Abstract
The phenotypic and strain-related properties of human prion diseases are, according to the prion hypothesis, proposed to reside in the physicochemical properties of the conformationally altered, disease-associated isoform of the prion protein (PrPSc), which accumulates in the brains of patients suffering from Creutzfeldt-Jakob disease and related conditions, such as Gerstmann-Straussler-Scheinker disease. Molecular strain typing of human prion diseases has focused extensively on differences in the fragment size and glycosylation site occupancy of the protease-resistant prion protein (PrPres) in conjunction with the presence of mutations and polymorphisms in the prion protein gene (PRNP). Here we report the results of employing an alternative strategy that specifically addresses the conformational stability of PrPSc and that has been used previously to characterize animal prion strains transmitted to rodents. The results show that there are at least two distinct conformation stability states in human prion diseases, neither of which appears to correlate fully with the PrPres type, as judged by fragment size or glycosylation, the PRNP codon 129 status, or the presence or absence of mutations in PRNP. These results suggest that conformational stability represents a further dimension to a complete description of potentially phenotype-related properties of PrPSc in human prion diseases.
Transmissible spongiform encephalopathies (TSE) are fatal neurological diseases that occur in farmed and free-ranging animals, including scrapie in sheep and goats, transmissible mink encephalopathy, bovine spongiform encephalopathy in cattle, and chronic wasting disease in deer and elk (4, 52). Sheep scrapie is the prototypic animal TSE and has been investigated for many years in the form of rodent-adapted isolates that have given rise to distinct stable laboratory strains (6). Despite intensive investigation, there is little or no direct evidence for a unique foreign nucleic acid genome associated with scrapie infectivity, and the paradigm most frequently invoked to explain TSE is that of transmissible amyloidosis (evidence reviewed in references 8 and 46). This protein-only or prion hypothesis proposes that the infectious agent or prion is an altered form (termed PrPSc [disease-associated isoform of the prion protein]) of the host-encoded prion protein (PrPC), which is produced by a posttranslational conversion involving rearrangements of secondary (increased β-sheet content) and quaternary (self-aggregation) structure (41). Polymorphisms in the host prion protein gene in part determine susceptibility to infection and modify the resultant disease phenotype, but in order to explain the existence of distinct strains, PrPSc has been proposed to exist in different, stable, and replicative biophysical forms or “conformers” that “encipher” strain-like properties (reviewed in references 1 and 11). Recent reports of the successful production of prion infectivity with distinct structural and biological properties, from recombinant PrP, strongly support the prion hypothesis in general and the structural variant hypothesis in particular (9, 23-28, 51).
The concept of molecular strain typing has acquired considerable currency in the identification and diagnosis of human prion diseases. Three major clinicopathological phenotypes of human prion disease are recognized (reviewed in reference 17): Creutzfeldt-Jakob disease (CJD), Gerstmann-Straussler-Scheinker disease (GSS), and fatal insomnia (FI). CJD may be acquired, in the case of variant CJD (vCJD) obtained from bovine spongiform encephalopathy (BSE)-infected cattle, may occur in association with mutations in the human prion protein gene (PRNP) as familial CJD, or may occur in a sporadic fashion (sporadic CJD [sCJD]), perhaps due to rare stochastic events leading to PrPSc formation and accumulation. FI occurs sporadically (sporadic FI [sFI]) or in association with the D178N PRNP mutation (fatal familial insomnia [FFI]), and GSS is associated with a series of pathogenic mutations in PRNP, the most common of which is P102L (reviewed in reference 17).
PrPSc analysis in this context has relied heavily on the biophysical differences in PrPSc, as determined by limited proteolysis of postmortem human prion disease brain samples and analysis of the protease-resistant core of PrPSc (termed PrPres or PrP27-30) size by Western blotting (10, 32). Three major conformational states have been inferred from the observation of 21-kDa (type 1), 19-kDa (type 2), or 7- to 8-kDa PrPres. Variable glycosylation site occupancy adds to this diversity (10, 14, 33), as follows: PrPres types are given the suffix “A” if mono- or nonglycosylated PrPres predominates or “B” if diglycosylated PrPres predominates (33). These parameters have diagnostic value in that sCJD cases are marked by type 1 and/or type 2A PrPres, vCJD has type 2B PrPres, and two different pathological phenotypes in GSS are associated with either type 1 or 7- to 8-kDa PrPres (13, 34, 35, 39). Moreover, the clinicopathological variation between different cases of sCJD appears to correlate to a large extent with the relative mixture of type 1 and type 2A PrPres in combination with the patient's PRNP codon 129 polymorphic status (MM, MV, or VV) (7, 36).
These observations provide a correlation between PrPSc types and human disease phenotype, consistent with different prions underlying different prion diseases, but the Western blotting approach is indirect, of relatively low sensitivity, and likely to be an incomplete description of PrPSc. An alternative approach to defining PrPSc, termed the conformation-dependent immunoassay (CDI), depends upon the effect of increasing concentrations of the chaotropic salt guanidine hydrochloride (GdnHCl) to unmask epitopes in PrP that become hidden during the structural rearrangements involved in PrPSc formation (43). CDI has shown that up to 90% of the PrPSc present in a sCJD brain is susceptible to proteolysis and would therefore not figure in conventional Western blot typing assays (44). Moreover, the stability profiles of PrPSc, as defined by unmasking of the monoclonal antibody 3F4 epitope, have been reported to be unique for a series of well-characterized scrapie strains in hamsters (43). A related assay, termed the conformation stability assay (CSA), has also been developed (37), in which conformational stability is measured as a function of the resistance to proteolysis following exposure to increasing concentrations of GdnHCl. Rodent-adapted prion strains that were indistinguishable by electrophoretical mobility of PrPres can be distinguished by CSA when the stability of PrPres is expressed as the concentration of GdnHCl required to make half of the protein susceptible to proteolytic degradation (GdnHCl1/2) (37, 38). Given the growing realization that PrPSc stability may be a strain-related property, we have asked whether there are consistent differences in human PrPSc stability between different human prion diseases, and whether any such differences correlate with currently used parameters, such as PrPres fragment size, PrPres glycosylation site occupancy, and codon 129 status.
MATERIALS AND METHODS
Human brain materials.
Thirteen variant CJD (vCJD) cases, fourteen sporadic CJD (sCJD) cases, two GSS cases with P102L mutations, and five control (non-CJD) cases with other neurological diseases were analyzed in this study. Cases were selected based on the availability of autopsy-collected frozen brain specimens with consent for research use. Appropriate ethical approval was obtained (LREC 2000/4/157). All cases used were of United Kingdom origin. Brain tissue from each case had been examined both histopathologically and biochemically, and a definite diagnosis had been reached by established criteria (5, 16). The codon 129 polymorphism of the PRNP of each case was determined by restriction fragment length polymorphism (30). The protease-resistant core fragments of PrPSc (PrPres) found in brain was classified as type 1, 2A, or 2B as previously described according to the nomenclature of others (13, 35). Among the 14 sporadic CJD cases, 13 cases were of the MM1 subtype, and 1 case had a VV2 subtype. One GSS case showed the typical three PrPres bands (type 1), whereas the other GSS case had only proteinase K (PK)-resistant fragments of ∼8 kDa. Among the five neurological control cases, three cases were of the MM genotype at codon 129 of PRNP (with diagnoses of vascular dementia, motor neurone disease, and B-cell lymphoma), one case was of the MV genotype (with a diagnosis of vascular dementia), and one case was of the VV genotype (with a diagnosis of dementia with Lewy bodies). In all instances, the tissue analyzed was gray matter-enriched frontal cortex (FC), and additionally, cerebellar cortex (Cb) and thalamus (Th) in vCJD cases, taken from frozen half-brain specimens.
