Abstract
Evasion of interferon (IFN)-mediated antiviral signaling is a common defense strategy for pathogenic RNA viruses. To date, research on IFN antagonism by hantaviruses is limited and has focused on only a subset of the numerous recognized hantavirus species. The host IFN response has two phases, an initiation phase, resulting in the induction of alpha/beta IFN (IFN-α/β), and an amplification phase, whereby IFN-α/β signals through the Jak/STAT pathway, resulting in the establishment of the cellular antiviral state. We examined interactions between these critical host responses and the New World hantaviruses. We observed delayed cellular responses in both Andes virus (ANDV)- and Sin Nombre virus (SNV)-infected A549 and Huh7-TLR3 cells. We found that IFN-β induction is inhibited by coexpression of ANDV nucleocapsid protein (NP) and glycoprotein precursor (GPC) and is robustly inhibited by SNV GPC alone. Downstream amplification by Jak/STAT signaling is also inhibited by SNV GPC and by either NP or GPC of ANDV. Therefore, ANDV- and SNV-encoded proteins have the potential for inhibiting both IFN-β induction and signaling, with SNV exhibiting the more potent antagonism ability. Herein we identify ANDV NP, a previously unrecognized inhibitor of Jak/STAT signaling, and show that IFN antagonism by ANDV relies on expression of both the glycoproteins and NP, whereas the glycoproteins appear to be sufficient for antagonism by SNV. These data suggest that IFN antagonism strategies by hantaviruses are quite variable, even between species with similar disease phenotypes, and may help to better elucidate species-specific pathogenesis.
Hantavirus is a genus of rodent-borne trisegmented negative-strand RNA viruses in the family Bunyaviridae. The three segments, L (large), M (medium), and S (small), encode four proteins: an RNA-dependent RNA polymerase, a glycoprotein precursor (GPC), which is cotranslationally cleaved into Gn and Gc (formerly G1 and G2) surface glycoproteins, and the nucleocapsid protein (NP), respectively (53). Hantaviruses are broadly classified into New World or Old World based on geographic location. Pathogenic New World hantaviruses cause a hantavirus cardiopulmonary syndrome (HCPS) in the Americas, whereas pathogenic Old World hantaviruses cause a hemorrhagic fever with renal syndrome (HFRS) in Europe and Asia (4, 17, 29, 41, 58). Andes virus (ANDV) and Sin Nombre virus (SNV) cause HCPS and are the most pathogenic hantavirus species found in South and North America, respectively, with a case fatality rate between 20 to 40% (32).
Following infection of a susceptible host, virus recognition is mediated by pattern recognition receptors (PRRs), which are predominantly comprised of Toll-like receptors (TLRs) and retinoic acid-inducible gene I-like RNA helicases (RLHs) (i.e., retinoic acid-inducible gene I helicase [RIG-I] and melanoma differentiation-associated gene 5 helicase [MDA-5]) (56). Virus recognition and signaling through these PRRs initiate induction of alpha interferon (IFN-α) and IFN-β and antiviral immune defenses (52). TLRs are type I integral membrane glycoproteins characterized by extracellular domains containing various numbers of leucine-rich repeat (LRR) motifs and a cytoplasmic signaling domain. TLRs can be divided into several subfamilies, each of which recognizes related pathogen-associated molecular patterns (PAMPs). TLR2, TLR3, TLR4, TLR7, TLR8, and TLR9 have various functions in virus recognition, with TLR3 specific for recognition of double-stranded RNA (dsRNA), a common intermediate of virus infection (1). RIG-I and MDA-5 are closely related members of the DExD/H box-containing helicase family, and while both activate cellular responses by signaling through IPS-1 (also called MAVS, VISA, and Cardif) and IRF-3, they are not functionally equivalent, recognizing 5′ triphosphate-RNA and dsRNA, respectively (26, 48, 57). RIG-I, in particular, has been implicated as the PRR for a number of negative-strand RNA viruses (36). However, the PRR responsible for recognition of hantavirus RNA and initiation of early IFN responses is not known.
Type I IFNs (IFN-α/β) are crucial regulators of immune cell activation, development of an antiviral state, cell growth, and apoptosis (21). Named after the component Janus kinases (Jaks) and signal transducers and activators of transcription (STATs), the Jak/STAT pathway transduces a signal initiated by IFNs that bind to the ubiquitously expressed IFN-α/β receptor (IFNAR). Almost immediately after formation of the receptor-ligand complex, the IFNAR-associated tyrosine kinases Jak1 and Tyk2 become auto- and transphosphorylated. STAT-1 and STAT-2 are subsequently recruited and activated via tyrosine phosphorylation. STAT-1 and STAT-2 form a heterotrimeric complex with IFN regulatory factor 9 (IRF9), known as the interferon-stimulated gene factor 3 (ISGF3) transcription factor. ISGF3 translocates to the nucleus, where it binds IFN-stimulated response element (ISRE) sequences in the promoters of IFN-α/β-regulated genes and initiates their transcription (15).
The hantavirus-encoded elements responsible for evasion of host immune responses remain largely uncharacterized. IFN antagonism has been recognized in several species of hantaviruses, both Old and New World. New World Sigmodontinae-associated hantaviruses, ANDV, and New York-1 virus (NY-1V) have been shown to inhibit induction of IFN-β (2, 54). In contrast, Prospect Hill virus (PHV), a nonpathogenic Arvicolinae-borne hantavirus, has been shown to induce IFN-β, indicating a potential link between different pathogenicities of hantaviruses in humans and the virus's ability to antagonize innate immune responses (54). However, when IFN-mediated signaling was investigated, the association between species pathogenicity and antagonism became less clear. One group reported lower Jak/STAT-dependent myxovirus resistance protein A (MxA) RNA levels in NY-1V-infected cells than in PHV-infected cells, suggesting that PHV was less efficient than NY-1V at antagonizing IFN-dependent responses (2). However, a second study suggested that ANDV and PHV were both able to inhibit Jak/STAT signaling (54). Thus, the role of IFN antagonism in virus pathogenicity is unclear, and further research is required to investigate interspecies variation in IFN antagonism and the associated mechanisms of suppression.
The hantavirus glycoproteins have been implicated as mediators of antagonism, responsible for suppression of both IFN-β induction and signaling. A glycoprotein of NY-1V, specifically the Gn cytoplasmic tail, was found to be responsible for inhibition of RIG-I- and TANK-binding kinase 1 (TBK-1)-dependent IFN responses (2, 3). The glycoproteins of both ANDV and PHV were shown to inhibit nuclear translocation of STAT-1 (54). However, it is unknown if the glycoproteins are the sole mediators of IFN antagonism and if they are the primary antagonists encoded by all hantaviruses. Furthermore, the IFN antagonism function of the authentically expressed and matured glycoproteins Gn and Gc, which are cotranslationally cleaved in infected cells, has not been fully explored.
To better understand the mechanism of IFN antagonism by New World hantaviruses, we have examined the modulation of IFN induction and signaling by ANDV and SNV, the most important HCPS-causing pathogens. Here, we report that SNV proteins antagonize virus recognition more efficiently than ANDV proteins; however, SNV and ANDV proteins suppress IFN-dependent Jak/STAT signaling to similar extents. Despite the ability of proteins from both viruses to inhibit amplification of IFN responses, interestingly, ANDV utilizes NP and GPC, whereas SNV uses GPC alone. These results provide evidence for a previously unrecognized hantavirus Jak/STAT antagonist in ANDV NP. Furthermore, our data suggest that New World hantavirus species differ in both the ability to mediate and mechanism of IFN antagonism and that these characteristics may be independent of virus pathogenicity in humans.
MATERIALS AND METHODS
Cells and viruses.
A549 cells (human lung carcinoma cells), human embryonic kidney (HEK) 293 cells, and Vero E6 cells (African green monkey kidney) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, 1% l-glutamine, and 1% penicillin-streptomycin. Huh7 cells stably transfected with TLR3 (Huh7-TLR3), a kind gift from Kui Li, University of Tennessee Health Science Center, were cultured in DMEM supplemented with 8% fetal calf serum, 1% l-glutamine, 1% penicillin-streptomycin, and blasticidin (2 μg/ml) (59).
The ANDV Chile-9717869 strain was kindly provided by Connie Schmaljohn, U.S. Army Medical Research Institute of Infectious Diseases, Ft. Detrick, MD (38). ANDV CHI-7913 and SNV-77734 were kindly provided by Brian Hjelle, University of New Mexico Health Sciences Center (HSC), Albuquerque, NM. Laguna Negra virus (LNV; 510B strain) and Maporal virus (MAPV; HV-97021050 strain) were kindly provided by Tom Ksiazek, Centers for Disease Control and Prevention, Atlanta, GA (20, 28). ANDV, SNV, LNV, and MAPV were propagated in a biosafety level 3 laboratory in Vero E6 cells maintained in DMEM supplemented with 2% fetal bovine serum (FBS). Sendai virus (SeV), strain Cantell, was obtained from Charles River Laboratories (Wilmington, MA).
Virus titration.
