Abstract
The National Institutes of Health has proposed a roadmap for clinical research. Test projects of this roadmap include centralized data management for distributed research, the harmonization of clinical and research data, and the use of data standards throughout the research process. In 2003, RxNorm was named as a standard for codifying clinical drugs. Clinical researchers looking to implement RxNorm have few template implementation plans. Epidemiological studies and clinical trials (types of clinical research) have different requirements for model standards and best implementation tools. This paper highlights two different (epidemiological and intervention) clinical research projects, their unique requirements for a medication standard, the suitability of RxNorm as a standard for each, and application and process requirements for implementation. It is hoped that our experience of selecting and implementing the RxNorm standard to address varying study requirements in both domestic and international settings will be of value to other efforts.
Keywords: Biomedical research, Data collection, Terminology, Medication systems
INTRODUCTION
Standards
The National Institutes of Health has proposed a roadmap for research [1], including a restructuring of the clinical research enterprise. Test projects of this roadmap include centralized data management for distributed research, the harmonization of clinical and research data, and the use of data standards throughout the research process. The need for data standards in clinical research has been cited.[2–4]
The U.S. agencies involved in the Consolidated Health Informatics (CHI) initiative have worked together to identify and recommend the use of the “best” data standards in a variety of areas (e.g., anatomy, problem lists, laboratory, test names, and clinical drugs).[5] All federally funded clinical research has recently been charged to share data following NIH guidelines.[6] Although the CHI has named RxNorm as the standard for clinical drug names [5], there is little experience in its use, and scarce implementation guidance. Different research designs and settings dictate unique standards requirements, and different data collection processes call for customized applications to successfully implement any standard. In this paper we use the term “medication data” to include drug products (defined as drug + dosage + form), clinical drugs (defined as drug + dosage) and active ingredients (drug names with no associated dosage or formulary information).
Despite the lack of experience and guidance in implementing data standards in clinical research, organizations are pressuring their research staff to demonstrate standards compliance. This paper describes two large and very different research efforts that have adopted RxNorm as a CHI-recommended standard for medication data. Because of RxNorm’s youth and its recent selection as a standard, there are few tools to assist with the implementation of RxNorm into existing data collection procedures. We describe these diverse large-scale research projects and their requirements for an ideal representation for medication data, and then name the reasons RxNorm was selected as the standard. Further, we describe the unique operational features of each study and describe the application and process requirements for implementing RxNorm.
RxNorm
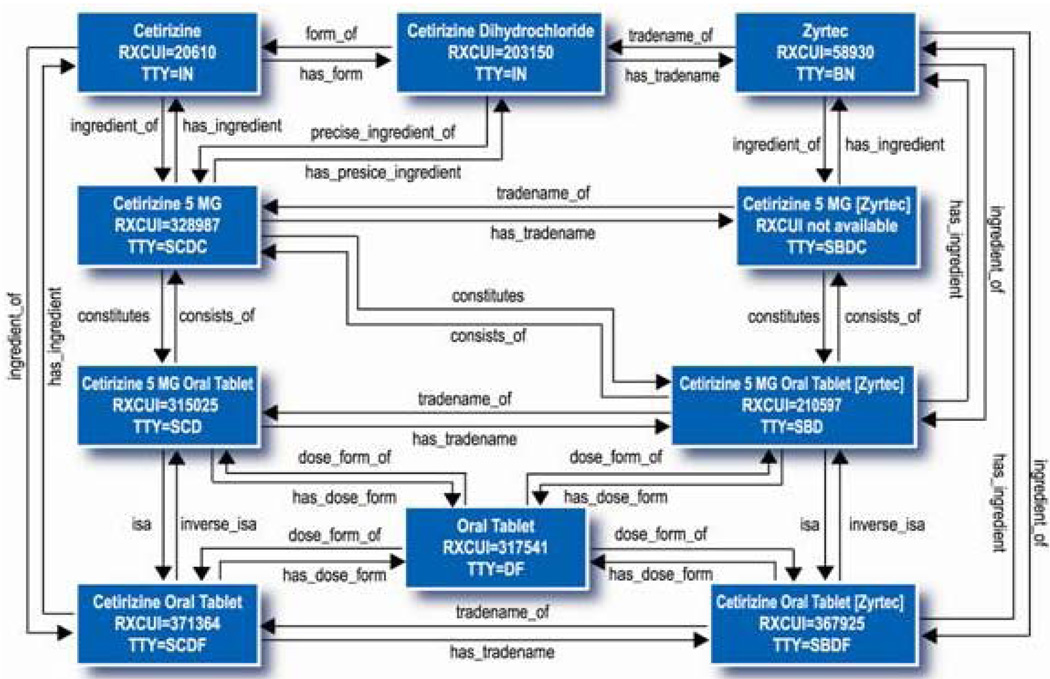
RxNorm, developed and maintained by the National Library of Medicine (NLM), is a controlled nomenclature of medications at varying levels of detail in order to express what a clinician might order for a patient and the type of order a pharmacy might recieve.[7] RxNorm has a simple relational model that relates multiple dosages, forms, and packaging of medications to their component ingredients (as generic or non-proprietary brand names).[8] (See Figure 1.)