Preparation of brain tissue.
Brain tissue homogenates were prepared in nine volumes (wt/vol) of phosphate-buffered saline (PBS), pH 7.4, containing 2% Sarkosyl by two cycles of homogenization in the FastPrep instrument (Qbiogene). The homogenized brain samples were stored at −80°C until use.
Enrichment of disease-associated PrP.
The 10% homogenates were diluted to 5% (wt/vol) using PBS containing 2% (wt/vol) Sarkosyl and were incubated for 10 min at room temperature on a shaking platform. The samples were then centrifuged at 500 × g for 5 min at 20°C, and the supernatants were collected. For the enrichment of the disease-associated isoform of PrP, 100-μl aliquots of cleared brain homogenates in PBS containing 2% Sarkosyl were centrifuged at 20,800 × g for 1 h at 4°C as described previously (48, 50). After careful aspiration of the supernatants, pellets containing a detergent-insoluble fraction of PrP were used for GdnHCl-induced denaturation. In some cases, pellets were resuspended at the starting volume using homogenization buffer in order to investigate the effect of this method of enriching PrPSc.
CDI.
The CDI method used in this study was based on an enzyme-linked immunosorbent assay (ELISA)-formatted, time-resolved fluorometry CDI as described previously (2, 43). Samples prepared in 2% Sarkosyl in PBS were divided into the following two parts: one part was mixed with the same volume of PBS containing Complete EDTA-free protease inhibitors (native sample [N]), and the other part was mixed with the same volume of 8 M GdnHCl and incubated for 6 min at 81°C (denatured sample [D]). Both N and D were adjusted using distilled water containing EDTA-free protease inhibitors to a final concentration of GdnHCl of 0.35 M. A 96-well black polystyrene plate (Fisher) was coated with 2 μg/well anti-PrP antibody MAR-1 (CSL Behring, Marburg, Germany) overnight at room temperature. MAR-1 specifically recognizes human PrP with a correctly formed disulfide bridge. After removal of excess antibody, the plate was washed four times with wash buffer (PerkinElmer) and then saturated with 0.5% (wt/vol) bovine serum albumin (BSA) and 6% (wt/vol) sorbitol in wash buffer for 1 h at room temperature on a plate shaker. After the plate was washed four times with wash buffer, prepared samples were loaded in duplicate or triplicate onto the plate (200 μl/well). After incubation of the plate for 2 h at room temperature with shaking, the plate was washed as done previously. The detection of bound PrP was achieved by the Europium (Eu)-labeled MAb 3F4, recognizing amino acids 109 to 112 of human PrP. The detector antibody diluted in assay buffer (PerkinElmer) at 50 ng/ml was added to each well, followed by incubation for 2 h at room temperature on a plate shaker. The plate was developed after six washes by 5 min of incubation in enhancement solution (PerkinElmer) at room temperature. The time-resolved fluorescence (TRF) signal, measured by cps (counts per seconds), was made using a Victor 2 fluorometer (PerkinElmer).
Western blot analysis.
In this study, polyacrylamide gel electrophoresis was performed using NuPAGE Novex gel system (Invitrogen) as described previously (54). Samples were digested with PK at 37°C for 1 h, and the PK activity was terminated by adding Pefabloc to a final concentration of 1 mM. Undigested or PK-digested samples were mixed with NuPAGE LDS sample buffer to a final concentration of 1×. In some cases, pellets were resuspended with 2× sample buffer. The samples were incubated for 10 min at 100°C and then separated on 10% Bis-Tris NuPAGE gels. The separated proteins were transferred to polyvinylidene difluoride (PVDF) membranes and then were blocked for 1 h with 5% (wt/vol) nonfat dry milk in TBS-T (20 mM Tris-HCl, pH 7.4; 150 mM NaCl; 0.1% Tween 20). The immunodetection of PrP was achieved using the MAb 3F4 (Dako) at a final concentration of 75 ng/ml IgG for 1 h. The membrane was washed three times with TBS-T and incubated with horseradish peroxidase-conjugated anti-mouse IgG F(ab′)2 fragments (Amersham) at a dilution of 1:40,000 for 1 h. Following five washes in TBS-T, blots were developed with ECL Plus reagent (Amersham) and then exposed to Hyperfilm ECL (Amersham) for periods of 30 s to 30 min. In some cases, developed membranes were scanned on a Storm 860 scanning fluorometer. The molecular weight of PrP was determined by reference to IgG-binding MagicMark XP Western protein standards (Invitrogen). Quantitative analysis of the blots was performed using a GS-800 imaging densitometer and Quantity One software (Bio-Rad Laboratories). For the quantification of images visualized by Storm 860, ImageQuant software (Molecular Dynamics) was used.
Denaturation transition of PrPC and PrPSc in CDI.
The stability of PrPSc in brain homogenates or the detergent-insoluble pellet fractions of the homogenates was determined by treatment with GdnHCl at different concentrations as described previously (37, 43). The 10% brain homogenates were mixed with an equal volume of 2% Sarkosyl in PBS and incubated for 10 min at room temperature. After clarification at 500 × g for 5 min, aliquots of the supernatant were mixed with GdnHCl, with final concentrations ranging from 0 M to 7 M. Alternatively, the detergent-insoluble pellets were resuspended in a solution containing GdnHCl at various concentrations (range, 0 to 7 M). Samples containing different amounts of GdnHCl were incubated overnight at room temperature on a shaking platform and were then adjusted using distilled water containing EDTA-free protease inhibitors to a final concentration of GdnHCl of 0.35 M. The extent of unfolding of PrPSc exposed to different concentrations of GdnHCl was measured in triplicate by CDI.
CDI D/N ratio.
TRF counts obtained with CDI were used to generate a D/N ratio (2, 43). The folding state of PrPSc induced by overnight incubation at 7 M GdnHCl or by incubation at 81°C in the presence of 4 M GdnHCl was taken to be full denaturation (42, 43). The D/N ratios were obtained by dividing TRF counts of denatured samples (D) by the counts of the corresponding native samples (N; no GdnHCl).
Determination of the denatured fraction of PrPSc.
CDI results obtained with PrPSc-enriched pellets were used to determine the denatured fraction of PrPSc after an overnight incubation with particular concentrations of GdnHCl. For this, TRF counts obtained from an aliquot incubated in the absence of GdnHCl (no GdnHCl) were firstly subtracted from those obtained from aliquots treated with GdnHCl at various concentrations. Then, the denatured fraction of PrPSc at a particular concentration of GdnHCl was expressed as a relative value (percentage) of TRF counts at that concentration against the counts at 7 M GdnHCl and was plotted against the corresponding concentrations of GdnHCl. The denatured fraction of PrPSc (percentage) was calculated as follows: (TRFx M GdnHCl − TRF0 M GdnHCl/TRF7 M GdnHCl − TRF0 M GdnHCl) × 100.
Conventional CSA.
For the study of the stability of PrPres against GdnHCl-induced denaturation, aliquots of the 10% brain homogenates prepared in PBS were mixed with an equal volume of GdnHCl, giving a range of guanidine concentrations (0.0 M to 4.0 M) (37). After 2 h of incubation of the mixtures at room temperature with shaking, samples were adjusted to a final concentration of 0.4 M GdnHCl in Tris buffer (10 mM Tris-HCl [pH 7.4], 0.5% NP-40, and 0.5% sodium deoxycholate) and then digested with PK at 20 μg/ml for 1 h at 37°C. PK activity was terminated by adding Pefabloc at a final concentration of 1 mM. Proteins were then precipitated by mixing the samples with 5 volumes of prechilled methanol and incubated at −80°C overnight. Samples were then centrifuged at 16,000 × g for 30 min at 4°C, and the supernatants were carefully aspirated. The remaining pellets were resuspended with 25 μl of 2× LDS buffer and boiled for 10 min. Proteins were analyzed by Western blotting as described above. The conformation stability assay (CSA) included samples processed in parallel but unexposed to the chaotropic salt and analyzed by Western blotting with and without prior PK treatment.