Virus infectivity was measured by titration and calculated as focus forming units (FFU) by use of an indirect immunofluorescent assay (IFA), as previously described (51). In brief, ANDV and SNV were adsorbed onto Vero E6 cells and overlaid with 1.2% carboxyl methylcellulose in Eagle's minimum essential medium (EMEM) supplemented with 2% FBS (Sigma-Aldrich) and antibiotics. Cells were fixed 7 to 10 days postinfection (dpi) with 100% ice-cold methanol and dried, and antigen-positive foci were detected with a rabbit anti-SNV N hyperimmune serum (diluted 1:1,000 in phosphate-buffered saline; a gift from Brian Hjelle) for 1 h. Cells were washed and incubated with peroxidase-conjugated goat anti-rabbit IgG antibodies (KPL) for 1 h and then washed, and foci were visualized using a metal-enhanced DAB substrate kit (Thermo Scientific).
Antibodies and cytokines.
The following antibodies were used in this study: anti-phospho-STAT-1 (Tyr701) (Cell Signaling Technology, Beverly, MA); antinucleoprotein of Andes virus IgG fraction (clone 8F3/F8), anti-glycoprotein 1 of Andes virus IgG fraction (clone 6B9/F5), anti-glycoprotein 2 of Andes virus IgG fraction (clone 2H4/F6), and antinucleoprotein of Sin Nombre (Four Corners) virus IgG fraction, epitope e (clone 5F1/F7) (Austral Biologicals, San Ramon, CA); monoclonal β-actin antibody (AC-15) (Santa Cruz Biotechnology, Santa Cruz, CA); Alexa Fluor 488-conjugated goat anti-rabbit IgG(H+L) and Alexa Fluor 594-conjugated goat anti-mouse IgG(H+L) (Life Technologies); and peroxidase-labeled affinity-purified antibody to mouse IgG(H+L) and peroxidase-labeled affinity-purified antibody to rabbit IgG(H+L) (KPL). SNV N rabbit polyclonal antibody was kindly provided by Brian Hjelle, University of New Mexico HSC, Albuquerque, NM. The monoclonal antibody to Zaire ebolavirus (ZEBOV) VP24 was kindly provided by Yoshihiro Kawaoka, University of Wisconsin—Madison, Madison, WI. Recombinant human IFN-β was purchased from PBL Interferon Source (Piscataway, NJ).
Hantavirus and ebolavirus expression plasmids.
To construct plasmids encoding recombinant hantavirus proteins, corresponding open reading frames (ORFs) were either subcloned from existing plasmids or inserted based on cDNA derived by Superscript III (Life Technologies)-mediated reverse transcription-PCRs (RT-PCRs) using 3 μl of purified RNA extracted from Vero E6 cells infected with the corresponding virus. All PCRs described below were performed with iProof high-fidelity DNA polymerase (Bio-Rad) according the manufacturer's recommendations.
The ANDV GPC expression plasmid was generated by PCR amplification of the ANDV M segment from cDNAs derived from an ANDV isolate (Chile-9717869; GenBank accession number AF291703). The entire GPC ORF was inserted into KpnI and NheI sites in pCAGGS/MCS, possessing the chicken beta-actin promoter. The ANDV Gn and Gc expression plasmids were constructed by PCR amplification of regions of the ANDV GPC expression plasmid ORF. Two stop codons were added to the downstream Gn primer, corresponding to position 1952 of the GPC ORF, to terminate expression immediately prior to the WASSA cleavage site. A start codon and Kozak sequence were added to the upstream Gc primer, corresponding to position 1902 of the GPC ORF, 50 nucleotides upstream of the cleavage site to allow correct processing of the N terminus of the Gc protein. Primers generated for creating the GPC ORF expression plasmid containing a Kozak sequence in the upstream primer and an additional stop codon in the downstream primer were used as the upstream Gn and downstream Gc primers, respectively. ANDV Gn and Gc ORFs were inserted into pCAGGS expression plasmids using KpnI and NheI restriction sites. The ANDV NP expression plasmid was generated by PCR amplification of the cDNA derived from ANDV (Chile-9717869; GenBank accession number AF291702). A Kozak sequence and an additional stop codon were added to the upstream and downstream primers, respectively. The NP ORF was subsequently inserted into pCAGGS/MCS using EcoRI and XhoI restriction sites. For construction of the V5-tagged ANDV NP expression plasmid, ANDV NP was PCR amplified and directionally cloned into a Gateway entry vector (Invitrogen, Carlsbad, CA), followed by subcloning into pcDNA3.1-nV5-DEST (Invitrogen) to generate an N-terminal V5 epitope-tagged ANDV NP.
SNV NP and GPC ORF were subcloned into pCAGGS/MCS from NP and GPC ORF-containing pET vectors, kind gifts from Brian Hjelle (62). The SNV NP ORF was inserted into pCAGGS/MCS by PCR amplification using forward and reverse primers to insert a leading KpnI restriction site and Kozak sequence and a trailing XhoI restriction site. LNV and MAPV expression clones were generated as follows. The LNV NP ORF (LNV 510B strain; GenBank accession number AF005727) was subcloned into pCAGGS/MCS from pATX-LNVNP by PCR amplification using forward and reverse primers to insert a leading KpnI restriction site and Kozak sequence and a trailing XhoI restriction site. To generate pCAGGS-MAPVNP, the MAPV NP ORF was initially cloned into a pATX plasmid by producing cDNA of the open reading frame from the MAPV S segment (MAPV HV-97021050; GenBank accession number AY267347) using the forward and reverse primers to insert novel BlnI and XhoI restriction sites. The open reading frame was then subcloned into pCAGGS/MCS by PCR amplification to insert a leading KpnI restriction site and Kozak sequence and a trailing XhoI restriction site. The primers used are detailed in Table 1. ZEBOV (Mayinga-76 strain; GenBank accession number AF086833)-derived pCAGGS-ZEBOV VP24 and pCAGGS-ZEBOV VP35 were kindly provided by Yoshihiro Kawaoka, University of Wisconsin—Madison, Madison, WI (60). V5 epitope-tagged LGTV was constructed as previously described (7). The authenticity of all plasmid constructs was confirmed by nucleotide sequence analysis.
TABLE 1.
Sequences used to generate hantavirus expression clones
| Purpose and protein | Primer sequence (5′-3′) |
|
|---|---|---|
| Forward | Reverse | |
| RT-PCR to generate cDNA and clone in pATX | ||
| ANDV NP | CGATGAATTCCACCATGAGCACCCTCCAAGAATTGCAGGAAAAC | CGATCTCGAGTTACTACAACTTAAGTGGCTCTTGGTTGGAGAT |
| LNV NP | CTCGAGATGAGCAACCTCCAAGAAGTACAG | ATCGATCGGAAAGCCCTCACATACCAC |
| MAPV NP | CATGGCTCGAGTGATGAGCAACCTCCAAG | CATGGGAGCTCACTACCAGTCTGACTCACA |
| PCR to amplify and subclone genes into pCAGGS/MCS | ||
| ANDV NP | CGATGAATTCCACCATGAGCACCCTCCAAGAATTGCAGGAAAAC | CGATCTCGAGTTACTACAACTTAAGTGGCTCTTGGTTGGAGAT |
| ANDV GPC | AGTCGGTACCCACCATGGAAGGGTGGTATCTGGTTGTTC | AGTCGCTAGCCTATTAGACAGTTTTCTTGTGCCCTCTC |
| ANDV Gn | AGTCGGTACCCACCATGGAAGGGTGGTATCTGGTTGTTC | AGTCGCTAGCCTACTATGCACTTGCGGCCCAAATAACA |
| ANDV Gc | AGTCGGTACCCACCATGGTATGGTGCCTATTGTTGACAT | AGTCGCTAGCCTATTAGACAGTTTTCTTGTGCCCTCTC |
| SNV NP | GATCGGTACCCACCATGAGCACCCTCAAAGAAGTG | CATGCTCGAGTTAAAGTTTAAGTGGTTCTTGGTT |
| LNV NP | GATCGGTACCCACCATGAGCAACCTCCAAGAAGT | CATGCTCGAGTTAGAGTTTTAGGGGTTCC |
| MAPV NP | GATCGGTACCCACCATGAGCAACCTCCAAGAAATA | CATGCTCGAGTCACAACTTCAAAGGCTCTTG |
| PCR to amplify and subclone genes into pENTR | ||
| ANDV NP | CACCATGAGCACCCTCCAAG | CTACAACTTAAGTGGCTCT |
Transfection.
For transfection of A549 and HEK 293 cells in 24-well plates, Lipofectamine LTX and Plus reagent (Life Technologies) were used according to the manufacturer's recommendations. For ISRE assays, our transfection mix was as follows: 0.5 ng of pCAGGS/MCS expression clone, 0.5 ng p-luc (either pISRE-luc or pIFN-β-luc), 0.1 ng pRL-TK, 1.1 μl Plus reagent, and 3 μl of Lipofectamine LTX. The transfection mix for wells transfected with two plasmids of interest was adjusted as follows: 0.5 ng of each pCAGGS/MCS expression clone, 0.5 ng of either pIFN-β-luc or pISRE-luc, 0.1 ng pRL-TK, 1.6 μl Plus reagent, 3.5 μl of Lipofectamine LTX, and 100 μl Opti-MEM 1 reduced serum liquid (Life Technologies).
For transfection of Vero E6 cells in 8-well Nunc Lab-Tek II chamber slide systems (Sigma), Lipofectamine LTX and Plus reagent (Life Technologies) were used according to the manufacturer's recommendations. Cells (∼50 to 60% confluent) were transfected with a mix of 200 ng DNA, 0.2 μl Plus reagent, 0.5 μl of Lipofectamine LTX, and 40 μl of Opti-MEM per well.
