Abstract
The proteolytic activities of a disintegrin and metalloproteinase (ADAM); a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS), and matrix metalloproteinase (MMP) families play important roles in normal and multiple pathological conditions. These metalloproteases have potential roles in the degradation of the ECM and in the processing of bioactive molecules. In the present study, RNA was isolated from multiple normal fibroblast and metastatic melanoma cell lines, as well as the isogenic normal tissue and tumor samples, and the gene expression levels of six ADAMs, eight MMPs, and four ADAMTSs were analyzed by real-time PCR. This approach allowed for detected changes in mRNA expression of the individual metalloproteinase genes to be compared between normal and metastatic states and also between tissue and cultured cells. Increased gene expression of several ADAM and MMP family members (MMP1, MMP8, MMP15, and ADAM15) occurred in melanoma tissue and was replicated in tissue cultures. In general, the level of ADAM and MMP mRNA expression was several-fold higher in cultured cells compared with the isogenic tissue from which they were derived. Passage-dependent expression patterns were observed for MMP8 and MMP9 in in-house-derived metastatic melanoma cell lines. This reiterates earlier suggestions that experiments using cells that have been maintained in culture should be interpreted with great care.
Keywords: ADAM, MMP, tissue
INTRODUCTION
Melanoma, the malignancy of the melanocytes primarly found in the skin, is one of the most highly invasive and metastatic tumors. As a result of the incidence of melanoma doubling every 15 years,1 it is becoming an increasingly common malignancy. According to the World Health Organization, approximately 65,000 melanoma-related deaths occur per annum, contributing 0.1% to total global mortality.2
Despite many experimental and clinical efforts aimed at improving the criteria for melanoma diagnosis and treatment, the most reliable prognostic factor is still the tumor thickness.3 There is no accepted histopathological, molecular, or immunohistochemical marker that clearly defines subsets of melanoma. The production of melanoma metastases depends on the completion of a multistep process involving selection, migration, survival, and growth of a unique subpopulation of cells with very specific properties.4 Therefore, information regarding the changes in the genomic and expression level exhibited during melanoma tumorigenesis and progression could be used for classification purposes.5,6
Gene expression profiles provide a snapshot of cell functions and processes at the time of sample preparation. The analysis of expression patterns of numerous genes at once in tumor cells and the comparison of the expression profile obtained with healthy cells should provide insights concerning consistent changes in gene expression that are associated with tumor cellular function.7 Members of the adamalysin [a disintegrin and metalloproteinase (ADAM) and a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS)] and the matrixin [matrix metalloproteinase (MMP)] subfamilies of metalloendopeptidases have all been shown to take part in processes crucial to tumorigenesis and cancer progression, such as the remodeling of the ECM, processing bioactive molecules, and ectodomain shedding.8,9 Roughly half of the ADAMs possesses an active catalytic domain and shows a sheddase activity as well as affinity toward other substrates in the extracellular space, such as cell-adhesion molecules or components of the ECM and the plasma.10 Two ADAMs have been shown to possess in vitro proteolytic activity against ECM components: ADAM9 hydrolyzes fibronectin and gelatin,11 and ADAM10 has type IV collagenase activity.12
The ADAMTSs are extracellular, multidomain enzymes similar to the ADAM proteins. Their known functions include collagen processing, cleaving matrix proteoglycans, inhibition of angiogenesis, and modulating blood coagulation homeostasis.13,14 The members of the MMP family are collectively capable of digesting all known ECM macromolecules.15 Their role in angiogenesis, tumor growth, and metastasis has been investigated and described extensively.15
There are very few studies about collagenolytic metalloenzyme expression profiling in melanoma.16–18 The first aim of the current study, therefore, was to compare the expression of metalloproteinases with previously shown or presumed collagenolytic properties in melanoma tissue and cultured cells, as well as normal tissue and fibroblasts.