Data presentation and statistics.
All graphs were constructed in Microsoft Office Excel, and statistical analysis employed the Student t test (TTest worksheet function) in this same program. P values of less than 0.05 were considered significant.
RESULTS
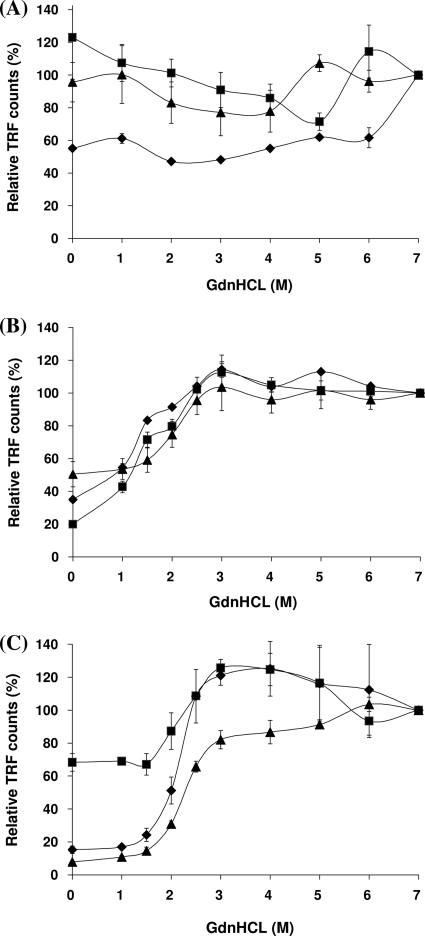
The effect on 3F4 epitope exposure by concentration-dependent guanidine denaturation was assayed in homogenates of the frontal cortex obtained from three cases of variant CJD, three cases of sporadic CJD (MM1 subtype), and three other neurological disease (non-CJD) cases. As expected, the results showed progressive 3F4 epitope exposure by increasing the guanidine concentration in all CJD cases but full or nearly full exposure in the non-CJD cases, even at low guanidine concentrations. The results were found to vary between cases, irrespective of whether they were normalized using the 7 M guanidine value, which was regarded as providing complete exposure (100%), with the counts presented as relative TRF (percentage) (Fig. 1A to C), or whether the data were presented as D/N ratios, with the 0 M guanidine reading given a value of 1.0 (data not shown). The reason for this variability was not clear. However, a possible cause was considered to be that autopsy human brain specimens vary in the absolute and relative abundances of PrPC and PrPSc and that this has a confounding effect on CDI. We reasoned that the removal of PrPC might be necessary before meaningful guanidine-induced denaturation curves could be obtained for autopsy human brain specimens.
FIG. 1.
Change in CDI TRF counts following exposure to increasing concentrations of GdnHCl. Brain homogenates from the frontal cortex were treated with increasing concentrations of GdnHCl (range, 0 to 7 M) and measured by CDI. Three cases each from neurological control (A), vCJD (B), and sCJD subtype MM1 (C) were analyzed. Each symbol (squares, diamonds, and triangles) represents individual cases of each phenotype. TRF counts at particular concentrations of GdnHCl were expressed as relative values (percentages) to those at 7 M GdnHCl. Data shown represent the average values ± SD for triplicate wells except for one control case (diamonds in panel A, duplicate wells). The non-CJD cases presented were all of the PRNP codon 129 MM genotype.
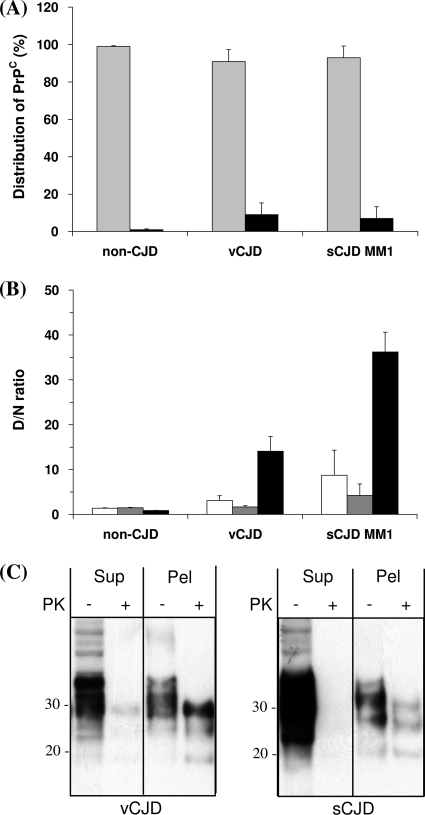
In order to explore this possibility and more directly investigate the guanidine-induced structural transition of PrPSc in the CJD brain, we exploited the known Sarkosyl insolubility of PrPSc. Preliminary characterization of the Sarkosyl-soluble supernatant and Sarkosyl-insoluble pellet fractions showed that >90% of PrP detected by 3F4 CDI under nondenaturing conditions was in the supernatant fraction (Fig. 2A), whereas elevated D/N ratios were largely a feature of the variant and sporadic CJD Sarkosyl-insoluble fractions (Fig. 2B). The characterizations of the supernatant comprised of PrPC and the pellet comprised of PrPSc (as determined by 3F4 accessibility/inaccessibility) were not absolute. The Sarkosyl-insoluble pellet in variant and sporadic CJD cases had higher levels of 3F4-accessible PrP than the non-CJD cases (Fig. 2A), and there was an elevated D/N ratio in the sporadic CJD supernatant compared to those in the supernatants of the non-CJD cases (Fig. 2B). Further investigation of PrP partitioning by Sarkosyl solubility showed that Sarkosyl-soluble PrP in variant and sporadic CJD cases was largely protease sensitive and that the Sarkosyl-insoluble material contained readily detectable protease-resistant PrP (Fig. 2C). These studies confirmed that the form of PrP in the Sarkosyl-insoluble pellet was predominantly abnormal prion protein, as determined by the criteria of conformation (PrPSc) and protease resistance (PrPres).
FIG. 2.
Distribution of PrPC and PrPSc after Sarkosyl extraction and centrifugation. Brain homogenates from the frontal cortex were prepared, and the supernatants and resuspended pellets were analyzed by CDI (A and B) and by Western blotting (C). (A) Distribution of PrPC in supernatants and pellets. The amounts of PrPC in the native state were determined by CDI, and the relative distribution of PrPC between supernatants (gray bars) and pellets (black bars) was expressed as a percentage. (B) Comparison of D/N ratios among initial brain homogenates, supernatants, and pellets. TRF counts of denatured samples were divided by the counts in the corresponding native samples to give D/N ratios. White bars, brain homogenates; gray bars, supernatants; black bars, pellets. (A and B) Data shown represent the average values ± SD obtained for three different cases. The vCJD and sCJD cases were all of the PRNP codon 129 MM genotype, whereas the three non-CJD cases are MM, MV, and VV at PRNP codon 129, respectively. (C) The supernatants (Sup) and pellets (Pel) obtained after Sarkosyl extraction and centrifugation were treated with proteinase K, and the distribution of PrPres was investigated by Western blotting, using MAb 3F4.