Immunofluorescence.
Virus protein expression and STAT-1 phosphorylation in cells were examined by methods previously described (7). In short, Vero E6 cells expressing each protein were treated with 2,000 U/ml of human IFN-β for 15 min at 37°C, fixed in ice-cold 100% methanol for 10 min, and stained overnight at 4°C using anti-phosphotyrosine 701-STAT-1 (Cell Signal Technology, Danvers, MA) (1:100 dilution) and the appropriate viral protein antibody, anti-nucleocapsid protein or antiglycoprotein antibodies (1:400 dilution) or anti-ZEBOV VP24 (1:10,000 dilution). Coverslips were mounted using ProLong Gold antifade reagent with DAPI (4′,6-diamidino-2-phenylindole) (Life Technologies). Images were captured using a Zeiss Axio Scope with Axiovision software.
Flow cytometry.
Virus protein expression and STAT-1 phosphorylation in cells were examined by methods previously described (7). In short, Vero E6 cells were transfected with V5-tagged viral protein constructs. Cells were treated for 15 min at 37°C with 1,000 U/ml of IFN-β 1 day posttransfection. Cells were labeled with anti-V5-fluorescein isothiocyanate (FITC) (Invitrogen) diluted 1:1,000 in anti-Stat1 (pY701)-Alexa 647 (BD Bioscience). Mouse IgG2a, κ-FITC (BD Bioscience) diluted 1:200 in mouse IgG2a, κ-Alexa 647 (BD Bioscience) was used for isotype control samples. Samples were analyzed by flow cytometry using the BD FACS LSR II cell sorting system. Two-dimensional (2D) scatter plots were used to differentiate populations of cells expressing the V5 antigen and/or pSTAT-1.
Western blot analysis.
Cells were lysed in sample buffer (Tris-HCl [pH 6.8], 4% SDS, 35% glycerol, 0.5% bromophenol blue, and 20% β-mercaptoethanol) and boiled. Equal volumes of cell lysate were separated by SDS-polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride (PVDF) membrane (GE Healthcare, Piscataway, NJ). Membranes were blocked overnight at 4°C in Tris-buffered saline (TBS) containing 5% skim milk and 0.1% Tween 20. The membrane was then incubated for 1 h in this buffer containing SNV N rabbit polyclonal antibody (diluted 1:10,000) or β-actin monoclonal antibody (diluted 1:2,000). Membranes were washed, incubated with horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature, washed, and finally developed with ECL (Amersham Biosciences).
Reporter gene assays.
The pISRE-Luc cis reporter plasmid (Stratagene) expresses the firefly luciferase gene under the control of five direct repeats of the interferon-stimulated response element (ISRE) found in the promoter of the 54-kDa interferon-stimulated gene (ISG54). The pIFN-β-luc reporter plasmid, kindly provided by Michael Gale (University of Washington, Seattle, WA), expresses firefly luciferase under the control of the entire human IFN-β promoter (19). The pRL-TK plasmid (Promega) contains a herpes simplex virus thymidine kinase (HSV TK) promoter-driven Rluc encoding Renilla luciferase, used to control for transfection efficiency.
For the ISRE activity assay, HEK 293 cells were cotransfected with pCAGGS plasmids encoding various viral proteins, the IFN-inducible firefly luciferase reporter (pISRE-luc) plasmid, and a plasmid constitutively expressing the Renilla luciferase protein (pRL-TK). At 24 h posttransfection, cells were treated with 1,000 U/ml of human IFN-β. At 6 to 8 h posttreatment, cells were lysed and measured for luciferase activity on a Turner Biosystems Modulus 96-well microplate reader using the Dual-Luciferase reporter assay system according to the manufacturer's instructions (Promega). The firefly luciferase activity of the IFN-treated sample was normalized to the Renilla luciferase value for that sample to control for transfection efficiency. Fold induction for each sample was then determined relative to the normalized luciferase activity value for untreated cells transfected with the same viral protein expression plasmid.
For the IFN-β promoter assay, A549 cells were cotransfected with pCAGGS plasmids encoding various viral proteins, the IRF-3-dependent firefly luciferase reporter (pIFN-β-luc) plasmid, and a plasmid constitutively expressing the Renilla luciferase protein (pRL-TK). At 24 h posttransfection, cells were induced with 150 hemagglutinin (HA) units/ml of SeV. At 8 to 10 h postinfection, cells were lysed and measured for luciferase activity as described above. The firefly luciferase activity of the SeV-induced sample was normalized to the Renilla luciferase value for that sample to control for transfection efficiency. Fold induction for each sample was then determined relative to the normalized luciferase activity value for uninduced cells transfected with the same viral protein expression plasmid.
ISG56, MxA, and viral qRT-PCR.
To investigate cellular innate immune responses to hantavirus infection, A549 or HuH7-TLR3 cells were infected with ANDV or SNV (multiplicity of infection [MOI] of 0.03). Cell lysates were collected 1, 2, and 3 dpi. RNA was isolated using the RNeasy minikit (Qiagen) and analyzed by a SYBR green-based two-step RT-PCR, using β-actin as a cell lysate control as previously reported (45).
ANDV and SNV S-segment copy numbers were determined as previously described (46, 51). In brief, RNA was isolated using the RNeasy minikit (Qiagen). One-step quantitative RT-PCR (qRT-PCR) was conducted on RNA extracts using a Corbett Rotor-gene 6000 system with either ANDV S-segment-specific primers or SNV S-segment-specific primers and a dually labeled fluorescent probe (all from TIB Molbiol, Adelphia, NJ). A 162-nucleotide ANDV fragment or 66-nucleotide SNV fragment was amplified in triplicate for each sample using TaqMan One-Step RT-PCR master mix (ABI) according to the manufacturer's instructions.
Statistical analysis.
Data from ISRE and pIFN-β luciferase assays were analyzed by one-way analysis of variance (ANOVA) with a Bonferroni multiple-comparison post hoc test to determine significant differences (P < 0.05) between samples.
RESULTS
ANDV and SNV infection elicits minimal or delayed expression of ISG56 and MxA in A549 and Huh7-TLR3 cells.
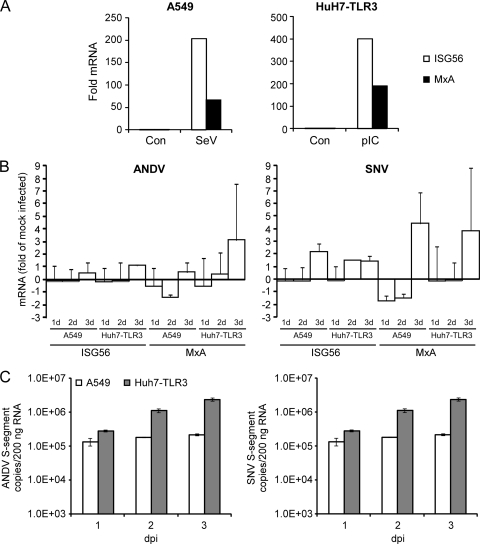
To date, the PRR used by cells to detect New World hantavirus infection remains elusive. A recent publication by Handke et al. suggests that Hantaan virus (HTNV), an Old World hantavirus, may be recognized via TLR3-dependent mechanisms (23). To this end, we sought to gain a better understanding of induction of innate immune responses by pathogenic New World hantaviruses using cell lines that are competent for the two most common RNA virus-sensing PRR pathways, RLH-mediated signaling (A549) and TLR3-mediated signaling (Huh7-TLR3) (59). Activation of RLH/IPS-1-dependent signaling in A549 cells and TLR3-dependent signaling in Huh7-TLR3 cells was confirmed by infection with SeV and treatment with poly(I:C), respectively (Fig. 1 A). A549 cells and Huh7-TLR3 cells were infected with ANDV or SNV (MOI of 0.03). Using qRT-PCR, we measured the transcription of genes encoding IRF-3-dependent ISG56 and MxA, a gene product specific to type I IFN Jak/STAT signaling, at 1, 2, and 3 dpi (Fig. 1B). In both A549 and Huh7-TLR3 cells there was no notable upregulation of ISG56 by ANDV compared to expression in mock-infected cells. Similarly, up to 2 dpi we did not observe any prominent induction of MxA. The only increase in transcription of the MxA gene was observed in Huh7-TLR3 cells at 3 dpi. Infection of A549 or Huh7-TLR3 cells by SNV elicited minimal and/or delayed induction of ISG56, with a modest induction of ISG56, maximum of 2-fold, observed in A549 and Huh7-TLR3 cells 2 to 3 dpi. Similarly, induction of MxA by SNV was delayed and was first detected in both A549 and Huh7-TLR3 cells at 3 dpi. These results are not attributed to low virus inoculum; we have used an MOI as high as 1.0 with comparable results (data not shown).
FIG. 1.
