies of individual vector integrants were markedly different, ranging from 0.002% to 3.2% of total and averaging 0.37 to 0.53%, depending on the time points (Fig. 5c). Furthermore, the relative frequencies of individual vector integrants changed across the four time points. Of the 401 analyzed vector integrants, 239 (60%) were identified at multiple time points and were relatively frequent (average, 0.16%) compared to those that appeared at only one time point (162 vector integrants with a 0.02% frequency average), suggesting that large numbers of clonal populations were circulating in the blood, frequently under the detection limit, with dynamic and heterogeneous kinetics. Thus, the new assay can detect subtle clonal frequency changes over time and analyze a large number of clones in parallel.
To further validate the assay, seven clones were chosen for clone-specific real-time PCR (Fig. 5c and d). Copy numbers of the left or the right vector-host junctions were determined and normalized by total vector integrant at each time point. The majority of clones had a strong correlation between the new assay and the real-time PCR result (γ greater than 0.9) (Fig. 5d), demonstrating accurate clonal frequency detection by the quantitative VIS sequencing assay.
DISCUSSION
We have described a new system for accurate and high-throughput, parallel quantification of individual clones in a complex polyclonal cell population. Hundreds of repopulating clones in a rhesus macaque, transplanted with lentivirus vector-transduced autologous HPSC, were quantified at four time points over 9 years posttransplant. The novel bidirectional VIS PCR method enabled concurrent analysis of both the left and the right vector-host junctions. Through a relatively simple process, we increased the capacity of clonal analysis by nearly 2-fold. Furthermore, this method significantly improved the sequence enumeration process during VIS sequence analysis by using the two data sets from each junction to validate each other, enabling identification of the correct VIS sequences among error-containing similar sequences. Under our experimental conditions, PCR amplification and sequencing were efficient for VIS DNA of <500 bp. Hence, we focused on the portion of the vector integrants that can be detected without the DNA length-associated bias. Using the described analysis, we were able to detect vector-marked clones with frequencies of ≥0.002%, equivalent to 1/150,000 PBC of the 108-month sample, with a vector marking frequency of 32% (data not shown). Due to the sensitivity of the assay, we were able to detect a wide variety of different frequencies of repopulating clones in the circulating PBC. Clonal frequencies detected at each time point were confirmed by clone-specific real-time PCR.
Compared to Southern blot analysis, VIS sequencing is more sensitive and allows for a higher-throughput clonality analysis by counting VIS sequences. With conventional chain termination sequencing, there are many experimental steps that may have different efficiencies for different lengths of target DNA, including PCR, cloning of VIS DNA, and transformation for plasmid DNA purification. Simple VIS sequence enumeration without considering these factors may have caused the poor representation of clonal frequencies reported by Harkey et al. (15). 454 Pyrosequencing significantly simplified the VIS analysis procedure by circumventing cloning and bacterial culture steps using the innovative emPCR step, in which individual VIS DNA can be captured and amplified separately in a water-in-oil mixture prior to pyrosequencing. When ligation-mediated PCR (LM-PCR or INV-PCR) and 454 pyrosequencing are used, PCR is the major limiting step determining the efficiency of competitive sequencing of multiple VIS DNA. We did not observe the DNA length-associated bias at the sequencing step. Hence, application of simple analysis criteria, by which unreliable sequence frequency data are removed from the analysis, significantly improved the reliability of VIS quantification. In addition, we also observed that the amount of sample DNA and the intensity of sequencing were important factors influencing the sensitivity and the reliability of quantitative measurement. In our analysis, a high reproducibility was observed when 5 to 10 μg of genomic DNA (containing 2 × 105 to 5 × 105 copies of vector integrants) was used for the analysis. Considering that generally 1 ng to 1 μg of genomic DNA was used in previous studies (5, 15, 20, 31, 36), the amount we used may have allowed more reliable analysis of vector integrants. After accounting for these factors, we were able to reliably determine the relative quantities of 398 clones, corresponding to 40% of the total expected vector integrants, at four posttransplant time points.