Figure 1. RxNorm Data Structures.
Source: http://www.nlm.nih.gov/research/umls/rxnorm/overview.html
RxNorm provides standard names for clinical drugs (active ingredient + strength + dose form) as administered to patients. It provides links from clinical drugs to their active ingredients, drug products (active ingredient + dosage + form), and related brand names. NDCs (National Drug Codes) for specific drug products (where there are often many NDC codes for a single product) are linked to that product in RxNorm. RxNorm also links its names to drug lists commonly used in pharmacy management and drug interaction software, including those of First Databank, Micromedex, MediSpan, and Multum. By providing links between these vocabularies, RxNorm can mediate messages between systems not using the same software and vocabulary. RxNorm identifiers have recently been used as a translation or mediation of disparate drug vocabularies between the U.S. Department of Veterans Affairs and the Department of Defense.[9] The focus of RxNorm (at this time) is on U.S. drugs to support the representation of administered drug products for hospital information systems.[7]
SETTING
The Pediatrics Epidemiology Center (PEC) is located within the University of South Florida, College of Medicine, Department of Pediatrics. The PEC acts as a Data and Technology Coordinating Center to several large multi-site and multinational epidemiologic and clinical intervention studies, including The Environmental Determinants of Diabetes in the Young (TEDDY) [10, 11] study and the Rare Disease Clinical Research Network (RDCRN) [12] of observational and intervention trials in over 40 rare diseases. As a center, the PEC has a need for a single standard for medication data, although the requirements for a standard vary widely over the projects that the PEC supports. This paper explores the use of RxNorm as a standard in these two large projects (TEDDY and RDCRN) whose data coordination operations are located at the PEC (although they are geographically distributed projects).
TEDDY is an international study that is exploring genetic-environmental interactions in relation to the development of Type I Diabetes Mellitus (T1DM). Newborns identified to be at genetic risk for T1DM will be followed for 15 years for the appearance of various auto-antibodies and diabetes, with documentation of early childhood diet, child and maternal medications, infections, vaccinations, and psychosocial stressors. The projected sample of over 7,000 newborns is being recruited across six clinical centers worldwide (Finland, Germany, Sweden and three in North America).
The RDCRN [13] consists of ten clinical research consortia, distributed throughout the country, and each one focused on a group of rare diseases. Collectively, the RDCRN has over 50 active research protocols representing many study designs (e.g., observational, phase 1 & 2 clinical trials), with many more in development. The PEC coordinates the electronic collection of clinical research data for the network, applies data standards where necessary, and conducts the analysis for all network studies. Almost all RDCRN studies under development collect medication data – either as independent variables in observational study designs, or as intervention and control variables in clinical trials.