In addition, there are only a few reports in the literature describing the effect of the cell-culturing process on the expression pattern changes in metalloproteinases in any system. Cell lines derived from human cancers are the most widely used resource in laboratory-based cancer research.19 Although in vitro experiments can never reproduce the complexity of a whole organism, the simplicity of cell lines provides the ability to manipulate and analyze individual parameters specifically.20 Although most cell-based cancer research methods use cell lines, the vast majority of these cell lines was purchased from commercial sources and thus, has already undergone numerous cell divisions [usually passage 50 (P50)–P80 and above]. The long-term culturing, as well as the inherent genetic instability21 may contribute to phenotypic changes or subtle genomic modifications occurring during the culture process.22 Passage-dependent expression changes in cultured melanoma cells have not yet been examined. Thus, the second aim of the current study was to evaluate the effect of the culturing process on the temporal expression of metalloproteinases.
Quantitative real-time PCR (qPCR) was used to analyze the mRNA expression levels of eight ADAMs, four ADAMTSs, and seven MMPs in isogenic melanoma tissue, primary and passaged melanoma cells, normal tissue, and passaged fibroblasts. This experimental setup allowed for detected changes in expression of the individual metalloproteinase genes to be compared between tissue and cultured cells and also between the subsequent passages in culture.
MATERIALS AND METHODS
Cell Culture
All reagents were obtained from Invitrogen (Carlsbad, CA, USA) unless otherwise stated. The in-house fibroblast and melanoma cells were derived as follows (see also23,24). The metastatic melanoma and normal adjacent tissue samples were obtained from the Cooperative Human Tissue Network (CHTN; Bethesda, MD, USA). Necrotic areas, connective tissue, fatty tissue, and blood vessels were removed using sterile forceps and scissors. The samples were cut into 1 mm3-sized pieces and incubated in 100 U/mL collagenase type I in HBSS at 37°C for 1–2 h. Cells were collected by centrifugation at 1000 g for 8 min and resuspended in DMEM supplemented with 10% FBS and 2.5 μg/mL Amphotericin B (Fisher BioReagents, Fair Lawn, NJ, USA). After 24 h, 50% of the media was replaced. After another 24 h, media were fully replaced with DMEM supplemented with 2% FBS, 5 ng/mL EGF, 5 μg/mL insulin, 40 μg/mL bovine pituitary extract, 50 units/ml penicillin, and 0.05 mg/ml streptomycin sulfate.23 All cells were maintained at 37°C in a humidified incubator containing 5% CO2.
PCR
Nucleic acid concentrations were determined as described previously.24 Total RNA was isolated using TRIzol reagent with in-solution DNase treatment using the RNase-Free DNase set (Qiagen, Valencia, CA, USA), followed by a clean-up using the spin columns from the PureLink Micro-to-Midi Total RNA purification system. First-strand cDNA was reverse-transcribed using SuperScript III RT with random hexamers (Fisher BioReagents). The qPCR primers were manually designed or with the aid of PerlPrimer.25 The qPCR assays had been preoptimized with regard to MgCl2 and primer concentration, as well as annealing temperature. The reactions were carried out two times, each in triplicate on an ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA, USA) using Platinum SYBR Green qPCR SuperMix-UDG with ROX with 200 nM each forward and reverse primers and 2 μL cDNA in a final volume of 20 μL. The PCR products were identified based on their unique melting curve and confirmed by agarose gel electrophoresis. Absolute gene expression levels were determined by the aid of qPCR standards; “copy number” refers to the absolute number of copies/5 ng extracted total RNA.
Generation and Analysis of qPCR Standards
PCR products were formed using Tfi DNA polymerase as per the manufacturer's recommendations and subsequently purified by using the PureLink PCR purification kit. Standards were created by transforming chemically competent JM109 cells with the pGEM-T plasmid (Promega, Madison, WI, USA) carrying a single copy of each respective target PCR product. Plasmids were purified using the PureLink HQ mini plasmid DNA purification kit. The experimental samples were calibrated to the external standard curves generated by performing the qPCR reaction using a tenfold dilution series (102–107 copies/reaction). The amplification efficiency was close to 100% in the case of all primer pairs used. The difference in amplification efficiency between two reactions was never higher than 0.8%.