The system we demonstrate here is a sensitive, accurate, and high-throughput assay. It can perform parallel analyses of multiple samples, generating data that may be equivalent to hundreds of clone-specific real-time PCRs. Here, our primary purpose was validation of the methodology, so only a few samples from one animal were analyzed. With accumulation of sufficient data at multiple time points in more animals, in-depth characterization of primate HPSC engraftment and repopulation kinetics would be achievable at the clonal level. Furthermore, this system is also a useful tool for the development of efficient and safe retrovirus vectors to test the effects of insertional mutagenesis or the effects of the vector components, such as therapeutic transgenes, promoters, enhancers, and other noncellular elements. The assay can be further developed and easily adapted for use in the clinic as the efficiency and the safety of stem cell-based gene therapies rely heavily on the understanding and monitoring of normal and abnormal growth of engineered stem cells in the recipient.
Supplementary Material
Acknowledgments
This study was partly funded by the NIH (AI055281-06A2 and CA68859), the California Institute of Regenerative Medicine (RS1-00172-01), and the UCLA AIDS Institute/Center for AIDS Research (AI28697) and was supported in part by the Intramural Research Program of the National Heart, Lung, and Blood Institute, NIH.
We thank Cynthia Dunbar and her research group for support and for helpful discussions. We thank the NIH veterinary and animal support staff at 5 Research Court and the animal facility in Poolesville, MD, for maintaining animals over the years. We thank Rina Lee (University of California—Los Angeles) for editorial support.
Footnotes
Published ahead of print on 15 September 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Aiuti, A., F. Cattaneo, S. Galimberti, U. Benninghoff, B. Cassani, L. Callegaro, S. Scaramuzza, G. Andolfi, M. Mirolo, I. Brigida, A. Tabucchi, F. Carlucci, M. Eibl, M. Aker, S. Slavin, H. Al-Mousa, A. Al Ghonaium, A. Ferster, A. Duppenthaler, L. Notarangelo, U. Wintergerst, R. H. Buckley, M. Bregni, S. Marktel, M. G. Valsecchi, P. Rossi, F. Ciceri, R. Miniero, C. Bordignon, and M.-G. Roncarolo. 2009. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N. Engl. J. Med. 360:447-458. [DOI] [PubMed] [Google Scholar]
- 2.Aiuti, A., S. Slavin, M. Aker, F. Ficara, S. Deola, A. Mortellaro, S. Morecki, G. Andolfi, A. Tabucchi, F. Carlucci, E. Marinello, F. Cattaneo, S. Vai, P. Servida, R. Miniero, M. G. Roncarolo, and C. Bordignon. 2002. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science 296:2410-2413. [DOI] [PubMed] [Google Scholar]
- 3.An, D. S., S. K. P. Kung, A. Bonifacino, R. P. Wersto, M. E. Metzger, B. A. Agricola, S. H. Mao, I. S. Y. Chen, and R. E. Donahue. 2001. Lentivirus vector-mediated hematopoietic stem cell gene transfer of common gamma-chain cytokine receptor in rhesus macaques. J. Virol. 75:3547-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An, D. S., R. P. Wersto, B. A. Agricola, M. E. Metzger, S. Lu, R. G. Amado, I. S. Y. Chen, and R. E. Donahue. 2000. Marking and gene expression by a lentivirus vector in transplanted human and nonhuman primate CD34+ cells. J. Virol. 74:1286-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cartier, N., S. Hacein-Bey-Abina, C. C. Bartholomae, G. Veres, M. Schmidt, I. Kutschera, M. Vidaud, U. Abel, L. Dal-Cortivo, L. Caccavelli, N. Mahlaoui, V. Kiermer, D. Mittelstaedt, C. Bellesme, N. Lahlou, F. Lefrere, S. Blanche, M. Audit, E. Payen, P. Leboulch, B. l'Homme, P. Bougneres, C. Von Kalle, A. Fischer, M. Cavazzana-Calvo, and P. Aubourg. 2009. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science 326:818-823. [DOI] [PubMed] [Google Scholar]