TEDDY: REQUIREMENTS AND DATA COLLECTION
Medication Data Requirements
The TEDDY project is a large epidemiologic study that collects medication data in the context of periodic self-report of maternal and child medications (names only, not dosages or drug form). The data for the TEDDY study is collected by distributed research staff using scannable automated forms, and the data are transmitted electronically through scanning or manually entered on online forms to the PEC. There were three major requirements for “standard” medication names in the TEDDY project. First, because of the use of scannable automated forms and the various languages involved, there was a need for a code to support each reported medication. (The use of free text is not feasible because of length restrictions for the automated form fields, the high likelihood of spelling errors on original forms or scanning errors due to multiple handwriting sources.[14]) Given the need for an alphanumeric code for each medication, the ideal standard should adhere to best-practice criteria [15] regarding the management of numerical codes. The codes should be non-sensical, unique, non-ambiguous, permanent, and should never be recycled to encode other concepts. Additionally, the ideal standard for a long-term project, such as TEDDY, should include a defined process to add codes as new medications or gaps in the standard are discovered.[16]
The second major requirement in identifying a standard for medication names in the TEDDY project is that the study is international, and therefore the ideal standard would be something in routine use in all of the member countries. However, there is currently no international standard coding scheme for medications. While the World Health Organization’s International Nonproprietary Names (INN) provides a well-adopted global standard naming for non-proprietary (generic) names, there is no accessible numerical code associated with unique drug names. Additionally, the INN does not maintain data on the many different brand names (across countries) for each generic drug.
The third requirement for a standard for medication names in the TEDDY project is the need to store generic-named medication data that is often reported by study participants by brand names. Given that the analysis of medication data for TEDDY will be at the level generically-named active ingredients, the ideal coding scheme should include relationships between brand names and generic ingredient names. There are no comprehensive electronic sources that capture brand name – generic relationships globally, although individual countries have reputable sources for this.
A minor consideration is cost and access, because of multiple sites using the standard in this project. While the use of standards is important enough to warrant licensing fees, the number of data collection sites (n=12) in the 6 study clinical centers would potentially create logistical problems for updating and distributing a commercial standard. As a rule, the PEC strives to select free and open access standards wherever possible.
RxNorm was selected as the ideal standard for the TEDDY project because it meets all of the requirements outlined above. While it is not international in mission, the U.S. National Library of Medicine does attempt to address international needs in all of its activities, and generally ensures free and global access to its products. The NLM guarantees “best” terminology practices, and the numerical codes in RxNorm will never be changed or reassigned. These code numbers are easily accessible through the NLM-developed UMLS and the RxNav [17] interfaces. While the medication data in the TEDDY project is limited to medication names (not dosage or drug form) and a set of corresponding unique codes to support data collection, the PEC operates other studies that require a more detailed standard (which can represent dosage and drug form), and as a Center, the PEC wanted one standard for all medication data. While other standards could have addressed the needs of the TEDDY project, requirements for another PEC project, the RDCRN, entailed a flexible medication standard to encompass unique codes for medication names only, as well as medication name + dosage + drug form.
RxNorm was implemented in the TEDDY project in May 2005, and since then we have found 99% coverage (at the level of active ingredients) of all the medications reported in the TEDDY study. Of the 5,832 participants enrolled as of December 1, 2008, over 742 unique medications have been reported representing 686 unique active ingredients or combinations of active ingredients.
Process for Collecting Medication Data in TEDDY Project
The process identified for the coding of medications in TEDDY includes reporting medications by generic-named ingredients to the PEC, where the RxNorm codes are centrally assigned and managed for the project. If the reported medication has been collected previously in the TEDDY study, it is included in an on-line codebook, where research staff can access the correct code. If the medication name has not been previously collected, field researchers contact the PEC, whose staff determines the correct RxNorm code using the NLM’s RxNav tool.[17] Because there is no single source for international brand name – generic name medications, each TEDDY site is responsible for determining generic (active) ingredients from any brand name reported medications (when the brand name cannot be found in RxNorm). The PEC developed an on-line codebook (linking multiple proprietary names to unique generic ingredients, or unique combinations of generic ingredients) to facilitate user access to a subset of (common) RxNorm codes remotely and on demand. Further, the PEC has an internal system in place to deal with generic drug names that might not be in RxNorm. This process includes the assignment of a temporary ID number while the PEC contacts NLM about including the medication in RxNorm.