RESULTS AND DISCUSSION
Sample Distribution
A panel of 23 samples was analyzed. It included four fibroblast and eight melanoma cell lines and five normal and six melanoma tissue samples (Table 1). The normal tissue samples were all normal, adjacent samples to the melanoma tissue samples. Grouping the samples as such provides the opportunity to compare the normal samples with the melanoma samples, as well as the tissue samples with the cultured cells. As a result of the derivation of the in-house melanoma cells from individual patient samples, as well as aiming to only look at general trends, the sample pool was kept to a limited number. Samples are categorized based on pathology reports, as well as the presence or absence of two widely accepted melanoma markers, tyrosinase and MelanA (primers given in Table 2).
TABLE 1.
Samples used for expression profiling
| Abbreviation | Type | Origin |
|---|---|---|
| NAT56WF | f | CHTN, lung fibroblasts |
| NAT56UM | f | CHTN, lung fibroblasts |
| NAT58WM | f | CHTN, skeletal muscle fibroblasts |
| BJ | f | ATCC, foreskin fibroblasts |
| Mel58WM | m | CHTN |
| Mel56UM | m | CHTN |
| Mel93WF | m | CHTN |
| Mel80WM | m | CHTN |
| SK-2-MEL | m | ATCC, derived from skin metastasis |
| M14 clone#5 | m | a |
| WM-266-4 | m | ATCC, derived from skin metastasis |
| Hs 895.T | m | ATCC, derived from lung metastasis |
| tNAT56WF | NT | CHTN, lung tissue |
| tNAT56UM | NT | CHTN, lung tissue |
| tNAT58WM | NT | CHTN, skeletal muscle |
| tNAT46WM | NT | CHTN, skeletal muscle |
| tNT77WM | NT | CHTN, skeletal muscle |
| tMel58WM | MMT | CHTN, muscle metastasis |
| tMel56UM | MMT | CHTN, lung metastasis |
| tMel93WF | MMT | CHTN, unknown |
| tMel46WM | MMT | CHTN, unknown |
| tMel77WM | MMT | CHTN, muscle metastasis |
| tMel80WM | MMT | CHTN, lymph node metastasis |
WM, Wistar melanoma; f, fibroblast; m, melanoma; NT, normal tissue; MMT, metastatic melanoma tissue; ATCC, American Type Culture Collection (Manassas, VA, USA).
aObtained as a generous gift from Dr. Barbara M. Mueller (La Jolla Institute for Molecular Medicine, La Jolla, CA, USA).
TABLE 2.
Primers used in this study
| Target | Sequence |
|---|---|
| HTYR | F-TGACCCAATATGAATCTGGT |
| R-GGACTAGCAAATCCTTCCAG | |
| MelanA | F-CTCATTAAGGAAGGTGTCCTG |
| R-GGGTAACCATAGATGAAGTGAG | |
| ADAM08 | F-ATCATGGCAGGCAGCATT |
| R-CCAAAAAGCTCTCCAGGTAGG | |
| ADAM09 | F-GCGGGATTAATGTGTTTGGA |
| R-TGCTCCACAGGAACAATCTCT | |
| ADAM10 | F-CAGAACTATGGGTCTCATGTACCTC |
| R-CTCTGTTCCAGAATCATGTGG | |
| ADAM12 | F-GTCAAATGGCGGTTGAGAAA |
| R-AACACCATGGGAAATGGGTA | |