- 6.Cavazzana-Calvo, M., S. Hacein-Bey, G. de Saint Basile, F. Gross, E. Yvon, P. Nusbaum, F. Selz, C. Hue, S. Certain, J.-L. Casanova, P. Bousso, F. L. Deist, and A. Fischer. 2000. Gene therapy of human severe combined immunodeficiency (SCID)-X1 dis. Science 288:669-672. [DOI] [PubMed] [Google Scholar]
- 7.Chun, T.-W., L. Carruth, D. Finzi, X. Shen, J. A. DiGiuseppe, H. Taylor, M. Hermankova, K. Chadwick, J. Margolick, T. C. Quinn, Y.-H. Kuo, R. Brookmeyer, M. A. Zeiger, P. Barditch-Crovo, and R. F. Siliciano. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183-188. [DOI] [PubMed] [Google Scholar]
- 8.Deichmann, A., S. Hacein-Bey-Abina, M. Schmidt, A. Garrigue, M. H. Brugman, J. Hu, H. Glimm, G. Gyapay, B. Prum, C. C. Fraser, N. Fischer, K. Schwarzwaelder, M.-L. Siegler, D. de Ridder, K. Pike-Overzet, S. J. Howe, A. J. Thrasher, G. Wagemaker, U. Abel, F. J. T. Staal, E. Delabesse, J.-L. Villeval, B. Aronow, C. Hue, C. Prinz, M. Wissler, C. Klanke, J. Weissenbach, I. Alexander, A. Fischer, C. von Kalle, and M. Cavazzana-Calvo. 2007. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J. Clin. Invest. 117:2225-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dick, J. E., M. C. Magli, D. Huszar, R. A. Phillips, and A. Bernstein. 1985. Introduction of a selectable gene into primitive stem cells capable of long-term reconstitution of the hemopoietic system of W/Wv mice. Cell 42:71-79. [DOI] [PubMed] [Google Scholar]
- 10.Gabriel, R., R. Eckenberg, A. Paruzynski, C. C. Bartholomae, A. Nowrouzi, A. Arens, S. J. Howe, A. Recchia, C. Cattoglio, W. Wang, K. Faber, K. Schwarzwaelder, R. Kirsten, A. Deichmann, C. R. Ball, K. S. Balaggan, R. J. Yanez-Munoz, R. R. Ali, H. B. Gaspar, L. Biasco, A. Aiuti, D. Cesana, E. Montini, L. Naldini, O. Cohen-Haguenauer, F. Mavilio, A. J. Thrasher, H. Glimm, C. von Kalle, W. Saurin, and M. Schmidt. 2009. Comprehensive genomic access to vector integration in clinical gene therapy. Nat. Med. 15:1431-1436. [DOI] [PubMed] [Google Scholar]
- 11.Gerrits, A., B. Dykstra, O. J. Kalmykowa, K. Klauke, E. Verovskaya, M. J. C. Broekhuis, G. de Haan, and L. V. Bystrykh. 2010. Cellular barcoding tool for clonal analysis in the hematopoietic system. Blood 115:2610-2618. [DOI] [PubMed] [Google Scholar]
- 12.Hacein-Bey-Abina, S., A. Garrigue, G. P. Wang, J. Soulier, A. Lim, E. Morillon, E. Clappier, L. Caccavelli, E. Delabesse, K. Beldjord, V. Asnafi, E. MacIntyre, L. Dal Cortivo, I. Radford, N. Brousse, F. Sigaux, D. Moshous, J. Hauer, A. Borkhardt, B. H. Belohradsky, U. Wintergerst, M. C. Velez, L. Leiva, R. Sorensen, N. Wulffraat, S. Blanche, F. D. Bushman, A. Fischer, and M. Cavazzana-Calvo. 2008. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 118:3132-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hacein-Bey-Abina, S., F. Le Deist, F. Carlier, C. Bouneaud, C. Hue, J.-P. De Villartay, A. J. Thrasher, N. Wulffraat, R. Sorensen, S. Dupuis-Girod, A. Fischer, E. G. Davies, W. Kuis, L. Leiva, and M. Cavazzana-Calvo. 2002. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N. Engl. J. Med. 346:1185-1193. [DOI] [PubMed] [Google Scholar]