Because of the length of RxNorm numerical codes, and because multiple RxNorm ingredient codes are required for medication products containing multiple active ingredients, research staff in the field requested the PEC provide shorter identifiers for recording reported medication data on Teleforms. The PEC has developed an internal system to assign and maintain unique medication ID #s to support data entry. These ID#s are called “TEDDY Medication Codes” and are 8 digits long. They fit within the field length limits of the scannable form, and are shorter than the RxNorm code, which eases data entry and prevents coding errors. Within the TEDDY data repository, the TEDDY codes are tied to specific RxNorm codes for generic drug names. The RxNorm code is the standard coding scheme used by TEDDY and is how the data are retrieved for analysis. Because the PEC codebook is a smaller list of TEDDY-reported medications using the shorter TEDDY medication codes, the researchers do not view or navigate RxNorm codes. Because the study population is well-defined, after 18 months of study, the codebook has reached a saturation point and new requests for TEDDY medication codes are infrequent.
The only link from reported brand name medication to the ingredient (generic) names entered in the PEC’s data repository is the original source data and the TEDDY medication code. The online TEDDY codebook allows distributed research personnel to search for previously used medications. Users can search the online codebook by generic or brand name, but the TEDDY medication codes retrieved are tied to the generic ingredient(s). Often multiple brand names (up to 15) are associated with a single active ingredient or combination of active ingredients in the codebook. Single medications with multiple active ingredients are stored in the TEDDY database as (multiple) instances of separate drugs.
RARE DISEASE NETWORK: REQUIREMENTS AND DATA COLLECTION
Medication Data Standards Requirements Specific to the Rare Disease Network
Because the RDCRN is funded by the NIH, and all primary sites are in the U.S., the selection of a standard was a bit easier, since the domestic CHI standards apply. Despite the CHI recommendation, the PEC laid out the requirements for an RDCRN standard and assessed the performance of RxNorm against these. First, the RDCRN has high regard for accepted terminology standards [15] and RxNorm was designed to meet these standards. (It should be noted, however, that these same requirements also constituted the criteria for standards selection by CHI.) Additionally, clinical research data standards should be compatible with clinical care data standards, to facilitate the re-use of clinical data for research purposes. The studies of the RDCRN are “closer” to the clinical care arena than epidemiological studies such as TEDDY, and the desire to adopt clinical data standards, such as RxNorm, that are likely to be incorporated into Electronic Medical Record Systems is particularly strong in the RDCRN project.
Because of the variety of studies involved in the RDCRN, there are different needs for a medication data standard across the component studies. The variety of study designs operating within the RDCRN dictated a single standard that could collect medication data at different levels of specificity (e.g., medication name only, vs. medication + dosage + form). Almost all studies in the RDCRN collect medication data in some form. For many observational studies collecting self-reported medication use, only the names of drugs are collected, as in the TEDDY project. Often this information is gathered by patient report, and can be in the form of generic name or brand name. For clinical trials and some observational studies, the medication data collected includes dosage and formulary information. A standard that represents the relationship between the different granularities of medication data collection across RDCRN studies was required. For later analysis and data mining activities, a desirable feature of a data standard would be one with explicit relationships between the different granularities of medication data (e.g., “Acetaminophen” is related to “Acetaminophen 100mg” is related to “Acetaminophen 100mg Extended Release Tablet”). The ideal representation for RDCRN medication data needs a structure that will allow unique codes for these varying levels of detail, yet capture the relationships between them. Therefore, the RDCRN can use different tables from the relational RxNorm model to capture drug information across studies, but relationships between the data of differing levels of specificity are explicit and can later be exploited.