| ADAM15 | F-TCATTCTCTGGGCCTACGG |
| R-TCCATGTTCACACCTCCTGA | |
| ADAM17 | F-CAAAGGAAGCTGACCTGGTT |
| R-TATTTCCCTCCCTGGTCCTC | |
| ADAM28 | F-CCTTATTCTGTTGGCGTTGTTCA |
| R-CCAAAGTTGTGGCCCATTTC | |
| ADAM33 | F-ACCGAAACTTGAACCACACC |
| R-GTCCTGAGAAGCTGGTCCAC | |
| MMP1 | F-GACTTAGTCCAGAAATACCTG |
| R-CAAAGAATTCCTGCATTTGC | |
| MMP2 | F-GAATACCATCGAGACCATGC |
| R-AGCCAATGATCCTGTATGTG | |
| MMP3 | F-CTGTTGATTCTGCTGTTGAG |
| R-AAGTCTCCATGTTCTCTAACTG | |
| MMP8 | F-CCAGCAAGAACATTTCTTCC |
| R-CAGTTAAGCCATTTATTGCCTC | |
| MMP9 | F-GGTAAGGAGTACTCGACCTG |
| R-ACAAACTGTATCCTTGGTCC | |
| MMP13 | F-AAATTATGGAGGAGATGCCC |
| R-AACAAGTTGTAGCCTTTGGA | |
| MMP14 | F-TACCGACAAGATTGATGCTG |
| R-AACGGTAGTACTTGTTTCCAC | |
| MMP15 | F-CATGGAAACAACCTCTTCCT |
| R-CTTGAAGTTGTCAACGTCCT | |
| ADAMTS2 | F-TATGGAAAGTCCATGAGCCT |
| R-GATCGTGGTATTCATCGTGG | |
| ADAMTS3 | F-ATATGCAAAGTCCATCAGCC |
| R-GTTCAGAGTGGTTGAGATCAG | |
| ADAMTS13 | F-AACCTTCTACAGAGAATGTGAC |
| R-GGAGCCACATTAATGAAGAG | |
| ADAMTS14 | F-GGAGGAAATCACCAGAACTC |
| R-TCTTGTTAATGGGATGTAGGG |
HTYR, Human tyrosinase; F, forward; R, reverse.
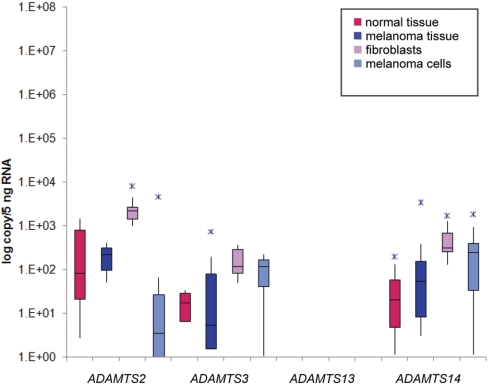
Expression of ADAMTS Family Members
Four ADAMTS genes were chosen for the present study (Table 2; Fig. 1). ADAMTS13 was included, as it was cloned originally from a metastatic melanoma cell line.26 This protease, however, proved to be undetectable on the mRNA level in all of the samples. All three procollagen N-propeptidases (ADAMTS2, ADAMTS3, and ADAMTS14) showed very low baseline expression. ADAMTS2 showed the highest overall expression in the studied samples; however, this was still on the order of only 103–104 copies/5 ng RNA. The fibroblast samples expressed the highest copy numbers for these targets. The expression of the ADAMTS genes in all other samples was between 0 and 102 copies/5 ng RNA.
FIGURE 1.
Expression of ADAMTS2, ADAMTS3, ADAMTS13, and ADAMTS14 in normal and melanoma tissue samples, fibroblasts, and melanoma cells.
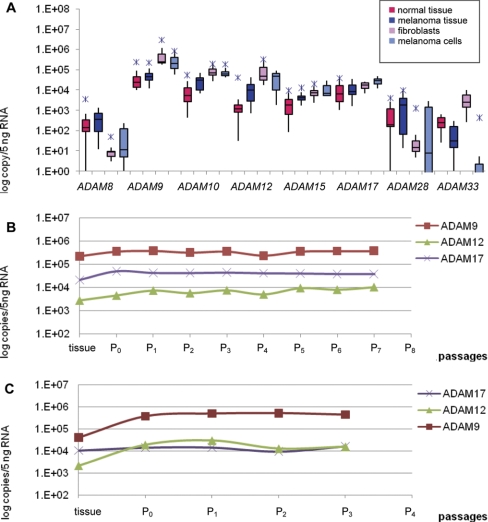
Expression of ADAM Family Members
Eight ADAM family members were examined herein (Table 2; Fig. 2A). ADAM8, ADAM28, and ADAM33 showed equally low baseline expression in melanoma and normal tissues, which all but disappeared in cultured cells. The one exception was ADAM33, where expression was increased over tenfold in fibroblasts. ADAM33 causes rapid induction of endothelial cell differentiation in vitro and angiogenesis ex vivo and in vivo.27 This proliferative property might be one induced based on microenvironmental effects (see below).