- 14.Hacein-Bey-Abina, S., C. Von Kalle, M. Schmidt, M. P. McCormack, N. Wulffraat, P. Leboulch, A. Lim, C. S. Osborne, R. Pawliuk, E. Morillon, R. Sorensen, A. Forster, P. Fraser, J. I. Cohen, G. de Saint Basile, I. Alexander, U. Wintergerst, T. Frebourg, A. Aurias, D. Stoppa-Lyonnet, S. Romana, I. Radford-Weiss, F. Gross, F. Valensi, E. Delabesse, E. Macintyre, F. Sigaux, J. Soulier, L. E. Leiva, M. Wissler, C. Prinz, T. H. Rabbitts, F. Le Deist, A. Fischer, and M. Cavazzana-Calvo. 2003. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302:415-419. [DOI] [PubMed] [Google Scholar]
- 15.Harkey, M. A., R. Kaul, M. A. Jacobs, P. Kurre, D. Bovee, R. Levy, and C. A. Blau. 2007. Multiarm high-throughput integration site detection: limitations of LAM-PCR technology and optimization for clonal analysis. Stem Cells Dev. 16:381-392. [DOI] [PubMed] [Google Scholar]
- 16.Howe, S. J., M. R. Mansour, K. Schwarzwaelder, C. Bartholomae, M. Hubank, H. Kempski, M. H. Brugman, K. Pike-Overzet, S. J. Chatters, D. de Ridder, K. C. Gilmour, S. Adams, S. I. Thornhill, K. L. Parsley, F. J. T. Staal, R. E. Gale, D. C. Linch, J. Bayford, L. Brown, M. Quaye, C. Kinnon, P. Ancliff, D. K. Webb, M. Schmidt, C. von Kalle, H. B. Gaspar, and A. J. Thrasher. 2008. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 118:3143-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jordan, C. T., and I. R. Lemischka. 1990. Clonal and systemic analysis of long-term hematopoiesis in the mouse. Genes Dev. 4:220-232. [DOI] [PubMed] [Google Scholar]
- 18.Keller, G., C. Paige, E. Gilboa, and E. F. Wagner. 1985. Expression of a foreign gene in myeloid and lymphoid cells derived from multipotent haematopoietic precursors. Nature 318:149-154. [DOI] [PubMed] [Google Scholar]
- 19.Kim, S., Y. Kim, T. Liang, J. S. Sinsheimer, and S. A. Chow. 2006. A high-throughput method for cloning and sequencing human immunodeficiency virus type 1 integration sites. J. Virol. 80:11313-11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuramoto, K., D. A. Follmann, P. Hematti, S. Sellers, B. A. Agricola, M. E. Metzger, R. E. Donahue, C. von Kalle, and C. E. Dunbar. 2004. Effect of chronic cytokine therapy on clonal dynamics in nonhuman primates. Blood 103:4070-4077. [DOI] [PubMed] [Google Scholar]
- 21.Margulies, M., M. Egholm, W. E. Altman, S. Attiya, J. S. Bader, L. A. Bemben, J. Berka, M. S. Braverman, Y.-J. Chen, Z. Chen, S. B. Dewell, L. Du, J. M. Fierro, X. V. Gomes, B. C. Godwin, W. He, S. Helgesen, C. H. Ho, G. P. Irzyk, S. C. Jando, M. L. I. Alenquer, T. P. Jarvie, K. B. Jirage, J.-B. Kim, J. R. Knight, J. R. Lanza, J. H. Leamon, S. M. Lefkowitz, M. Lei, J. Li, K. L. Lohman, H. Lu, V. B. Makhijani, K. E. McDade, M. P. McKenna, E. W. Myers, E. Nickerson, J. R. Nobile, R. Plant, B. P. Puc, M. T. Ronan, G. T. Roth, G. J. Sarkis, J. F. Simons, J. W. Simpson, M. Srinivasan, K. R. Tartaro, A. Tomasz, K. A. Vogt, G. A. Volkmer, S. H. Wang, Y. Wang, M. P. Weiner, P. Yu, R. F. Begley, and J. M. Rothberg. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazurier, F., O. I. Gan, J. L. McKenzie, M. Doedens, and J. E. Dick. 2004. Lentivector-mediated clonal tracking reveals intrinsic heterogeneity in the human hematopoietic stem cell compartment and culture-induced stem cell impairment. Blood 103:545-552. [DOI] [PubMed] [Google Scholar]
- 23.McKenzie, J. L., O. I. Gan, M. Doedens, J. C. Y. Wang, and J. E. Dick. 2006. Individual stem cells with highly variable proliferation and self-renewal properties comprise the human hematopoietic stem cell compartment. Nat. Immunol. 7:1225-1233. [DOI] [PubMed] [Google Scholar]