ANDV and SNV elicit minimal and/or delayed ISG56 and MxA gene transcription in infected A549 and Huh7-TLR3 cells. (A) Induction of ISGs in A549 cells infected with Sendai virus (SeV) 4 h postinfection and Huh7-TLR3 treated with poly(I:C) 4 h posttreatment. (B) A549 and Huh7-TLR3 cells were infected with ANDV or SNV (MOI = 0.03). Cellular RNA was collected at 1, 2, and 3 dpi and analyzed for ISG56 and MxA mRNA levels by qRT-PCR using a SYBR green-based 2-step RT-PCR on a Corbett Rotor-gene 6000 system (values are means ± standard errors of the means [SEM] for two experiments). (C) TaqMan detection of intracellular levels of ANDV or SNV S segment in A549 and Huh7-TLR3 cells using TaqMan probes on a Corbett Rotor-gene 6000 system. Total RNA was collected at 1, 2, and 3 dpi. Standards were based on serial dilutions of known ANDV or SNV NP plasmid copy numbers, and samples were standardized by normalizing total input RNA per sample to 200 ng.
To confirm viral replication in A549 and Huh7-TLR3 cells, ANDV and SNV S-segment copy numbers were determined using TaqMan qRT-PCR. An increase in S-segment genomic RNA over time was observed for ANDV- and SNV-infected A549 and Huh7-TLR3 cells; however, with both viruses, replication efficiency appeared to be much higher in Huh7-TLR3 (Fig. 1C). Viral replication kinetics in A549 cells, as well as Vero E6 cells that lack type I IFNs (data not shown), were consistent with previously published observations (31). Our data show that, in A549 and Huh7-TLR3 cells, ANDV and SNV infection induces negligible and/or delayed ISG56 and MxA cellular responses. This suggests that virus is simply not being recognized by cells during replication due to the lack of the appropriate PRR or the inaccessibility of PAMPs, or that IFN responses, either induction or amplification, are antagonized by ANDV and SNV.
ANDV and SNV differ in their mechanisms of antagonizing SeV-induced IFN-β promoter activity.
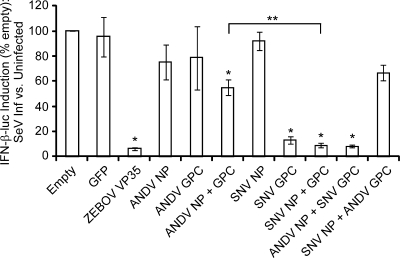
To investigate whether delayed cellular responses to pathogenic New World hantavirus infection are potentially due to virus-mediated IFN antagonism, we investigated the effect of viral protein expression on SeV-induced IFN-β promoter activity. Using a luciferase expression construct under the control of the IFN-β promoter, we compared the levels of luciferase activity in A549 cells expressing ANDV NP and/or GPC, SNV NP and/or GPC, or control proteins in response to infection with SeV. ZEBOV VP35, a well-characterized antagonist of type I IFN induction, was used as a positive control to validate the assay (5, 6, 47). Expression of constitutively expressed luciferase was not found to be selectively inhibited by any viral or control protein (data not shown). The expression of ANDV NP or GPC alone did not result in reduction of IFN-β promoter activity (Fig. 2). However, coexpression of ANDV NP and GPC had a statistically significant (P < 0.05) inhibitory effect on IFN-β promoter activity compared to results for the empty vector and green fluorescent protein (GFP) control plasmids. Similar to ANDV NP, SNV NP, expressed alone, did not inhibit IFN-β-luc activity. In contrast to results for ANDV, expression of SNV GPC or coexpression of NP and GPC resulted in potent inhibition of IFN-β-luc activity (P < 0.0001), comparable to that seen with ZEBOV VP35. Coexpression of heterologous NP and GPC confirmed the noted ability of SNV GPC to inhibit SeV-induced IFN-β-luc activity, as, even in the presence of ANDV NP, SNV GPC expression significantly reduced luciferase activity (P < 0.0001). Consistent with levels observed in the presence of ANDV GPC alone, ANDV GPC was able to reduce the activity of luciferase in the presence of SNV NP; however, the reduction was not significant compared to empty vector or GFP expression (Fig. 2). Thus, of all viral proteins investigated, SNV GPC was found to be a potent inhibitor of SeV-induced IFN-β promoter activity.
FIG. 2.
Inhibition of SeV-induced IFN-β-luc promoter activity by ANDV and SNV proteins. A549 cells were transfected with plasmids expressing ANDV NP and/or GPC, SNV NP and/or GPC, or ZEBOV VP35 (as a control). Cells were infected with 150 HA units/ml of Sendai virus 24 h posttransfection and harvested 8 to 10 h postinfection. The effect of protein expression on IFN-β promoter activity was calculated as fold induction of IFN-β-luciferase in SeV-induced cells versus that in uninduced cells (means ± SEM for a minimum of three biological replicates, comprising three experimental replicates for each construct and each experimental condition; *, P < 0.05). SNV NP and GPC expression resulted in significantly lower levels of IFN-β promoter activity than ANDV NP and GPC expression (**, P < 0.05).
ANDV NP and GPC partially inhibit STAT-1 activation and nuclear translocation in response to exogenous IFN-β.
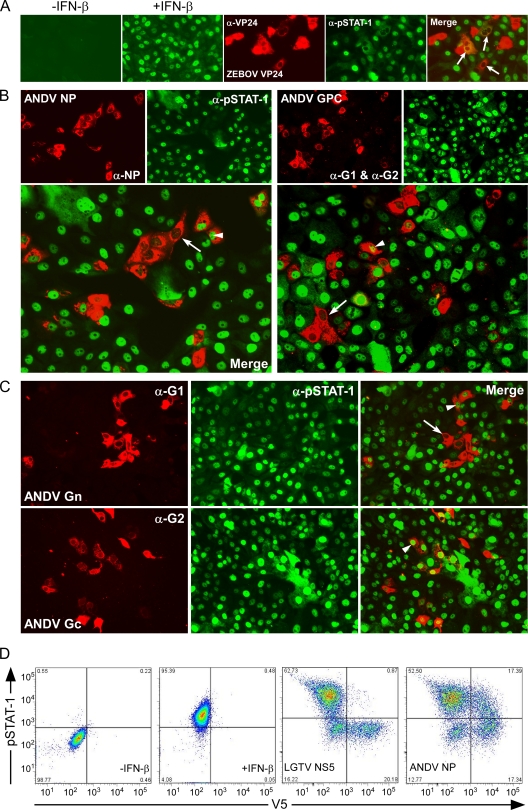
In contrast to SNV GPC, we did not find ANDV proteins to be highly potent antagonists of IFN-β expression, despite a lack of IFN responses in infected cells (Fig. 1). To investigate if antagonism by ANDV may target amplification of IFN responses rather than induction, the effect of ANDV NP, Gn, Gc, and GPC expression on tyrosine phosphorylation and hence activation of STAT-1 was tested in Vero E6 cells. Cells were treated at 24 h posttransfection with 2,000 U/ml of IFN-β, resulting in phosphorylation and nuclear translocation of STAT-1 (Fig. 3A, +IFN-β). As a positive control, we used ZEBOV VP24, which does not interfere with activation of STAT-1 but completely inhibits nuclear translocation of pSTAT-1 (Fig. 3A, merge). STAT-1 phosphorylation in response to IFN-β was inhibited in at least 50% of cells expressing either ANDV NP or GPC, suggesting that, in addition to previous reports of a role for GPC, ANDV NP may also play a role in inhibition of IFN-β-mediated Jak/STAT signaling. The inhibition of STAT-1 phosphorylation and subsequent nuclear translocation by NP or GPC was not complete; in a subset of cells expressing either protein, partial inhibition or an apparent lack of inhibition was observed (Fig. 3B). Furthermore, inhibition of STAT-1 phosphorylation in response to IFN-β by ANDV Gn also appeared to occur in at least 50% of the cells expressing viral protein. In contrast to the other ANDV proteins, Gc did not inhibit STAT-1 activation or nuclear translocation in response to IFN-β (Fig. 3C). To further support these findings, the effect of protein expression on STAT-1 phosphorylation was quantified using flow cytometry. pSTAT-1 was quantified in IFN-β-induced Vero E6 cells expressing V5-tagged ANDV NP or V5-tagged Langat virus (LGTV) NS5 as a positive control for inhibition of STAT-1 phosphorylation (7). ANDV NP expression resulted in inhibition of STAT-1 activation in 49.9% of cells, similar to results obtained by IFA.
FIG. 3.
Suppression of IFN-dependent activation and nuclear translocation of STAT-1. (A) Vero cells stained for pSTAT-1. Cells treated with 2,000 U/ml of IFN-β demonstrate the nuclear location of activated STAT-1, pSTAT-1 (+IFN-β). ZEBOV VP24, used as a positive control, inhibits nuclear translocation of pSTAT-1 in IFN-treated Vero cells at 1 day posttransfection but is unable to inhibit activation of STAT-1, as evidenced by cytoplasmic detection of STAT-1 in cells expressing viral protein (merge). α, anti. (B) Vero cells were transfected with plasmids expressing ANDV NP or GPC. At 1day posttransfection, cells were treated with 2,000 U/ml of IFN-β for 15 min, fixed, and stained using antibodies detecting ANDV nucleocapsid protein or Gn and Gc proteins (top left panels) and pSTAT-1 (top right panels). The corresponding panels below show the merged images. (C) Vero cells were transfected with plasmids expressing ANDV Gn or Gc. At 1 day posttransfection, cells were treated with 2,000 U/ml of IFN-β for 15 min, fixed, and stained using antibodies detecting ANDV Gn or Gc protein (left) and pSTAT-1 (middle). The merged images are shown at the right. Arrows indicate inhibition of STAT-1 activation and subsequent nuclear translocation; arrowheads indicate cells in which STAT-1 activation and nuclear translocation were not inhibited. (D) Flow cytometry investigating the effect of V5-tagged viral protein on STAT-1 activation using LGTV NS5 as a positive control confirms inhibition of STAT-1 activation by ANDV NP.