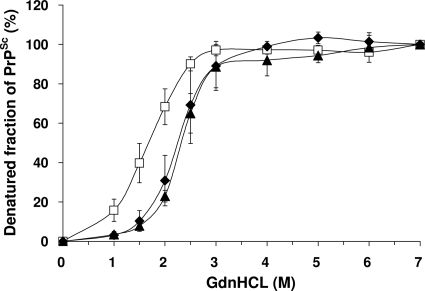
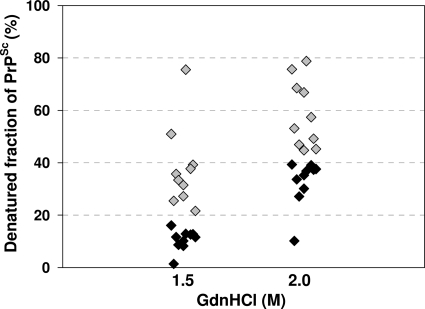
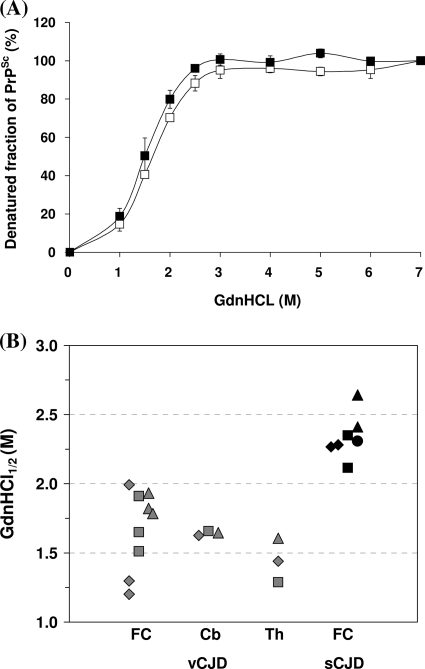
This Sarkosyl-insoluble fraction was then used for CDI to investigate whether there were any differences in the guanidine-induced transition of PrP between native and denatured states in variant and sporadic CJD PrPSc. In contrast to the more complex curves obtained using the original Sarkosyl extracts (Fig. 1), CDI of Sarkosyl-insoluble fractions from the CJD brain produced simple sigmoidal curves, with the major transition occurring between guanidine concentrations of 1 M and 3 M (Fig. 3). Comparison of three cases of variant CJD to three cases of sporadic CJD (MM1 subtype) showed that the curves were of the same general shape, but the curve for variant CJD was displaced to the left, indicating that variant CJD PrPSc is less stable than sporadic CJD of the MM1 subtype. Differences between the variant CJD and sporadic CJD MM1 subtype curves were significant for the 1.5 M and 2.0 M readings, with P values of <0.001. Similar analysis of the curve for a single typical case of sporadic CJD (VV2 subtype) showed close similarity to the curve for the MM1 subtype (Fig. 3). The guanidine-induced transition can be usefully expressed as a GdnHCl1/2 value, the GdnHCl concentration required to denature half of the PrPSc molecules present in a sample (21, 37). Analysis of replicate samples obtained from three different cases gave values of 2.344 (standard deviation [SD], ±0.159) for sporadic CJD (MM1 subtype) and 1.678 (SD, ±0.175) for variant CJD. Further analysis of a larger group of variant and sporadic CJD cases (n = 10) using the guanidine concentrations where the major transition occurred (1.5 M and 2.0 M) showed that the stability difference is a robust general phenomenon (at 1.5 M, P value of <0.001; at 2.0 M, P value of <0.0001), although case-to-case variation is evident (Fig. 4).
FIG. 3.
GdnHCl-induced denaturation of PrPSc from the frontal cortices of vCJD and sCJD brains. The extent of unfolding of PrPSc between native (0 M) and denatured (7 M) states was investigated by CDI. The denatured fraction of PrPSc in each concentration of GdnHCl was calculated. In vCJD (white squares) and sCJD subtype MM1 (black diamonds), data represent the average values ± SD for three individuals (two to three samples obtained from each individual). Data for VV2 sCJD (black triangles) was based on the examination of one sample in one case. The result for each sample was obtained by use of one to four independent experiments performed in triplicate.
FIG. 4.
Denatured fractions of PrPSc after treatment with 1.5 M or 2 M GdnHCl. Frontal cortex samples obtained from 10 vCJD and 10 sCJD MM1 subtype cases were treated with different concentrations of GdnHCl (0 M, 1.5 M, 2 M, and 7 M) and tested by CDI. The denatured fraction of PrPSc in each concentration of GdnHCl was calculated, and the results for individual cases were plotted against the concentration of GdnHCl. The results for the vCJD cases are depicted as gray diamonds, and the results for the sCJD MM1 cases are depicted as black diamonds.
The structural transition of PrPSc may, in principle, be an intrinsic property of PrP or may be affected by other components present in the samples under testing. Analysis of the vCJD cerebellum and thalamus (Fig. 5A) showed that PrPSc present in these regions produced curves similar in shape to those obtained when analyzing the frontal cortex (Fig. 3). The GdnHCl1/2 value of the cerebellum (1.643; SD, ±0.016) was very close to that of frontal cortex (1.678; SD, ±0.175); however, the value of the thalamus was somewhat lower (1.445; SD, ±0.159). Statistical analysis of the values obtained at the 1.5 M and 2.0 M guanidine readings confirmed that none of these pairwise comparisons achieved significance (P values > 0.05), although differences between the thalamus and the other two regions approached significance at the 2.0 M reading, with P values for the frontal cortex compared to those for the thalamus and cerebellum of 0.079 and 0.056, respectively. Plotting these individual values shows that although the mean values obtained between the vCJD frontal cortex and the vCJD thalamus differ, the thalamic values fall within the range defined by the values of the frontal cortex samples as a whole (Fig. 5B). Pairwise comparison of these groups confirmed that any differences between values obtained for different regions of the variant CJD brain were not significant (P value > 0.1), whereas the comparison of the variant CJD frontal cortex and sporadic CJD (MM1 subtype or MM1 subtype plus VV2 subtype) frontal cortex values was significantly different (P value < 0.0001).
FIG. 5.
GdnHCl-induced denaturation of PrPSc from the cerebellums and thalami of vCJD cases. (A) Extracts of cerebellums and thalami from vCJD cases were centrifuged, and the resultant pellets were exposed to increasing concentrations of GdnHCl. The degree of unfolding of PrPSc was measured by CDI, and the denatured fraction of PrPSc was calculated (empty squares, cerebellum; filled squares, thalamus). Data shown represent the average values ± SD for three individuals. The result for the cerebellum was an average value from three independent experiments in triplicate, and the result for the thalamus was an average value from a one-time experiment in triplicate. (B) Based on denaturation profiles shown in panel A and Fig. 3, the GdnHCl1/2 values were interpolated using the best fit of a second-order polynomial curve in Excel. Each symbol represents the value of an individual sample (vCJD, gray; sCJD, black). The results for three vCJD cases (diamonds, vCJD1; squares, vCJD2; triangles, vCJD3) and four sCJD cases (diamonds, MM1-1; squares, MM1-2; triangles, MM1-3; circles, VV2) are shown. The GdnHCl1/2 values of five samples (three FC samples, one cerebellum [Cb] sample, and one thalamus [Th] sample) in each vCJD case, two FC samples in each case of sCJD MM1 subtype, and one FC sample of a sCJD subtype VV2 case are depicted.