Unlike TEDDY, all of the studies in the RDCRN capture data on electronic, online Case Report Forms (CRFs). Because of the scope of RDCRN studies, the space of drugs that could conceivably be collected over the RDCRN lifespan is impossible to predefine (unlike the TEDDY study, which has a nearly saturated codebook of medications used only in pregnant and mothering women and young children). The assortment of study designs in the RDCRN introduces variance in how data questions are structured. There are two types of medication “questions” on the CRFs for RDCRN studies. First, pre-defined medications of interest can be put on CRFs as the “questions” with answers related to the use, history dosage, etc. Because data using these medication names is collected on all study subjects, it is coded once in the study data repository for the entire life of the study (the research questions and forms should not change during the study). The second type of medication items are open-ended questions, meaning the “answer space” is where the entirety of RxNorm codes are needed, and is unknown at each administration of the CRF. The “answers” to open-ended type questions (e.g., “List concomitant medications”) are not static for the study or a subject, and cannot be predicted before the start of the study. The coding of answers from these open-ended questions (using any standard) would have to occur frequently throughout the study as new data are collected. The size, distributed nature, and diverse content of the network made this an unappealing decision for the PEC. The ideal strategy for collecting data for open-ended questions is to provide tools for research staff to access the RxNorm collection and code their own research data at the time of data entry.
Process for Collecting Medication Data in RDCRN Studies
Because of the vast number of medications conceivably used by all RDCRN subjects, the implementation of any medication standard for the RDCRN requires remote and instant access to the standard by all (distributed) research staff. Because of the number and turnover and distributed location of RDCRN researchers, a web-based application was ideal. Because of logistical and resource limitations to extensive training, the interface had to be intuitive. To ensure acceptance of the coding application, access had to be fast and on-demand, and searching for desired concepts should be easy.[18] The application for RxNorm coding needed to allow the PEC to control the level of specificity of the coding by study. (E.g., if the study only examines medication name, we do not want users to have any chance to select more detailed RxNorm clinical drug codes.)
The RDCRN uses electronic case report forms (CRFs). For questions structured ahead of time (meaning the medication of interest is “in the question”), the user does not need to be involved in coding. Questions inquiring about particular medications are easily placed on online CRFs, and coded into RxNorm by PEC at any time over the life of the study. The use of RxNorm is invisible to the user.
For (open-ended) questions that are not structured ahead of time (e.g., “List All Meds taken in past year”), picklists can not be generated because the potential answer space is infinite, and free-text data can be error-prone and have limited value for analysis.[14] In these cases, access to a complete terminology is ideal. Open-ended items require a way to browse RxNorm for relevant codes. Within these online CRFs, a tool is embedded to enable researchers to access a remote terminology host, which maintains RxNorm updates.[19]
Strategies can be used to subset RxNorm. Using a simple formula that examines keyword as a proportion of words in RxNorm term, calls to a terminology host return “certainty” ratings, which can be used to filter out lengthier terms, such as “Penicillin 250000 UNT Oral Tablet”, which is a simple strategy for presenting only RxNorm terms that reflect a desired level of specificity (e.g., “Penicillin”). The tool is flexible, and the PEC can allow users to search all of RxNorm, or can suppress codes that include dosage and form information when this level of specificity is not required by the protocol or where it can’t be universally obtained.
This tool allows distributed research staff to search the entire space of RxNorm, including generic and proprietary names. A wild card feature is also available. Although the implementation is too new for formal evaluation, we suspect that utility is enhanced because users can search for the codes they need, using the labels most familiar to them.[18]
DISCUSSION
The PEC acts as a representative for all TEDDY and RDCRN sites to request addition of new terms to the NLM’s RxNorm when needed, alleviating individual investigators of this burden. The PEC also developed customized tools for both projects. The motivation for all PEC-developed tools was the same: to facilitate the use of standards by enabling research staff to search most relevant views of the standard.
The TEDDY study had to deal with the issue of international standards, and lack of a global resource that links brand and generic names. Although RxNorm is solely a U.S. standard, at the ingredient level, most medications reported on the TEDDY study to date have been found in RxNorm. Unlike TEDDY, the broad scope of RDCRN studies make it impossible to pre-populate picklists or create manageable codebooks. Since many questions are open-ended, users require the ability to search all of RxNorm. The distinction between structured and open-ended questions can have important implications for the implementation of any data standard.