FIGURE 2.
ADAM expression profiling. (A) Expression of ADAM8, ADAM9, ADAM10, ADAM12, ADAM15, ADAM17, ADAM28, and ADAM33 in normal and melanoma tissue samples, fibroblasts, and melanoma cells. (B) ADAM9, ADAM12, and ADAM17 expression in Mel80WM cells as a function of passage number. (C) ADAM9, ADAM12, and ADAM17 expression in Mel65WM cells as a function of passage number. sd are small so that error bars are not visible when the y axis is logarithmic.
Three of the ADAMs (ADAM10, ADAM12, and ADAM15) showed more than a twofold increase in melanoma tissue compared with the normal tissue samples. ADAM10 was decreased slightly in the cultured melanoma cells compared with fibroblasts. This ADAM was over tenfold increased in the cell lines compared with the tissue samples. ADAM10 is involved in the shedding of many membrane-bound proteins, such as TNF-α, ErbB ligands betacellulin (BTC), and EGF, Notch, and E-cadherin.28–30 Consequently, changes in ADAM10 catalytic activity are implicated in carcinogenesis. Excess ADAM10 activity may promote cell growth in cancer proliferation assays, because of enhanced production of soluble, mature forms of BTC and EGF.31
ADAM12 was threefold decreased in the cultured melanoma cells compared with fibroblasts. However, this ADAM was approximately tenfold increased in the cell lines compared with the tissue samples, especially in the fibroblasts. ADAM12 functions as a sheddase, adhesion molecule, and ECM-degrading proteinase and is involved in cancer progression. Its cysteine-rich domain is known to support tumor cell adhesion through syndecans, which trigger signaling events and lead to β1 integrin-dependent cell spreading.32,33 ADAM12 also cleaves various ECM molecules including gelatin, type IV collagen, and fibronectin, suggesting a potential role for this enzyme in ECM digestion in cancer invasion and metastasis.34 ADAM12 is also up-regulated in cancers of stomach, liver, and colon, and ADAM12 levels in urine correlate with breast cancer progression.34
ADAM9 showed the highest overall expression in all sample types, with slightly higher copy numbers in the normal tissue samples and the fibroblasts. It also showed the greatest increase in cultured cells compared with the tissue samples from which they were derived. ADAM15 was more than twofold increased in melanoma tissue, as well as in the melanoma cells, compared with the normal tissue samples and fibroblasts. In a previous study, the expression of ADAM9 was analyzed in melanoma in vivo and in melanoma cell lines in vitro.17 The protein expression appeared to be restricted to the melanoma cells within the invading front. mRNA analysis showed ADAM9 expression in varying amounts in all cell lines, independent of their invasive and metastatic properties. ADAM9 is up-regulated in breast, pancreatic, and stomach cancers.35 It has been suggested that ADAM9 plays a role in tumorigenesis, invasion, and metastasis through modulation of growth factor activity and integrin function.