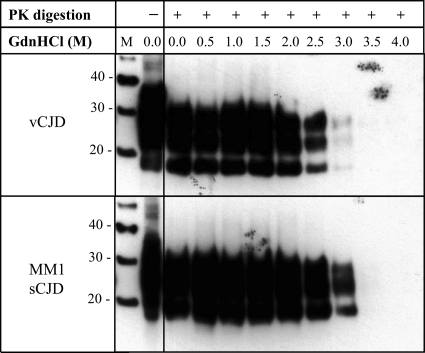
The conformation-dependent immunoassay as employed here specifically relates to conformational stability, as measured by access to the 3F4 epitope, whereas the conformation stability assay (CSA) employs concentration-dependent guanidine denaturation followed by proteinase K (PK) digestion, thus providing a measure of the stability of the protease-resistant core of PrP to guanidine-induced denaturation. CSA confirmed the reduced stability of PrP found in vCJD cases compared to that of PrP found in sporadic CJD (MM1) cases, with an estimated difference of approximately 0.5 M GdnHCl between the vCJD and sCJD values, as judged by Western blotting (Fig. 6).
FIG. 6.
Conformation stability assay of PrPres from frontal cortices of vCJD and MM1 sCJD cases. Western blot analysis of PrPres from vCJD and sCJD MM1 brain samples treated with increasing concentrations of GdnHCl (0 to 4 M, as indicated) and then subjected to proteinase K digestion at the concentration of 20 μg/ml for 1 h at 37°C (+) or allowed to remain undigested. After methanol precipitation of the protein, samples were analyzed by Western blotting using the anti-PrP MAb 3F4. Molecular mass markers (M) are shown, and they are labeled with their molecular mass in kDa. The result shown is representative of two independent experiments, with samples from two different cases of vCJD and sCJD (MM1).
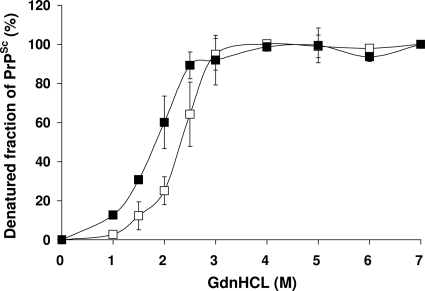
Lastly, we examined the CDI properties of PrPSc from two cases of Gerstmann-Straussler-Scheinker disease that differed in their PrPres Western blot profiles, although both carried the P102L mutation linked to methionine at codon 129. Both produced simple sigmoidal curves (Fig. 7), but the case that was characterized by type 1 PrPres had a more stable PrPSc than that which produced a predominantly ∼8-kDa PrPres, with GdnHCl1/2 values of 2.399 M and 1.877 M, respectively.
FIG. 7.
Comparison of denaturation profiles between two GSS cases with different PrPres patterns. The PrPSc denaturation profile from a GSS case with typical three-band PrPres (empty squares) was compared with that from a GSS case with lower-molecular-weight PrPres only (filled squares). The extent of unfolding of PrPSc between native (0 M) and denatured (7 M) states was determined by CDI. Data shown represent average values ± SD from three independent experiments in triplicate.
DISCUSSION
Definitions of PrPSc.
The accumulation of abnormal prion protein (PrPSc) is the only unambiguous marker of TSE or prion diseases established to date; however, the exact biochemical definitions of the forms of PrPSc most closely associated with infectivity and neurotoxicity and which might encipher the agent strain are incompletely defined at present. Operational definitions of PrPSc have included insolubility in nondenaturing detergents, affinity for particular ligands (including PrPSc-specific antibodies), and selective precipitation with polyoxometalates, in addition to resistance to proteolytic degradation (for examples, see references 19, 20, 22, 29, and 31). The last of these has been thoroughly exploited to the extent that the terms protease-resistant prion protein (PrPres) and disease-associated prion protein (PrPSc) are sometimes used interchangeably. The finding that a significant proportion of PrPSc found in the sCJD brain is protease sensitive (44) clearly implies that assays based on PrPres may be an incomplete description of the structural diversity of PrPSc.
Methodological considerations.
The conformation-dependent immunoassay (CDI) is an alternative approach to assessing conformational differences between PrPC and PrPSc and a tool to investigate the potentially strain-related properties of PrPSc. This approach avoids proteolysis, instead exploiting the chaotropic salt guanidine hydrochloride (GdnHCl) to unmask epitopes hidden in PrPSc. Chaotropic agents like GdnHCl disrupt hydrogen bonds that are responsible for the secondary structure (α-helices and β-sheets) of proteins (47). Since hydrogen bonding also helps maintain tertiary and quaternary structures of proteins, chaotropic agents can also affect these structural aspects. Thus, GdnHCl-induced denaturation of PrPSc molecules may occur at the following two different structural levels: loss of secondary structure (i.e., uncoiled into an irregularly coiled polypeptide) and loss of quaternary structure (i.e., dissociation of PrPSc aggregates).
In a previous study that examined the dissociation/unfolding pathways of PrPSc in hamster prion strains during exposures to ascending concentrations of GdnHCl (42), the GdnHCl-induced denaturation of PrPSc molecules was reported to consist of the following stepwise pathway: aggregates → dissociated folded monomer → partially unfolded intermediate → unfolded monomer. The dissociation of PrPSc aggregates into folded monomers occurred with the midpoint of transition at 1.5 to 2.0 M GdnHCl, followed by the state of stable intermediate (midpoint of transition, ∼3.5 M GdnHCl). Given that the hidden recognition site of MAb 3F4 becomes accessible in most of PrPSc molecules following incubation with 3 M GdnHCl, the degree of unfolding of PrPSc molecules measured by CDI most likely reflects the degree of dissociation of aggregated PrPSc. However, the loss of β-sheet structure following incubation with GdnHCl may also contribute to the CDI-based denaturation profiles, since the region with the 3F4 epitope has been proposed to acquire high β-sheet content during the conversion of PrPC into PrPSc (15), and the acquisition of β-sheet structure is thought to make the 3F4 epitope inaccessible (43). Despite these speculations, the exact mechanism by which GdnHCl denatures PrPSc remains elusive (25), and the presence of detergents such as N-lauroylsarcosinate sodium salt (Sarkosyl) may also interfere with the tertiary and quaternary structures of PrPSc aggregates by disrupting hydrophobic interactions (47). The detection monoclonal antibody used in CDI is MAb 3F4, which recognizes residues 109 to 112 of human PrP, and therefore, the conformational stability of PrPSc measured by CDI is thought to represent that of the N terminus of PrP (18, 43). The N-terminal region of PrPSc encompassing the 3F4 epitope was found to be critical in retaining infectivity in the Chandler mouse-adapted scrapie strain (45) and has also been implicated in relatively low infectivity associated with synthetic prions (3, 23).
Although the conformational stability assay (CSA) shares features with the CDI (notably, GdnHCl-induced denaturation and 3F4 detection), it is important to recognize that the CSA does not assess the stability of protease-sensitive PrPSc, although it does give an indication of the stability of the PrP molecule as a whole, by virtue of the size of the protease-resistant fragments seen on Western blots. For these reasons, we have primarily used the CDI (using the CSA as a confirmatory assay) to directly examine whether PrPSc associated with different human prion diseases differs in its conformational stability.
Stability differences identified by CDI.
Previous studies of the conformational stability of PrPSc using the CDI have been restricted to the analysis of animal prion diseases (scrapie) in the form of rodent-adapted strains in hamsters and mice (43, 49). To date, no equivalent studies have been published using human prion strains transmitted to rodents, nor has the potential been explored for the CDI to distinguish between the forms of PrPSc found in different human prion diseases.
Our results using postmortem brain tissue from representative cases of sCJD and vCJD show that PrPSc in the most commonly occurring MM1 subtype of sCJD is demonstrably more resistant to GdnHCl-induced denaturation than that in vCJD, as judged by CDI and the 3F4 epitope. This observation is supported by similar findings using the CSA, in which the method to measure conformational stability differs. In principle, such a conformation difference might be attributable to the same molecular property that is responsible for the Western blot type 1/type 2 PrPres difference; however, the sCJD VV2 subtype case examined here shared the conformational stability of the MM1 sCJD case, and not that of the vCJD (MM2B) case, suggesting that the conformational stability as measured by CDI is based on a property (or properties) of PrPSc that is different from the property of PrPres that determines the Western blot type.