Typically, terminologies and data standards are evaluated on inherent structural features [15] and on their ability to capture concepts as needed by the domain.[20–25] This ability to capture required concepts at the required level of detail – referred to as coverage - is by definition application-specific and measured at a point in time. The dynamic nature of medicine and biomedical research create a need for processes to address new terminology gaps quickly. Responsive terminology updating and maintenance processes will ensure high content coverage of clinical terminologies, including RxNorm. While we do not provide a formal evaluation of RxNorm content or implementation within the TEDDY and RDCRN projects, we estimate high “coverage” due to the nature of our implementations. In the 4 year implementation experience we describe, the PEC received only a handful of requests for terms (implying that users were successfully finding codes for the thousands of data entries that we report). When the PEC did receive requests for RxNorm term names, we provide the user with either the correct RxNorm code or a temporary code for use while we negotiated with RxNorm. We assume that, as the data center for both the TEDDY and RDCRN projects, we would have explicitly heard of requests for missing terms, although it is possible that researchers would not report the medication or alert us to missing RxNorm terms. In the future, concept coverage and assessment of structural features of RxNorm will obviously be important to assess. Of equal importance will be the development of other evaluation criteria, including those to assess complexity and the resources required for accurate and reliable use of RxNorm, which will provide helpful metrics with which to evaluate the use of any controlled terminology in medical data capture systems.[26]
CONCLUSION
An organization could easily become overwhelmed by the selection and the implementation of data standards. A fruitful way to dissect the problem is to explore first the standard requirements, and then deal with structural and process requirements for implementation. Varying standards requirements for a large epidemiological study and a network of clinical research studies both led to the selection of RxNorm as an ideal standard. These same factors affected the design and utility of applications to deliver the standards to the user. Applications that are customized to the standard, the study, or the data collection work flow have the potential to increase use of data standards.
ACKNOWLEDGMENTS
The authors wish to thank Cristina McCarthy, Wendy McCleod, Lori Ballard, and Kim Hunt for their contributions. The authors also wish to thank Dr. Stuart Nelson of the NLM for his support.
This research is funded by the National Institutes of Health DK63790 and RR019259, the National Center for Research Resources (NCRR). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH. We also thank the NIH Office of Rare Diseases for its support of the RDCRN. The terminology server hosted by Apelon, Inc. (http://www.apelon.com).
The TEDDY study is funded by DK 63829, 63861, 63821, 63865, 63863, 63836 and 63790 and Contract No. HHSN267200700014C from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Allergy and Infectious Diseases (NIAID), National Institute of Child Health and Human Development (NICHD), National Institute of Environmental Health Sciences (NIEHS), Juvenile Diabetes Research Foundation (JDRF), and Centers for Disease Control and Prevention (CDC).
REFERENCES
- 1.NIH. National Institutes of Health; NIH Roadmap. Accelerating Medical Discovery to Improve Health. 2005 doi: 10.1053/j.gastro.2003.10.027. http://nihroadmap.nih.gov/ [DOI] [PubMed]
- 2.Rindfleisch TC, Brutlag BL. Directions for Clinical Research and Genomic Research into the Next Decade. Implications for Informatics. Journal of the American Medical Association. 1998;5:404–411. doi: 10.1136/jamia.1998.0050404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zerhouni EA. American Medical Informatics Association Annual Symposium. Washington, D.C: 2005. Keynote Presentation. [Google Scholar]
- 4.Richesson RL, Krischer JP. Data Standards in Clinical Research: Gaps, Overlaps, Challenges and Future Directions. Journal of the American Medical Informatics Association. 2007;14(6):687–696. doi: 10.1197/jamia.M2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CHI. Consolidated Health Informatics; CHI Executive Summaries. 2004 http://www.whitehouse.gov/omb/egov/documents/CHIExecSummaries.doc.
- 6.NIH. National Institutes of Health; Final NIH Statement on Sharing Research Data. 2003 February 26; 2003 http://grants.nih.gov/grants/guide/notice-files/NOT-OD-03-032.html.
- 7.NLM. RxNorm. 2005 http://www.nlm.nih.gov/research/umls/rxnorm/index.html.