ADAM15 was increased fivefold in the cultured cells compared with the tissue samples. The data about the role of ADAM15 in cancers are inconsistent. Expression of ADAM15 is up-regulated in various cancers of the breast, prostate, stomach, and lung, and treatment of carcinoma cell lines with anti-ADAM15 antibodies reduces cell proliferation.36,37 However, the recombinant disintegrin domain of human ADAM15 is reported to be a potent intrinsic inhibitor of angiogenesis, tumor growth, and metastasis.38
ADAM17 showed the same expression for normal and melanoma tissues. It was increased, however, in melanoma cells compared with the melanoma tissue and the fibroblasts. ADAM17 is overexpressed in cancers of the breast, ovary, kidney, colon, and prostate.35 It was identified first as the proteinase responsible for the shedding of the important proinflammatory cytokine, TNF-α,39 hence earning the alternative name TACE. ADAM17 was also demonstrated to be the TGF-α sheddase and has since been reported to shed several additional, important cell surface proteins, including the EGFR ligands amphiregulin and heparin-binding (HB)-EGF, l-selectin, TNFRII, collagen XVII, and growth hormone receptor.40 The receptor tyrosine kinase EGFR and its downstream signaling pathway are key regulators of cell proliferation, and it is deregulated frequently in cancer. EGFR can be activated by several ligands processed by ADAM17, including amphiregulin, epigen, epiregulin, HB-EGF, and TGF-α, as well by substrates of ADAM10, including EGF and BTC.41,42 Overexpression of these ligands is a common event in many tumors and correlates frequently with poor prognosis.43
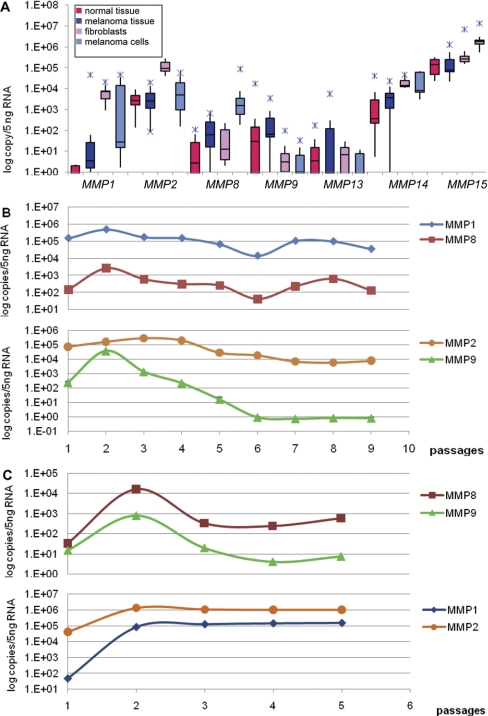
Expression of MMP Family Members
MMP8, MMP9, and MMP13 showed undetectable to very low expression (102 copies/5 ng DNA) in all samples studied (Fig. 3A). One exception was MMP8, which was clearly up-regulated in the cultured melanoma cells; it showed tenfold up-regulation compared with the melanoma tissue and 50- to 100-fold up-regulation compared with the fibroblasts. MMP8 is expressed in neural crest and adult melanoma cells but not in melanocytes.44 Several DNA microarray analyses demonstrated up-regulation of MMP8 in melanoma.18 WT MMP8 inhibits melanoma growth in vitro and tumor formation in vivo,45 indicating an inhibitory effect of MMP8 on tumorigenesis and metastasis. However, MMP8 is often mutated in melanoma, and the mutant MMP8 proteins have reduced enzyme activity and inhibitory effects on tumorigenesis.45 The low expression of MMP13 observed here was not surprising, as MMP13 induction in tumors occurs in the stroma rather than the tumor cells themselves.46
FIGURE 3.
MMP expression profiling. (A) Expression of MMP1, MMP2, MMP8, MMP9, MMP13, MMP14, and MMP15 in normal and melanoma tissue samples, fibroblasts, and melanoma cells. (B) MMP1, MMP2, MMP8, and MMP9 expression in Mel80WM cells. (C) MMP1, MMP2, MMP8, and MMP9 expression in Mel65WM cells. sd are small so that error bars are not visible when the y axis is logarithmic.