ANDV NP and GPC inhibit ISRE activity in response to exogenous IFN-β.
To quantify Jak/STAT antagonism by ANDV proteins and to investigate the effect of protein expression on Jak/STAT-dependent promoter activity, we monitored ISRE promoter activity using a luciferase expression construct under the control of a p54-ISRE promoter. In support of our IFA data, we found that ISRE activity was inhibited by expression of ANDV GPC or NP, compared to data for transfection of control constructs (empty vector, GFP) (Fig. 4). Expression of NP or GPC resulted in moderate levels of inhibition, similar to the inhibition observed in the IFA, and was not as potent as ZEBOV VP24 expression. ANDV NP was a stronger inhibitor of ISRE activity than GPC, although both were found to be significant (P < 0.05) compared to negative controls. Coexpression of ANDV NP and GPC inhibited ISRE expression more than any individual proteins and any other protein combinations investigated. Similar to our IFA results, individual expression of Gn had a statistically significant (P < 0.05) inhibitory effect on ISRE activity, whereas expression of Gc did not. To determine whether or not NP or GPC was primarily responsible for the inhibition seen with coexpression, we expressed NP or GPC with Gc. Both NP and GPC were able to reduce the induction levels seen with Gc alone (P < 0.05), suggesting that both NP and GPC play a role in antagonism of Jak/STAT signaling.
FIG. 4.
Inhibition of IFN-β-induced ISRE activity by ANDV proteins. (A) HEK 293 cells were transfected with plasmids expressing ANDV NP; ANDV GPC; ANDV Gn; ANDV Gc; a variety of combinations of ANDV NP, GPC, Gn, and Gc; or ZEBOV VP24 (as a control). Cells were treated with 1,000 U/ml of IFN-β 24 h posttransfection and harvested 6 to 8 h posttreatment. The effect of protein expression on ISRE activity was calculated as fold induction of ISRE-luciferase in treated cells versus untreated cells (means ± SEM from three to seven experiments; *, P < 0.05).
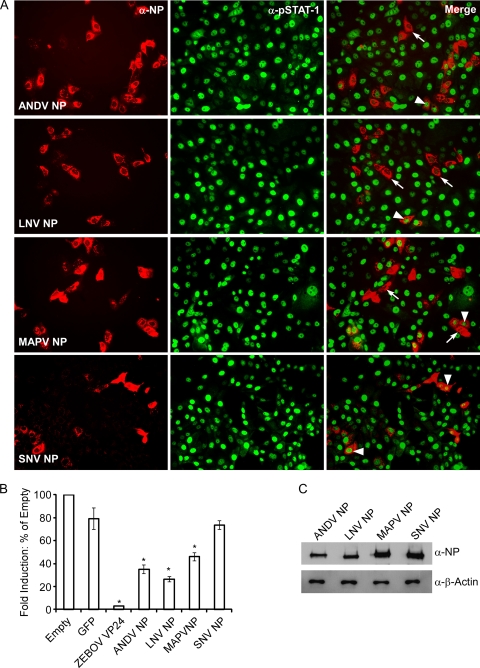
The nucleocapsid proteins of New World hantavirus species differ in their abilities to inhibit phosphorylation and nuclear translocation of STAT-1 and induction of ISRE in response to exogenous IFN-β.
Hantavirus is a diverse genus comprising over 20 recognized species and over 40 corresponding strains, with associated effects on humans ranging from the absence of detected disease to severe HCPS and HFRS (16, 18). We were interested in examining whether or not the inhibition of IFN-mediated Jak/STAT signaling observed in the presence of ANDV NP was specific to ANDV or a property of all New World hantaviruses. We tested inhibition of STAT-1 phosphorylation and nuclear translocation by IFA in cells transfected with the NP from pathogenic ANDV or SNV, less-pathogenic LNV, or apathogenic MAPV (10, 20, 28). Vero E6 cells were treated with IFN-β 24 h posttransfection, fixed, and double stained with anti-pSTAT-1 antibodies and either ANDV NP (used for ANDV, LNV, and MAPV NP)- or SNV NP-specific antibodies. The inhibition of STAT-1 phosphorylation and nuclear translocation by NP appeared to vary across species. Expression of NP from the South American species (ANDV, LNV, and MAPV) appeared to suppress STAT-1 phosphorylation and nuclear translocation in at least 50% of the cells. In contrast, the NP from SNV, a highly pathogenic HCPS-associated hantavirus, did not inhibit phosphorylation or nuclear translocation of pSTAT-1 (Fig. 5A, right panels). Of the South American species examined, LNV NP appeared to be the most potent antagonist, followed by ANDV NP and MAPV NP. Notably, similar to the effects seen with ANDV NP and GPC, inhibition of STAT-1 phosphorylation and nuclear translocation was not absolute, even in cells expressing LNV NP, the strongest inhibitor of the proteins tested.
FIG. 5.
The NPs from New World hantaviruses vary in their abilities to inhibit IFN-β-induced STAT-1 phosphorylation and nuclear translocation. (A) Vero cells were transfected with plasmids expressing ANDV NP, LNV NP, MAPV NP, or SNV NP. At 24 h posttransfection, cells were treated with 2,000 U/ml of IFN-β for 15 min, fixed, and stained using antibodies detecting ANDV NP, LNV NP, or MAPV NP (left panels) and pSTAT-1 (middle panels). The right panels show the merged images. Arrows indicate inhibition of phosphorylation and nuclear translocation; arrowheads indicate cells in which phosphorylation and nuclear translocation were not inhibited. α, anti. (B) New World hantavirus nucleocapsid proteins vary in their abilities to inhibit IFN-β-induced ISRE activity. HEK 293 cells were transfected with plasmids expressing ANDV NP, LNV NP, MAPV NP, or ZEBOV VP24 (as a control). Cells were treated with 1,000 U/ml of IFN-β 24 h posttransfection and harvested 6 to 8 h posttreatment. The effect of protein expression on ISRE activity was calculated as fold induction of ISRE-luciferase in treated cells versus that in untreated cells (means ± SEM for three experiments; *, P < 0.05 versus empty vector and GFP controls). (C) Expression of ANDV NP, LNV NP, MAPV NP, and SNV NP in HEK 293 cells from pCAGGS. Cell extracts were analyzed on a denaturing gel, followed by Western blotting with rabbit anti-SNV NP hyperimmune serum.
We then employed the ISRE-luciferase (ISRE-luc) assay and compared ISRE-promoter activities, as fold activities, in IFN-β-induced HEK 293 cells and uninduced cells. Activity in transfected cells expressing NP from ANDV, LNV, MAPV, or SNV was compared to that in cells transfected with either an empty vector or a vector expressing GFP. ZEBOV VP24 was used as a positive control. In accordance with the results of the STAT-1 phosphorylation and nuclear translocation assay, all New World hantavirus species' NPs tested significantly inhibited ISRE activity compared to empty vector and GFP control (P < 0.05), except SNV (Fig. 5B). Additionally, the interspecies variation noted in the IFA assay was also seen in ISRE activity; reduction in activity was strongest in the presence of LNV NP, followed by ANDV NP and then by MAPV NP. Once again, there was no evidence of Jak/STAT antagonism by SNV NP. To eliminate the possibility that the differences observed between species were due to differential protein expression, Western blotting was performed using SNV N polyclonal antibody, which confirmed comparable levels of expression of NP (Fig. 5C).
ANDV and SNV differ mechanistically in their antagonism of ISRE activity.
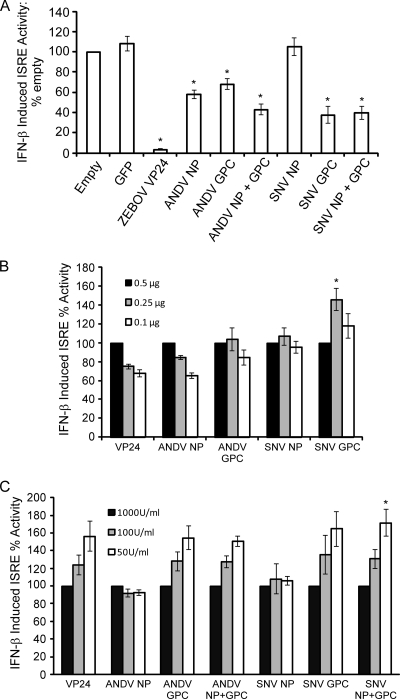
ANDV and SNV are considered the prototypic HCPS-associated hantaviruses. Of the species circulating in their respective geographical areas, both ANDV and SNV are associated with the highest number of human cases and the highest case fatality rates (10, 14). Our data suggest that ANDV NP functions as an antagonist of Jak/STAT signaling but that SNV NP does not. Reports have indicated that Gn (formerly G1) is the primary IFN antagonist of NY-1 virus, an SNV-like variant (2, 3). Given the evidence for antagonism by NY-1 G1 and our observations of potent inhibition of IFN induction by SNV GPC, we wanted to determine if the SNV GPC was able to antagonize Jak/STAT signaling similarly to ANDV GPC. To investigate the similarities and differences between antagonism by SNV and ANDV proteins, we used the ISRE-luc reporter assay in HEK 293 cells transfected with either ANDV NP and/or GPC or SNV NP and/or GPC. Surprisingly, in contrast to antagonism by ANDV, for which both NP and GPC appeared to have suppressive functions, antagonism by SNV appeared to be mediated solely by GPC (Fig. 6). Coexpression of SNV NP and GPC resulted in significantly reduced ISRE activity (P < 0.05), comparable to that seen with SNV GPC expression alone. Coexpression of SNV proteins, similar to coexpression of ANDV proteins, resulted in intermediate levels of ISRE response suppression. Taken together, results from this work demonstrate that the IFN antagonist function of NP is not shared between pathogenic hantaviruses, suggesting that New World hantaviruses may have evolved different mechanisms for IFN antagonism, independent of virulence in humans.