- 8.Liu S, et al. RxNorm: Prescription for Electronic Drug Information Exchange. IEEE, IT Pro. 2005 September – October;:17–23. 2005. [Google Scholar]
- 9.Bouhaddou O, et al. Exchange of computable patient data between the Department of Veterans Affairs (VA) and the Department of Defense (DoD): terminology mediation strategy. J Am Med Inform Assoc. 2008;15(2):174–183. doi: 10.1197/jamia.M2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.PEC. TEDDY Project Website. Pediatrics Epidemiology Center; 2006. http://teddy.epi.usf.edu/ [Google Scholar]
- 11.Hagopian WA, et al. TEDDY – The Environmental Determinants of Diabetes in the Young – An Observational Clinical Trial. Annals of the New York Academy of Science. 2006;(1079):320–326. doi: 10.1196/annals.1375.049. Immunology of Diabetes IV: Progression in Understanding. [DOI] [PubMed] [Google Scholar]
- 12.NIH. NIH Establishes Rare Diseases Clinical Research Network, NCRR; Press Release. 2003 Editor.
- 13.Hampton T. Rare Disease Research Gets Boost. JAMA. 2006;295:2836–2838. doi: 10.1001/jama.295.24.2836. [DOI] [PubMed] [Google Scholar]
- 14.Levin MA, et al. Extraction and mapping of drug names from free text to a standardized nomenclature. AMIA Annu Symp Proc. 2007:438–442. [PMC free article] [PubMed] [Google Scholar]
- 15.Cimino JJ. Desiderata for Controlled Medical Vocabularies in the Twenty-First Century. Methods of Information in Medicine. 1998;37(4–5):394–403. [PMC free article] [PubMed] [Google Scholar]
- 16.Tuttle MS, et al. The Semantic Web as "Perfection Seeking:" A View From Drug Terminology. 2001
- 17.NLM. National Library of Medicine; RxNav. A Semantic Navigation Tool for Clinical Drugs. 2005 http://mor.nlm.nih.gov/download/rxnav/
- 18.Tuttle MS, et al. Navigating to Knowledge. Meth Inform Med. 1995;34:214–231. [PubMed] [Google Scholar]
- 19.Richesson RL, et al. Paper: A Web-based SNOMED CT Browser: Distributed and Real-time Use of SNOMED CT During the Clinical Research Process. Medinfo 2007 Congress; Brisbane: 2007. [PubMed] [Google Scholar]
- 20.Campbell JR, et al. Phase II evaluation of clinical coding schemes: completeness, taxonomy, mapping, definitions, and clarity. CPRI Work Group on Codes and Structures. J Am Med Inform Assoc. 1997;4(3):238–251. doi: 10.1136/jamia.1997.0040238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bakken S, et al. An evaluation of ICNP intervention axes as terminology model components. Proc AMIA Symp. 2000:42–46. [PMC free article] [PubMed] [Google Scholar]
- 22.Penz JF, et al. Evaluation of SNOMED coverage of Veterans Health Administration terms. Medinfo. 2004;11(Pt 1):540–544. [PubMed] [Google Scholar]
- 23.Elkin PL, et al. Evaluation of the Content Coverage of SNOMED CT: Ability of SNOMED Clinical Terms to Represent Clinical Problem Lists. Mayo Clinic Procedings. 2006;81(6):741–748. doi: 10.4065/81.6.741. [DOI] [PubMed] [Google Scholar]
- 24.Warren JJ, Wilson RP. American Medical Informatics Association Annual Symposium. Washington, D.C: 2006. Representing Cardiovascular Concepts in an Electronic Health Record Using SNOMED CT®. [PMC free article] [PubMed] [Google Scholar]
- 25.Richesson RL, Fung KW, Krischer JP. Heterogeneous but "standard" coding systems for adverse events: Issues in achieving interoperability between apples and oranges. Contemp Clin Trials. 2008;29(5):635–645. doi: 10.1016/j.cct.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richesson RL, Andrews JE, Krischer JP. Use of SNOMED CT to Represent Clinical Research Data: A Semantic Characterization of Data Items on Case Report Forms in Vasculitis Research. Journal of the American Medical Informatics Association. 2006;13:536–546. doi: 10.1197/jamia.M2093. [DOI] [PMC free article] [PubMed] [Google Scholar]