Although the difference between sCJD and vCJD is clear and reproducible, we did observe a degree of intersample and intercase variability. This may reflect genuine microheterogeneity in PrPSc stability, or it may reflect confounding factors associated with cellular and molecular properties of the tissue sampled. Regional analysis of the vCJD brain showed that although the stability measurements of the frontal cortex and cerebellum were closely similar when expressed as average values, the equivalent measurement of the thalamus gave a lower GdnHCl1/2 value. This is intriguing because the pathology of the vCJD thalamus, including the PrP immunostaining pattern, is highly characteristic and differs from that of the cerebrum and cerebellum. However, the individual values for the vCJD thalamus fall within the range of individual values for the vCJD frontal cortex, so it is unclear at present what significance this observation may have.
The additional analysis of two GSS cases (both of the PRNP P102L codon 129 M genotype) that differ radically in their PrPres Western blot profiles showed that they too could be distinguished by the stability of PrPSc when measured by the access to the 3F4 epitope by CDI. The stability of PrPSc in the GSS case characterized by type 1 PrPres was similar to the stability of that in the sCJD cases, whereas the stability of PrPSc in the case in which proteinase K digestion and Western blotting produced bands of much smaller sizes (∼8 kDa) was found to be similar to the stability of that in the vCJD cases.
Although this study examined the stability of the “detergent-insoluble” fraction of PrP and was conducted in a limited number of disease phenotypes and/or cases, taken together, the results suggest that the structural stability of PrPSc as determined by CDI is not uniform among human prion diseases and that at least two stability states exist, which are at least partly independent of the conventional Western blot PrPres type (fragment size and glycoform ratio) and the PRNP genotype (pathogenic mutations and codon 129). Having now defined the GdnHCl concentration range at which the major structural transition occurs in CJD and GSS cases, it will be important to try to refine these methods to establish whether further meaningful distinctions can be made between examples of the full range of human prion diseases, including all of the subtypes of sCJD, fatal familial insomnia, and the more recently described protease-sensitive prionopathy (12).
Our proposal is not without precedent. Comparison of single cases of sCJD and vCJD using PrPSc-specific peptide capture followed by examination with sandwich ELISA has shown a higher stability for sCJD PrPSc (21). In their study, the difference in GdnHCl1/2 values was ∼0.6 M between the two forms of CJD (GdnHCl1/2 in vCJD, 1.85 M; GdnHCl1/2 in MM1 sCJD, 2.45 M), which resembles the values obtained using a greater number of vCJD and sCJD cases and the CDI in this study. CSA analysis of PrPres in sCJD of the MM genotype has shown that type 1 PrPres is more stable than type 2 (7), and this observation has been extended using a rapid dot blot variation of the CSA to encompass the MV and VV genotypes in sporadic CJD cases (53). Moreover, a recent conference abstract suggests that this higher structural stability of sCJD MM1 (compared to that of sCJD MM2) is conserved when these diseases are transmitted to bank voles (40). When GdnHCl1/2 values have been given in those papers, we have compared them with our own results (Table 1). Given that the experimental approaches differed markedly between those studies, there is a surprising degree of agreement between some of them. Where discrepancies exist, it is unclear whether these result from differences in the methodology employed (such as the use of CDI measurements of PrPSc or CSA measurement of PrPres), intersample or intercase variability, or some other factor.
TABLE 1.
Comparison of the GdnHCl1/2 values of PrPSc obtained in this study with those published previously
| Technique | GdnHCl1/2 value of PrPSc ina: |
Material analyzed | Reference | |||
|---|---|---|---|---|---|---|
| vCJD | sCJD MM1 | sCJD MM2A | sCJD VV2A | |||
| CDI | 1.678 | 2.344 | ND | 2.308 | Human brain | This study |
| PrPSc-specific capture | 1.850 | 2.450 | ND | ND | Human brain | 21 |
| CSA | ND | 2.760 | 1.420 | ND | Human brain | 7 |
| Detergent insolubility | ND | 2.950 | 1.460 | 2.690 | Bank vole brain | 40 |
ND = not determined.
Biological significance of PrPSc stability.
There is accumulating experimental evidence to suggest that the conformational stability of PrPSc has biological significance. Conformational stability was used as an argument to defend the original description of synthetic prions (23) against the criticism that they could have resulted from inadvertent contamination with common laboratory strains. The GdnHCl1/2 values of mouse PrPSc resulting from infection with synthetic prions differed from those of the commonly used RML strain (24). Further passage of synthetically derived prion resulted in two distinct strains that were found to differ in both incubation period and PrPSc stability, thus prompting an evaluation of the relationship between stability (as judged by PrPSc GdnHCl1/2 values measured by CSA) and incubation period in mice generally (25). A strong positive correlation between higher stability and longer incubation periods was found (25). The hypothesis that incubation time is a function of PrPSc stability (in addition to the known factors such as titer, route, and PrP expression level and sequence) was further tested by artificially producing an array of synthetic PrP amyloids differing in their conformational stability and then by testing their biological properties (9). The results showed that more labile PrPSc conformations correlated with faster replication, shorter incubation periods, and a degree of strain instability. How such considerations might relate to human prion diseases is unclear at this time. However, the finding that vCJD PrPSc is less intrinsically stable than that of sCJD may be interpreted as a further reason for concern regarding the potential for secondary transmission of the promiscuous BSE/vCJD prion strain.
Acknowledgments
The National CJD Surveillance Unit Brain & Tissue Bank is supported by the Medical Research Council (United Kingdom), and the National CJD Surveillance Unit as a whole is funded by the Department of Health, United Kingdom, and the Scottish Government. Albrecht Gröner is an employee of CSL Behring. The remaining authors have no relevant conflicts of interest to declare.
This is an independent report commissioned and funded by the Policy Research Programme in the Department of Health, United Kingdom. The views expressed in the publication are those of the authors and not necessarily those of the Department of Health.
Footnotes
Published ahead of print on 15 September 2010.