FIG. 6.
Inhibition of IFN-β-induced ISRE activity by ANDV and SNV proteins. HEK 293 cells were transfected with plasmids expressing ANDV NP and/or ANDV GPC, SNV NP and/or GPC, or ZEBOV VP24 (as a control). Cells were treated with 1,000 U/ml of IFN-β 24 h posttransfection and harvested 6 to 8 h posttreatment. The effect of protein expression on ISRE activity was calculated as fold induction of ISRE-luciferase in treated cells versus that in untreated cells (means ± SEM for four experiments, comprising two experimental replicates for each construct and each experimental condition; *, P < 0.05 versus empty vector and GFP controls). (B) Titration of plasmid concentration to 0.25 μg or 0.1 μg does not significantly affect the induction of ISRE activity compared to that at the original concentration used (set at 100% induction) of 0.5 μg. (C) Titration of IFN-β concentration down to 100 U/ml or 50 U/ml does not significantly affect the induction of ISRE activity compared to that at the original concentration used (set at 100% induction) of 1,000 U/ml.
To ensure inhibition was not a consequence of protein overexpression, we repeated the ISRE assay comparing plasmid levels 2- and 5-fold lower than the original concentration used in our assay. Percent induction of ISRE was compared to that at the original plasmid concentration (0.5 μg), set at 100%. Lower concentrations of plasmid did not generally result in significantly different levels of ISRE activity (Fig. 6B). IFN-β concentration was also investigated to ensure that inhibition was not affected by overwhelming levels of IFN-β stimulation. In almost every case, reduction of IFN-β by up to 20-fold did not significantly affect ISRE activity compared to that at the original concentration of 1,000 U/ml, set as 100% induction of ISRE (Fig. 6C). Thus, the inhibition mediated by hantavirus proteins was not due to artifacts of overexpression or overstimulation with IFN-β.
DISCUSSION
Suppression of host cellular IFN responses is a commonly employed survival strategy for viruses. In this report, we investigated antagonism of IFN responses by New World hantaviruses. We found that ANDV and SNV infection does not elicit robust cellular responses in A549 or Huh7-TLR3 cells, despite virus replication (Fig. 1). Our data suggest that the lack of cytokine induction in ANDV- and SNV-infected cells may not be explained by identical mechanisms, as these prototypic HCPS-associated hantaviruses differed in both ability and mechanism to antagonize IFN responses based on the effect of viral protein expression on both IFN-β induction and Jak/STAT signaling (Fig. 2 and 6). The GPC from SNV is a potent inhibitor of SeV-induced IFN-β reporter-dependent gene expression. In contrast, ANDV requires expression of both NP and GPC to antagonize IFN-β induction and does so at a level of around 50%, which is significantly less efficient than that by the SNV proteins (Fig. 2). In examining antagonism of IFN-dependent signaling by ANDV and SNV, we identify a novel role for ANDV NP as a functional IFN antagonist. Expression of ANDV NP alone resulted in a 50% inhibition of STAT-1 phosphorylation (Fig. 3) and of Jak/STAT-dependent promoter activity (Fig. 4), similar to that observed with GPC, which already has a recognized role in suppression of IFN responses (54). Inhibition seen with coexpression of NP and GPC suggests that the antagonism observed in the context of individual protein expression could work in the context of virus infection, in which both viral proteins would be present. Taken together, these data suggest that IFN antagonism by hantaviruses is species specific and may be independent of disease association in humans.
It has been suggested that pathogenic New World hantaviruses modulate the innate immune responses differently than nonpathogenic hantaviruses (54). The variation we observed between ANDV and SNV may be explained by species-specific cellular recognition, in that the viruses may process transcripts differently and thus may require different PRRs. Alternatively, these viruses may have simply evolved different mechanisms of antagonism. The PRR remains elusive for hantaviruses, and interspecies variation in hantavirus cellular detection has not been investigated. We were not able to detect any clear differences between cellular responses in A549 cells and Huh7-TRL3 cells that contain a member of one of the two major functional classes of PRRs, RNA helicases and TLRs, respectively (Fig. 1). Based on our findings we hypothesize that, in SNV-infected cells, the IFN gene is not transcribed due to the action of GPC, whereas in ANDV-infected cells IFN-β is made but amplification of IFN responses is dampened by inhibition of Jak/STAT signaling through the combined efforts of NP and GPC. The differential antagonism by these closely related viruses is clearly enticing and warrants further investigation to identify the PRR responsible for recognizing hantavirus infection. Future studies should focus on cell-type-dependent inhibition of host responses, in recognized primary target cells and in putative target cells, to investigate how these early host responses influence initial infection and subsequent amplification of virus in humans.
Gn was identified as an antagonist of IRF-mediated IFN-β induction in NY-1, a SNV-like variant (2, 3). We show that expression of the full SNV GPC suppresses IFN induction to levels as low as those seen with a well-characterized antagonist of RIG-I-mediated IFN induction, ZEBOV VP35 (Fig. 2). In addition, we provide new evidence that SNV GPC also functions as an antagonist of Jak/STAT signaling (Fig. 6). Thus, SNV appears to have evolved redundant mechanisms to evade host IFN responses. Encoding a protein able to target multiple aspects of the IFN response has been described for several RNA viruses, including influenza virus, rabies virus, and paramyxoviruses (9, 12, 22). However, while redundancy of IFN evasion by a single viral protein is not a novel strategy employed by viruses, many viruses evade IFN responses by encoding several viral protein antagonists with multiple corresponding cellular targets, which appears to be the strategy utilized by ANDV. Ebola virus encodes VP35 and VP24, paramyxoviruses encode V, C, and W proteins, and picornaviruses and coronaviruses encode a variety of IFN antagonists (6, 8, 9, 35, 42, 49, 50).
In contrast to that by SNV, antagonism of IFN-β induction by ANDV remains unclear. ANDV infection has been shown to inhibit IRF-3 dimerization, but expression of GPC alone was not sufficient to block nuclear translocation of IRF-3 (54). Our work suggests that perhaps more than one viral protein is necessary for antagonism by ANDV. Inhibition of IFN responses by ANDV also involves NP, a previously unrecognized IFN antagonist. Furthermore, we show that the role of NP is conserved in LNV and MAPV. The NPs of both LNV and MAPV were able to inhibit STAT-1 phosphorylation and nuclear translocation, and IFN-β-induced ISRE activity was reduced to 50% or less of levels seen in controls (Fig. 5). We found that antagonism by NP is not characteristic of all hantaviruses, as the NP of SNV had no effect on IFN-β-induced Jak/STAT signaling. ANDV, LNV, and MAPV are all South American hantaviruses, while SNV is endemic to North America. HCPS-associated and nonpathogenic New World hantaviruses may have evolved different strategies for IFN antagonism to optimize viral fitness based on species-specific rodent reservoirs and associated environmental pressures.
Interaction with the small ubiquitin-related modifier 1 (SUMO-1) and interference with importin-α proteins, such as karyopherin-α, have been identified as evasion strategies employed by well-recognized IFN antagonists ZEBOV VP35 and VP24, potent inhibitors of RIG-I-mediated IFN induction and Jak/STAT signaling, respectively (5, 6, 8, 11, 49, 50). The NPs of HTNV, Seoul virus, and Tula virus interact with proteins responsible for posttranslational modification and implicated in nuclear transport, regulation of transcription, and cell division, including SUMO-1 (24, 30, 34, 37). Additionally, HTNV interferes with the activation of NF-κB by binding to importin-α proteins, which are critical for nuclear transport (55). This suggests that the potential for functional interference with IFN signaling by hantavirus NP exists. However, the precise mechanism of inhibition by NP remains to be identified.
The ability of ANDV NP and GPC and SNV GPC to antagonize IFN-β responses is not as robust as that of other known inhibitors, such as ZEBOV VP35, ZEBOV VP24, and influenza virus NS1 (13, 33, 40, 49, 50). This difference may be a result of dramatic differences in virus ecology and evolution and may explain differences in disease progression. Hantavirus evolution is intimately associated with the rodent reservoirs of the virus; the incubation period for hantavirus disease is very long, ranging from 2 to over 6 weeks; and disease is considered to be predominantly immune mediated (25, 27, 63). The observed partial levels of inhibition may reflect the fact that hantaviruses are under selective pressure to evolve mechanisms of IFN antagonism that facilitate persistent infection, with negligible pathology, in host reservoirs. Only around one-half of the known Old and New World hantavirus species are known to cause human disease (39). Human infection is incidental to viral maintenance and is almost always a dead end in the infection chain, with the exception of isolated reports of human-to-human transmission of Andes virus. Thus, limited evolution of an efficient IFN antagonist in humans might have occurred (43, 44, 61). We speculate that partial inhibition by hantaviruses may be a result of coevolution between the virus and specific rodent hosts, which may lead to strategic inhibition that reduces virus replication to levels resulting in nonlethal pathogenesis in ideal host reservoirs.