- 24.Miller, A. D., and G. J. Rosman. 1989. Improved Retroviral vectors for gene transfer and expression. Biotechniques 7:980-990. [PMC free article] [PubMed] [Google Scholar]
- 25.Montini, E., D. Cesana, M. Schmidt, F. Sanvito, C. C. Bartholomae, M. Ranzani, F. Benedicenti, L. S. Sergi, A. Ambrosi, M. Ponzoni, C. Doglioni, C. Di Serio, C. von Kalle, and L. Naldini. 2009. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J. Clin. Invest. 119:964-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 27.Ott, M. G., M. Schmidt, K. Schwarzwaelder, S. Stein, U. Siler, U. Koehl, H. Glimm, K. Kuhlcke, A. Schilz, H. Kunkel, S. Naundorf, A. Brinkmann, A. Deichmann, M. Fischer, C. Ball, I. Pilz, C. Dunbar, Y. Du, N. A. Jenkins, N. G. Copeland, U. Luthi, M. Hassan, A. J. Thrasher, D. Hoelzer, C. von Kalle, R. Seger, and M. Grez. 2006. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat. Med. 12:401-409. [DOI] [PubMed] [Google Scholar]
- 28.Roche Diagnostics. 2007. Amplicon sequencing. Roche Diagnostics GmbH, Mannheim, Germany. http://www.454.com/downloads/protocols/5_AmpliconSequencing.pdf.
- 29.Schmidt, M., D. A. Carbonaro, C. Speckmann, M. Wissler, J. Bohnsack, M. Elder, B. J. Aronow, J. A. Nolta, D. B. Kohn, and C. von Kalle. 2003. Clonality analysis after retroviral-mediated gene transfer to CD34+ cells from the cord blood of ADA-deficient SCID neonates. Nat. Med. 9:463-468. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt, M., K. Schwarzwaelder, C. Bartholomae, K. Zaoui, C. Ball, I. Pilz, S. Braun, H. Glimm, and C. von Kalle. 2007. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR). Nat. Methods 4:1051-1057. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt, M., P. Zickler, G. Hoffmann, S. Haas, M. Wissler, A. Muessig, J. F. Tisdale, K. Kuramoto, R. G. Andrews, T. Wu, H.-P. Kiem, C. E. Dunbar, and C. von Kalle. 2002. Polyclonal long-term repopulating stem cell clones in a primate model. Blood 100:2737-2743. [DOI] [PubMed] [Google Scholar]
- 32.Schröder, A. R., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. 110:521-529. [DOI] [PubMed] [Google Scholar]
- 33.Silver, J., and V. Kerrikatte. 1989. Novel use of polymerase chain reaction to amplify cellular DNA adjacent to an integrated provirus. J. Virol. 63:1924-1928. (Erratum, 64:3150, 1990.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stein, S., M. G. Ott, S. Schultze-Strasser, A. Jauch, B. Burwinkel, A. Kinner, M. Schmidt, A. Kramer, J. Schwable, H. Glimm, U. Koehl, C. Preiss, C. Ball, H. Martin, G. Gohring, K. Schwarzwaelder, W.-K. Hofmann, K. Karakaya, S. Tchatchou, R. Yang, P. Reinecke, K. Kuhlcke, B. Schlegelberger, A. J. Thrasher, D. Hoelzer, R. Seger, C. von Kalle, and M. Grez. 2010. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat. Med. 16:198-204. [DOI] [PubMed] [Google Scholar]
- 35.Wang, G. P., C. C. Berry, N. Malani, P. Leboulch, A. Fischer, S. Hacein-Bey-Abina, M. Cavazzana-Calvo, and F. D. Bushman. Dynamics of gene-modified progenitor cells analyzed by tracking retroviral integration sites in a human SCID-X1 gene therapy trial. Blood 115:4356-4366. [DOI] [PMC free article] [PubMed]
- 36.Wang, G. P., A. Garrigue, A. Ciuffi, K. Ronen, J. Leipzig, C. Berry, C. Lagresle-Peyrou, F. Benjelloun, S. Hacein-Bey-Abina, A. Fischer, M. Cavazzana-Calvo, and F. D. Bushman. 2008. DNA bar coding and pyrosequencing to analyze adverse events in therapeutic gene transfer. Nucleic Acids Res. 36:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.