REFERENCES
- 1.Aguzzi, A., M. Heikenwalder, and M. Polymenidou. 2007. Insights into prion strains and neurotoxicity. Nat. Rev. Mol. Cell Biol. 8:552-561. [DOI] [PubMed] [Google Scholar]
- 2.Bellon, A., W. Seyfert-Brandt, W. Lang, H. Baron, A. Gröner, and M. Vey. 2003. Improved conformation-dependent immunoassay: suitability for human prion detection with enhanced sensitivity. J. Gen. Virol. 84:1921-1925. [DOI] [PubMed] [Google Scholar]
- 3.Bocharova, O. V., L. Breydo, V. V. Salnikov, A. C. Gill, and I. V. Baskakov. 2005. Synthetic prions generated in vitro are similar to a newly identified subpopulation of PrPSc from sporadic Creutzfeldt-Jakob disease. Protein Sci. 14:1222-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, P., and R. Bradley. 1989. 1755 and all that: a historical primer of transmissible spongiform encephalopathy. Br. Med. J. 317:1688-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Budka, H., A. Aguzzi, P. Brown, J. M. Brucher, O. Bugiani, F. Gullotta, M. Haltia, J. J. Hauw, J. W. Ironside, K. Jellinger, H. A. Kretzschmar, P. L. Lantos, C. Masullo, W. Schlotl, J. Tateishi, and R. O. Weller. 1995. Neuropathological diagnostic criteria for Creutzfeldt-Jakob disease (CJD) and other human spongiform encephalopathies (prion diseases). Brain Pathol. 5:459-466. [DOI] [PubMed] [Google Scholar]
- 6.Bruce, M. E. 2003. TSE strain variation. Br. Med. Bull. 66:99-108. [DOI] [PubMed] [Google Scholar]
- 7.Cali, I., R. Castellani, A. Alshekhlee, Y. Cohen, J. Blevins, J. Yuan, J. P. Langeveld, P. Parchi, J. G. Safar, W. Q. Zou, and P. Gambetti. 2009. Co-existence of scrapie prion protein types 1 and 2 in sporadic Creutzfeldt-Jakob disease: its effect on the phenotype and prion-type characteristics. Brain 132:2643-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caughey, B., and G. Baron. 2006. Prions and their partners in crime. Nature 443:803-810. [DOI] [PubMed] [Google Scholar]
- 9.Colby, D. W., K. Giles, G. Legname, H. Wille, I. V. Baskakov, S. J. DeArmond, and S. B. Prusiner. 2009. Design and construction of diverse mammalian prion strains. Proc. Natl. Acad. Sci. U. S. A. 106:20417-20422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collinge, J., K. C. L. Sidle, J. Meads, J. Ironside, and A. F. Hill. 1996. Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature 383:685-690. [DOI] [PubMed] [Google Scholar]
- 11.Collinge, J., and A. R. Clarke. 2007. A general model of prion strains and their pathogenicity. Science 318:930-936. [DOI] [PubMed] [Google Scholar]
- 12.Gambetti, P., Z. Dong, J. Yuan, X. Xiao, M. Zheng, A. Alshekhlee, R. Castellani, M. Cohen, M. A. Barria, D. Gonzalez-Romero, E. D. Belay, L. B. Schonberger, K. Marder, C. Harris, J. R. Burke, T. Montine, T. Wisniewski, D. W. Dickson, C. Soto, C. M. Hulette, J. A. Mastrianni, Q. Kong, and W. Q. Zou. 2008. A novel prion disease with abnormal prion protein sensitive to protease. Ann. Neurol. 63:697-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Head, M. W., T. J. R. Bunn, M. T. Bishop, V. McLoughlin, S. Lowrie, C. S. McKimmie, M. C. Williams, L. McCardle, J. MacKenzie, R. Knight, R. G. Will, and J. W. Ironside. 2004. Prion protein heterogeneity in sporadic but not variant Creutzfeldt-Jakob disease: U.K. cases 1991-2002. Ann. Neurol. 55:851-859. [DOI] [PubMed] [Google Scholar]
- 14.Hill, A. F., S. Joiner, J. A. Beck, T. A. Campbell, A. Dickinson, M. Poulter, J. D. F. Wadsworth, and J. Collinge. 2006. Distinct glycoform ratios of protease resistant prion protein associated with PRNP point mutations. Brain 129:676-685. [DOI] [PubMed] [Google Scholar]
- 15.Huang, Z., S. B. Prusiner, and F. E. Cohen. 1995. Scrapie prions: a three-dimensional model of an infectious fragment. Fold. Des. 1:13-19. [PubMed] [Google Scholar]
- 16.Ironside, J. W., M. W. Head, J. E. Bell, L. McCardle, and R. G. Will. 2000. Laboratory diagnosis of variant Creutzfeldt-Jakob disease. Histopathology 37:1-9. [DOI] [PubMed] [Google Scholar]
- 17.Ironside, J. W., B. Ghetti, M. W. Head, P. Piccardo, and R. G. Will. 2008. Prion diseases, p. 1197-1273. In S. Love, D. N. Lious, and D. Ellison (ed.), Greenfield's neuropathology, 8th ed., vol. 2. Hodder Arnold, London, United Kingdom. [Google Scholar]
- 18.Kascsak, R. J., R. Rubenstein, P. A. Merz, M. Tonna-DeMasi, R. Fersko, R. I. Carp, H. M. Wisniewski, and H. Diringer. 1987. Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J. Virol. 61:3688-3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kocisko, D. A., P. T. Landsbury, and B. Caughey. 1996. Partial unfolding and refolding of scrapie-associated prion protein: evidence for a critical 16-kDa C-terminal domain. Biochemistry 35:13434-13442. [DOI] [PubMed] [Google Scholar]
- 20.Korth, C., B. Stierli, P. Streit, M. Moser, O. Schaller, R. Fischer, W. Schulz-Schaeffer, H. Kretzschmar, A. Raeber, U. Braun, F. Ehrensperger, S. Hornemann, R. Glockshuber, R. Riek, M. Billiter, K. Wuethrich, and B. Oesch. 1997. Prion (PrPSc)-specific epitope defined by a monoclonal antibody. Nature 390:74-77. [DOI] [PubMed] [Google Scholar]
- 21.Lau, A. L., A. Y. Yam, M. M. Michelitsch, X. Wang, C. Gao, R. J. Goodson, R. Shimizu, G. Timoteo, J. Hall, A. Medina-Selby, D. Coit, C. McCoin, B. Phelps, P. Wu, C. Hu, D. Chien, and D. Peretz. 2007. Characterization of prion protein (PrP)-derived peptides that discriminate full-length PrPSc from PrPC. Proc. Natl. Acad. Sci. U. S. A. 104:11551-11556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, I. S., J. R. Long, S. B. Prusiner, and J. G. Safar. 2005. Selective precipitation of prions by polyoxometalate complexes. J. Am. Chem. Soc. 127:13802-13803. [DOI] [PubMed] [Google Scholar]
- 23.Legname, G., I. V. Baskakov, H. O. B. Nguyen, D. Riesner, F. E. Cohen, S. J. DeArmond, and S. B. Prusiner. 2004. Synthetic mammalian prions. Science 305:673-676. [DOI] [PubMed] [Google Scholar]
- 24.Legname, G., H. O. B. Nguyen, I. V. Baskakov, F. E. Cohen, S. J. DeArmond, and S. B. Prusiner. 2005. Strain-specified characteristics of mouse synthetic prions. Proc. Natl. Acad. Sci. U. S. A. 102:2168-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Legname, G., H. O. Nguyen, D. Peretz, F. E. Cohen, S. J. DeArmond, and S. Prusiner. 2006. Continuum of prion protein structures enciphers a multitude of prion isolate-specified phenotypes. Proc. Natl. Acad. Sci. U. S. A. 103:19105-19110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makarava, N., and I. V. Baskakov. 2008. The same primary structures of the prion protein yields two distinct self-propagating states. J. Biol. Chem. 283:15988-15996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makarava, N., V. G. Ostapchenko, R. Savchenko, and I. V. Baskakov. 2009. Conformational switching within individual amyloid fibrils. J. Biol. Chem. 284:14386-14395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makarava, N., G. G. Kovacs, O. Bocharova, R. Savtchenko, I. Alexeeva, H. Budka, R. G. Rohwer, and I. V. Baskakov. 2010. Recombinant prion protein induces a new transmissible prion disease in wild-type animals. Acta Neuropathol. 119:177-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKinley, M. P., R. K. Meyer, L. Kenaga, F. Rahbar, R. Cotter, A. Serban, and S. B. Prusiner. 1991. Scrapie prion rod formation in vitro requires both detergent extraction and limited proteolysis. J. Virol. 65:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nurmi, M. H., M. Bishop, L. Strain, F. Brett, C. McGuigan, M. Hutchison, M. Farrell, R. Tilvis, S. Erkkila, O. Simell, R. Knight, and M. Haltia. 2003. The normal population distribution of PRNP codon 129 polymorphism. Acta Neurol. Scand. 108:374-378. [DOI] [PubMed] [Google Scholar]