While our data provide evidence for interspecies variation in antagonism of IFN responses by New World hantaviruses, a clear understanding of the contributory role and various mechanisms of action of all viral proteins, including the RNA-dependent RNA polymerase, remains to be determined. To date, these studies have not been completed because of limitations in developing expression constructs, including the absence of antibodies and methods to confirm the function of expressed L protein. Ongoing research into both recognition and signaling will help to identify the pathways relevant to hantavirus infection and will provide insight into the species-specific mechanisms of IFN antagonism. Continued investigation into antagonism by hantaviruses will help to elucidate hantavirus pathogenesis and may identify new effective targets for therapeutic intervention.
Acknowledgments
We thank Michael Gale for the pIFN-β-luc reporter plasmid, Brian Hjelle for virus and antibodies, Yoshihiro Kawaoka for antibodies and plasmids, Tom Ksiazek and Connie Schmaljohn for virus, and Kui Li for Huh7-TLR3 cells. J.L. thanks Tilahun Yilma for his continued support during the completion of this work and Travis Taylor, Barry Rockx, and Andrea Marzi for helpful discussions.
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Disease, National Institutes of Health, and the Public Health Agency of Canada. J.L. was supported by funding from the Veterinary Scientist Training Program and the Students Training in Advanced Research programs of the University of California, Davis, School of Veterinary Medicine.
Footnotes
Published ahead of print on 15 September 2010.
REFERENCES
- 1.Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell 124:783-801. [DOI] [PubMed] [Google Scholar]
- 2.Alff, P. J., I. N. Gavrilovskaya, E. Gorbunova, K. Endriss, Y. Chong, E. Geimonen, N. Sen, N. C. Reich, and E. R. Mackow. 2006. The pathogenic NY-1 hantavirus G1 cytoplasmic tail inhibits RIG-I- and TBK-1-directed interferon responses. J. Virol. 80:9676-9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alff, P. J., N. Sen, E. Gorbunova, I. N. Gavrilovskaya, and E. R. Mackow. 2008. The NY-1 hantavirus Gn cytoplasmic tail coprecipitates TRAF3 and inhibits cellular interferon responses by disrupting TBK1-TRAF3 complex formation. J. Virol. 82:9115-9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arikawa, J., K. Yoshimatsu, and H. Kariwa. 2001. Epidemiology and epizootiology of hantavirus infection in Japan. Jpn. J. Infect. Dis. 54:95-102. [PubMed] [Google Scholar]
- 5.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Muhlberger, M. Bray, H. D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basler, C. F., X. Wang, E. Muhlberger, V. Volchkov, J. Paragas, H. D. Klenk, A. Garcia-Sastre, and P. Palese. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. U. S. A. 97:12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Best, S. M., K. L. Morris, J. G. Shannon, S. J. Robertson, D. N. Mitzel, G. S. Park, E. Boer, J. B. Wolfinbarger, and M. E. Bloom. 2005. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J. Virol. 79:12828-12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardenas, W. B., Y. M. Loo, M. Gale, Jr., A. L. Hartman, C. R. Kimberlin, L. Martinez-Sobrido, E. O. Saphire, and C. F. Basler. 2006. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J. Virol. 80:5168-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casola, A., X. Bao, A. Brasier, and R. Garofalo. 2009. Inhibition of antiviral signaling pathways by paramyxoviruses, p. 247-265. In A. R. Brasier, A. Garcia-Sastre, and S. M. Lemon (ed.), Cellular signaling and innate immune responses to RNA virus infections. ASM Press, Washington, DC.
- 10.Chang, B., M. Crowley, M. Campen, and F. Koster. 2007. Hantavirus cardiopulmonary syndrome. Semin. Respir. Crit. Care Med. 28:193-200. [DOI] [PubMed] [Google Scholar]
- 11.Chang, T. H., T. Kubota, M. Matsuoka, S. Jones, S. B. Bradfute, M. Bray, and K. Ozato. 2009. Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog. 5:e1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chelbi-Alix, M. K., A. Vidy, J. El Bougrini, and D. Blondel. 2006. Rabies viral mechanisms to escape the IFN system: the viral protein P interferes with IRF-3, Stat1, and PML nuclear bodies. J. Interferon Cytokine Res. 26:271-280. [DOI] [PubMed] [Google Scholar]
- 13.Donelan, N. R., B. Dauber, X. Wang, C. F. Basler, T. Wolff, and A. Garcia-Sastre. 2004. The N- and C-terminal domains of the NS1 protein of influenza B virus can independently inhibit IRF-3 and beta interferon promoter activation. J. Virol. 78:11574-11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duchin, J. S., F. T. Koster, C. J. Peters, G. L. Simpson, B. Tempest, S. R. Zaki, T. G. Ksiazek, P. E. Rollin, S. Nichol, E. T. Umland, et al. 1994. Hantavirus pulmonary syndrome: a clinical description of 17 patients with a newly recognized disease. The Hantavirus Study Group. N. Engl. J. Med. 330:949-955. [DOI] [PubMed] [Google Scholar]
- 15.Durbin, J. E. 2009. Jak-Stat pathway in response to virus infection, p. 75-90. In A. R. Brasier, A. Garcia-Sastre, and S. M. Lemon (ed.), Cellular signaling and innate immune responses to RNA virus infections. ASM Press, Washington, DC.
- 16.Enria, D. A., A. M. Briggiler, N. Pini, and S. Levis. 2001. Clinical manifestations of New World hantaviruses. Curr. Top. Microbiol. Immunol. 256:117-134. [DOI] [PubMed] [Google Scholar]
- 17.Fabbri, M., and M. J. Maslow. 2001. Hantavirus pulmonary syndrome in the United States. Curr. Infect. Dis. Rep. 3:258-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fauquet, C., M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.). 2005. Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses, 8th ed. Elsevier Academic Press, London, United Kingdom.
- 19.Fredericksen, B., G. R. Akkaraju, E. Foy, C. Wang, J. Pflugheber, Z. J. Chen, and M. Gale, Jr. 2002. Activation of the interferon-beta promoter during hepatitis C virus RNA replication. Viral Immunol. 15:29-40. [DOI] [PubMed] [Google Scholar]
- 20.Fulhorst, C. F., M. N. Cajimat, A. Utrera, M. L. Milazzo, and G. M. Duno. 2004. Maporal virus, a hantavirus associated with the fulvous pygmy rice rat (Oligoryzomys fulvescens) in western Venezuela. Virus Res. 104:139-144. [DOI] [PubMed] [Google Scholar]
- 21.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 22.Hale, B. G., R. E. Randall, J. Ortin, and D. Jackson. 2008. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89:2359-2376. [DOI] [PubMed] [Google Scholar]
- 23.Handke, W., R. Oelschlegel, R. Franke, D. H. Kruger, and A. Rang. 2009. Hantaan virus triggers TLR3-dependent innate immune responses. J. Immunol. 182:2849-2858. [DOI] [PubMed] [Google Scholar]
- 24.Hay, R. T. 2001. Protein modification by SUMO. Trends Biochem. Sci. 26:332-333. [DOI] [PubMed] [Google Scholar]
- 25.Hjelle, B., S. A. Jenison, D. E. Goade, W. B. Green, R. M. Feddersen, and A. A. Scott. 1995. Hantaviruses: clinical, microbiologic, and epidemiologic aspects. Crit. Rev. Clin. Lab. Sci. 32:469-508. [DOI] [PubMed] [Google Scholar]
- 26.Hornung, V., J. Ellegast, S. Kim, K. Brzozka, A. Jung, H. Kato, H. Poeck, S. Akira, K. K. Conzelmann, M. Schlee, S. Endres, and G. Hartmann. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994-997. [DOI] [PubMed] [Google Scholar]
- 27.Jenison, S., B. Hjelle, S. Simpson, G. Hallin, R. Feddersen, and F. Koster. 1995. Hantavirus pulmonary syndrome: clinical, diagnostic, and virologic aspects. Semin. Respir. Infect. 10:259-269. [PubMed] [Google Scholar]
- 28.Johnson, A. M., M. D. Bowen, T. G. Ksiazek, R. J. Williams, R. T. Bryan, J. N. Mills, C. J. Peters, and S. T. Nichol. 1997. Laguna Negra virus associated with HPS in western Paraguay and Bolivia. Virology 238:115-127. [DOI] [PubMed] [Google Scholar]
- 29.Kariwa, H., K. Yoshimatsu, and J. Arikawa. 2007. Hantavirus infection in East Asia. Comp. Immunol. Microbiol. Infect. Dis. 30:341-356. [DOI] [PubMed] [Google Scholar]
- 30.Kaukinen, P., A. Vaheri, and A. Plyusnin. 2003. Non-covalent interaction between nucleocapsid protein of Tula hantavirus and small ubiquitin-related modifier-1, SUMO-1. Virus Res. 92:37-45. [DOI] [PubMed] [Google Scholar]
- 31.Khaiboullina, S. F., A. A. Rizvanov, V. M. Deyde, and S. C. St. Jeor. 2005. Andes virus stimulates interferon-inducible MxA protein expression in endothelial cells. J. Med. Virol. 75:267-275. [DOI] [PubMed] [Google Scholar]
- 32.Khan, A. S., and J. C. Young. 2001. Hantavirus pulmonary syndrome: at the crossroads. Curr. Opin. Infect. Dis. 14:205-209. [DOI] [PubMed] [Google Scholar]
- 33.Kochs, G., A. Garcia-Sastre, and L. Martinez-Sobrido. 2007. Multiple anti-interferon actions of the influenza A virus NS1 protein. J. Virol. 81:7011-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, B. H., K. Yoshimatsu, A. Maeda, K. Ochiai, M. Morimatsu, K. Araki, M. Ogino, S. Morikawa, and J. Arikawa. 2003. Association of the nucleocapsid protein of the Seoul and Hantaan hantaviruses with small ubiquitin-like modifier-1-related molecules. Virus Res. 98:83-91. [DOI] [PubMed] [Google Scholar]
- 35.Lemon, S. M. 2009. Evasion of host antiviral defences by picornaviruses, p. 335-351. In A. R. Brasier, A. Garcia-Sastre, and S. M. Lemon (ed.), Cellular signaling and innate responses to RNA virus infections. ASM Press, Washington, DC.
- 36.Loo, Y. M., J. Fornek, N. Crochet, G. Bajwa, O. Perwitasari, L. Martinez-Sobrido, S. Akira, M. A. Gill, A. Garcia-Sastre, M. G. Katze, and M. Gale, Jr. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82:335-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maeda, A., B. H. Lee, K. Yoshimatsu, M. Saijo, I. Kurane, J. Arikawa, and S. Morikawa. 2003. The intracellular association of the nucleocapsid protein (NP) of hantaan virus (HTNV) with small ubiquitin-like modifier-1 (SUMO-1) conjugating enzyme 9 (Ubc9). Virology 305:288-297. [DOI] [PubMed] [Google Scholar]
- 38.Meissner, J. D., J. E. Rowe, M. K. Borucki, and S. C. St. Jeor. 2002. Complete nucleotide sequence of a Chilean hantavirus. Virus Res. 89:131-143. [DOI] [PubMed] [Google Scholar]
- 39.Mertz, G. J., B. Hjelle, M. Crowley, G. Iwamoto, V. Tomicic, and P. A. Vial. 2006. Diagnosis and treatment of new world hantavirus infections. Curr. Opin. Infect. Dis. 19:437-442. [DOI] [PubMed] [Google Scholar]
- 40.Mibayashi, M., L. Martinez-Sobrido, Y. M. Loo, W. B. Cardenas, M. Gale, Jr., and A. Garcia-Sastre. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 81:514-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monroe, M. C., S. P. Morzunov, A. M. Johnson, M. D. Bowen, H. Artsob, T. Yates, C. J. Peters, P. E. Rollin, T. G. Ksiazek, and S. T. Nichol. 1999. Genetic diversity and distribution of Peromyscus-borne hantaviruses in North America. Emerg. Infect. Dis. 5:75-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narayanan, K., and S. Makino. 2009. Coronaviruses and arteriviruses, p. 373-387. In A. R. Brasier, A. Garcia-Sastre, and S. M. Lemon (ed.), Cellular signaling and innate immune responses to RNA virus infections. ASM Press, Washington, DC.
- 43.Padula, P. J., A. Edelstein, S. D. Miguel, N. M. Lopez, C. M. Rossi, and R. D. Rabinovich. 1998. Hantavirus pulmonary syndrome outbreak in Argentina: molecular evidence for person-to-person transmission of Andes virus. Virology 241:323-330. [DOI] [PubMed] [Google Scholar]
- 44.Plyusnin, A., and S. P. Morzunov. 2001. Virus evolution and genetic diversity of hantaviruses and their rodent hosts. Curr. Top. Microbiol. Immunol. 256:47-75. [DOI] [PubMed] [Google Scholar]
- 45.Prescott, J., C. Ye, G. Sen, and B. Hjelle. 2005. Induction of innate immune response genes by Sin Nombre hantavirus does not require viral replication. J. Virol. 79:15007-15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prescott, J. B., P. R. Hall, V. S. Bondu-Hawkins, C. Ye, and B. Hjelle. 2007. Early innate immune responses to Sin Nombre hantavirus occur independently of IFN regulatory factor 3, characterized pattern recognition receptors, and viral entry. J. Immunol. 179:1796-1802. [DOI] [PubMed] [Google Scholar]
- 47.Prins, K. C., W. B. Cardenas, and C. F. Basler. 2009. Ebola virus protein VP35 impairs the function of interferon regulatory factor-activating kinases IKKɛ and TBK-1. J. Virol. 83:3069-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rehwinkel, J., C. P. Tan, D. Goubau, O. Schulz, A. Pichlmair, K. Bier, N. Robb, F. Vreede, W. Barclay, E. Fodor, and E. S. C. Reis. 2010. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 140:397-408. [DOI] [PubMed] [Google Scholar]
- 49.Reid, S. P., L. W. Leung, A. L. Hartman, O. Martinez, M. L. Shaw, C. Carbonnelle, V. E. Volchkov, S. T. Nichol, and C. F. Basler. 2006. Ebola virus VP24 binds karyopherin α1 and blocks STAT1 nuclear accumulation. J. Virol. 80:5156-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reid, S. P., C. Valmas, O. Martinez, F. M. Sanchez, and C. F. Basler. 2007. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin alpha proteins with activated STAT1. J. Virol. 81:13469-13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Safronetz, D., N. R. Hegde, H. Ebihara, M. Denton, G. P. Kobinger, S. St. Jeor, H. Feldmann, and D. C. Johnson. 2009. Adenovirus vectors expressing hantavirus proteins protect hamsters against lethal challenge with Andes virus. J. Virol. 83:7285-7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saito, T., and M. Gale, Jr. 2007. Principles of intracellular viral recognition. Curr. Opin. Immunol. 19:17-23. [DOI] [PubMed] [Google Scholar]
- 53.Schmaljohn, C. S., and S. T. Nichol. 2007. Bunyaviridae, p. 1742-1789. In D. M. Knipe and P. A. Howley (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 54.Spiropoulou, C. F., C. G. Albarino, T. G. Ksiazek, and P. E. Rollin. 2007. Andes and Prospect Hill hantaviruses differ in early induction of interferon although both can downregulate interferon signaling. J. Virol. 81:2769-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor, S. L., N. Frias-Staheli, A. Garcia-Sastre, and C. S. Schmaljohn. 2009. Hantaan virus nucleocapsid protein binds to importin alpha proteins and inhibits tumor necrosis factor alpha-induced activation of nuclear factor κB. J. Virol. 83:1271-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson, A. J., and S. A. Locarnini. 2007. Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol. Cell Biol. 85:435-445. [DOI] [PubMed] [Google Scholar]
- 57.Unterholzner, L., and A. G. Bowie. 2008. The interplay between viruses and innate immune signaling: recent insights and therapeutic opportunities. Biochem. Pharmacol. 75:589-602. [DOI] [PubMed] [Google Scholar]
- 58.Vapalahti, O., J. Mustonen, A. Lundkvist, H. Henttonen, A. Plyusnin, and A. Vaheri. 2003. Hantavirus infections in Europe. Lancet Infect. Dis. 3:653-661. [DOI] [PubMed] [Google Scholar]
- 59.Wang, N., Y. Liang, S. Devaraj, J. Wang, S. M. Lemon, and K. Li. 2009. Toll-like receptor 3 mediates establishment of an antiviral state against hepatitis C virus in hepatoma cells. J. Virol. 83:9824-9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Watanabe, S., T. Watanabe, T. Noda, A. Takada, H. Feldmann, L. D. Jasenosky, and Y. Kawaoka. 2004. Production of novel Ebola virus-like particles from cDNAs: an alternative to Ebola virus generation by reverse genetics. J. Virol. 78:999-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wells, R. M., S. Sosa Estani, Z. E. Yadon, D. Enria, P. Padula, N. Pini, J. N. Mills, C. J. Peters, and E. L. Segura. 1997. An unusual hantavirus outbreak in southern Argentina: person-to-person transmission? Hantavirus Pulmonary Syndrome Study Group for Patagonia. Emerg. Infect. Dis. 3:171-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yee, J., I. A. Wortman, R. A. Nofchissey, D. Goade, S. G. Bennett, J. P. Webb, W. Irwin, and B. Hjelle. 2003. Rapid and simple method for screening wild rodents for antibodies to Sin Nombre hantavirus. J. Wildl. Dis. 39:271-277. [DOI] [PubMed] [Google Scholar]
- 63.Young, J. C., G. R. Hansen, T. K. Graves, M. P. Deasy, J. G. Humphreys, C. L. Fritz, K. L. Gorham, A. S. Khan, T. G. Ksiazek, K. B. Metzger, and C. J. Peters. 2000. The incubation period of hantavirus pulmonary syndrome. Am. J. Trop. Med. Hyg. 62:714-717. [DOI] [PubMed] [Google Scholar]