- 31.Oesch, B., M. Jensen, P. Nilsson, and J. Fogh. 1994. Properties of the scrapie prion protein: quantitative analysis of protease resistance. Biochemistry 33:5926-5931. [DOI] [PubMed] [Google Scholar]
- 32.Parchi, P., R. Castellani, S. Capellari, B. Ghetti, K. Young, S. G. Chen, M. Farlow, D. W. Dickson, A. A. F. Sima, J. Q. Trojanowski, R. B. Petersen, and P. Gambetti. 1996. Molecular basis of the phenotypic variability in sporadic Creutzfeldt-Jakob disease. Ann. Neurol. 39:767-778. [DOI] [PubMed] [Google Scholar]
- 33.Parchi, P., S. Capellari, S. G. Chen, R. B. Petersen, P. Gambetti, N. Kopp, P. Brown, T. Kitamoto, J. Tateishi, A. Giese, and H. Kretzschmar. 1997. Typing prion isoforms. Nature 386:232-234. [DOI] [PubMed] [Google Scholar]
- 34.Parchi, P., S. G. Chen, P. Brown, W. Zou, S. Capellari, H. Budka, J. Hainfellner, P. F. Reyes, G. T. Golden, J. J. Hauw, D. C. Gajdusek, and P. Gambetti. 1998. Different pattern of truncated prion protein fragments correlated with distinct phenotypes in P102L Gerstmann-Straussler-Scheinker disease. Proc. Natl. Acad. Sci. U. S. A. 95:8322-8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parchi, P., A. Giese, S. Capellari, P. Brown, W. Schulz-Schaeffer, O. Windl, I. Zerr, H. Budka, N. Kopp, P. Piccardo, S. Poser, A. Rojiani, N. Streichemberger, J. Julien, C. Vital, B. Ghetti, P. Gambetti, and H. Kretzschmar. 1999. Classification of sporadic Creutzfeldt-Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann. Neurol. 46:224-233. [PubMed] [Google Scholar]
- 36.Parchi, P., R. Strammiello, S. Notari, A. Giese, J. P. M. Langeveld, A. Ladogana, I. Zerr, F. Roncaroli, P. Cras, B. Ghetti, M. Pocchiari, H. Kretzschmar, and S. Capellari. 2009. Incidence and spectrum of sporadic Creutzfeldt-Jakob disease variants and mixed phenotype and co-occurrence of PrPSc types: an updated classification. Acta Neuropathol. 118:659-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peretz, D., M. R. Scott, D. Groth, R. A. Williamson, D. R. Burton, F. E. Cohen, and S. B. Prusiner. 2001. Strain-specified relative conformational stability of the scrapie prion protein. Protein Sci. 10:854-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peretz, D., A. R. Williamson, G. Legname, Y. Matsunaga, J. Vergara, D. R. Burton, S. J. DeArmond, S. B. Prusiner, and M. R. Scott. 2002. A change in the conformation of prions accompanies the emergence of a new prion strain. Neuron 34:921-932. [DOI] [PubMed] [Google Scholar]
- 39.Piccardo, P., S. R. Dlouhy, P. M. J. Lievens, K. Young, T. D. Bird, D. Nochlin, D. W. Dickson, H. V. Vinters, T. R. Zimmerman, I. R. A. Mackenzie, S. J. Kish, L. C. Ang, C. De Carli, M. Pocchiari, P. Brown, C. J. Gibbs, D. C. Gajdusek, O. Bugiani, J. Ironside, F. Tagliavini, and B. Ghetti. 1998. Phenotypic variability of Gerstmann-Straussler-Scheinker disease is associated with prion protein heterogeneity. J. Neuropathol. Exp. Neurol. 57:979-988. [DOI] [PubMed] [Google Scholar]
- 40.Pirisinu, L., M. D. Bari, P. Fazzi, S. Marcon, C. D'Agostino, E. Esposito, S. Simon, P. Frassanito, G. Vaccari, U. Agrimi, and R. Nonno. 2009. Biochemical characterization of animal and human strains in bank voles (Myodes glareolus), abstr. P.10.8, p. 179. Abstr. Prion 2009.
- 41.Prusiner, S. B. 1998. Prions. Proc. Natl. Acad. Sci. U. S. A. 95:13363-13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Safar, J., P. P. Roller, D. C. Gajdusek, and C. J. Gibbs. 1993. Conformational transitions, dissociation, and unfolding of scrapie amyloid (prion) protein. J. Biol. Chem. 268:20276-20284. [PubMed] [Google Scholar]
- 43.Safar, J., H. Wille, V. Itri, D. Groth, H. Serban, M. Torchia, F. E. Cohen, and S. B. Prusiner. 1998. Eight prion strains have PrP(Sc) molecules with different conformations. Nat. Med. 4:1157-1165. [DOI] [PubMed] [Google Scholar]
- 44.Safar, J. G., M. D. Geschwind, C. Deering, S. Didorenko, M. Sattavat, H. Sanchez, A. Serban, M. Vey, H. Baron, K. Giles, B. L. Miller, S. J. DeArmond, and S. B. Prusiner. 2005. Diagnosis of human prion disease. Proc. Natl. Acad. Sci. U. S. A. 102:3501-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shindoh, R., C. L. Kim, C. H. Song, R. Hasebe, and M. Horiuchi. 2009. The region approximately between amino acids 81 and 137 of proteinase K-resistant PrPSc is critical for the infectivity of the Chandler prion strain. J. Virol. 83:3852-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Somerville, R. A. 2002. TSE agent strain and PrP: structure and function. Trends Biochem. Sci. 27:606-612. [DOI] [PubMed] [Google Scholar]
- 47.Tanford, C. 1968. Protein denaturation. Adv. Protein Chem. 23:121-282. [DOI] [PubMed] [Google Scholar]
- 48.Thackray, A. M., L. Hopkins, and R. Bujdoso. 2007. Proteinase K-sensitive disease-associated ovine prion protein revealed by conformation-dependent immunoassay. Biochem. J. 401:475-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thackray, A. M., L. Hopkins, M. A. Klein, and R. Bujdoso. 2007. Mouse-adapted ovine scrapie prion strains are characterized by different conformers of PrPSc. J. Virol. 81:12119-12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wadsworth, J. D., S. Joiner, J. M. Linehan, S. Cooper, C. Powell, G. Mallinson, J. Buckell, I. Gowland, E. A. Asante, H. Budka, S. Brandner, and J. Collinge. 2006. Phenotypic heterogeneity in inherited prion disease (P102L) is associated with differential propagation of protease-resistant wild-type and mutant prion protein. Brain 129:1557-1569. [DOI] [PubMed] [Google Scholar]
- 51.Wang, F., X. Wang, C. G. Yuan, and J. Ma. 2010. Generating a prion with bacterially expressed recombinant prion protein. Science 327:1132-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watts, J. C., A. Balachandran, and D. Westaway. 2006. The expanding universe of prion diseases. PLoS Pathog. 2:e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wemheuer, W. M., S. L. Benestad, A. Wrede, U. Schulze-Sturm, W. E. Wemheuer, U. Hahmann, J. Gawinecka, E. Schutz, I. Zerr, B. Brenig, B. Bratberg, O. Andreoletti, and W. Schulz-Schaeffer. 2009. Similarities between isoforms of sheep scrapie and Creutzfeldt-Jakob disease are encoded by distinct prion types. Am. J. Pathol. 175:2566-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yull, H. M., D. L. Ritchie, J. P. Langeveld, F. G. van Zijderveld, M. E. Bruce, J. W. Ironside, and M. W. Head. 2006. Detection of type 1 prion protein in variant Creutzfeldt-Jakob disease. Am. J. Pathol. 168:151-157. [DOI] [PMC free article] [PubMed] [Google Scholar]