MMP9 showed an overall low baseline expression in the tissue samples, highly variable copy numbers (0–104 copies/5 ng RNA) in the normal tissue types, and 103–102 copies/5 ng RNA in the melanoma samples. MMP9 was undetectable or showed very low expression in cultured cells. MMP9 production is not associated with the clinical outcome of melanoma.47
MMP1 mRNA was mostly undetectable in normal tissue samples, and the melanoma samples had very low baseline expression (10–100 copies/5 ng RNA; Fig. 3A). In cultured cells, however, MMP1 was up-regulated significantly. The expression levels of the mRNA were between 103 and 104 copies/5 ng RNA in the case of the fibroblasts and highly variable (102–3×104 copies/5 ng RNA) in the case of the melanoma cells. Clinical studies have shown the expression of MMP1 to be positively correlated with the malignancy and poor outcome of melanoma.48 MMP1 is produced by invasive but not noninvasive melanomas.49 There is evidence suggesting that MMP1 produced from melanoma cells plays a role in tumor progression by degrading matrix proteins and generating active growth factors such as TGF-β in vivo.49,50 MMP1 has been shown to be up-regulated in fibroblasts that are cocultured in fibrillar collagen with melanoma cells.16
MMP2 showed high basal expression (103–104 copies/5 ng RNA) in both tissue types, as well as the melanoma cells (Fig. 3A), presumably indicating constitutive expression levels. In fibroblasts, however, MMP2 was increased 100-fold compared with melanoma cells. MMP2 is believed to play an important role in skin and uveal melanoma progression. A significant correlation was found between MMP2 expression and clinical outcome in sinonasal and oral malignant melanoma. Greater overall survival was seen in patients with low MMP2 expression.51 The MMP2 gene is expressed constitutively in many cells; therefore, MMP2 is seen in many tumors and at several stages of tumor progression.52–56 Overexpression of MMP2 is associated with tumor progression in human melanoma, and its expression is even more pronounced in invasive melanomas.47,57–60 MMP2 was shown to be present by immunohistochemistry in malignant melanoma lesions but not in benign and atypical nevi.61 The overexpression of MMP2 has also been linked to hematogenous metastasis and to impaired survival in male melanoma patients, suggesting a difference according to sex.58 MMP2 shows multiple functions as an extracellular/cell surface enzyme and is broadly recognized for its matrix-degrading ability and involvement in cell motility.62 A great variety of nonmatrix MMP2 physiological targets has also been identified, including bFGF, FGFR1, and the CX3CL1 chemokine fractalkine.63–65
MMP14 showed equal baseline expression in both tissue types, 102–104 copies/5 ng RNA (Fig. 3A), again presumably showing constitutive expression levels. In cultured cells, MMP14 expression was increased eight- to tenfold in fibroblasts and melanoma cells. MMP14/membrane type 1 (MT1)-MMP is usually up-regulated in carcinoma cells.66 Expression levels of MT1-MMP correlate with the malignant nature of cells. Weakly tumorigenic and poorly invasive human breast carcinoma cell lines do not express MT1-MMP or MMP2, but invasive and metastatic ones do.67 A similar correlation was reported in human cervical cancer cell lines.68 Greater overall survival was seen in patients with low MT1-MMP in sinonasal and oral malignant melanomas.51 However, fibroblast-derived MT1-MMP promotes the invasion and growth of head and neck squamous cell carcinomas.69 MT1-MMP specifically cleaves native type I and type III collagens70 and a variety of cell surface-bound biomolecules71 and activates pro-MMP2 and pro-MMP13.72,73 As MMP2 has type IV collagenase activity, and stromal MMP13 is important in melanoma tumor growth and organ-specific metastasis,46 MT1-MMP is believed to be one of the key enzymes for invasion of the basement membrane in the metastatic process, as well as in migration, as it confers invasive activity to the cells.
MMP15 showed the highest overall expression in all of the samples used in this study (105–107 copies/5 ng RNA; Fig. 3A). This gene was increased threefold in melanoma tissue compared with the normal tissue types and increased fivefold in melanoma cells compared with the fibroblasts. When comparing MMP15 expression in the tissue samples and the cultured cells, this gene was increased tenfold in culture. Elevated expression of MMP15/MT2-MMP has been reported in glioblastomas, and it has also been reported to correlate with invasiveness.74 Ovarian,75 urothelial,76 and breast77,78 carcinomas also showed elevated levels of MT2-MMP. In a previous study, the expression of MT2-MMP was not detected in benign lesions, and in melanoma cells, MT2-MMP was moderately or strongly detected.79 It was also found to be colocalized with MMP2.79 The function of MT2-MMP is not as well described as that of MT1-MMP and is just beginning to be addressed. Studies using a cell line derived from a MT1-MMP knockout mouse have shown the important contribution of MT2-MMP to cell invasion of fibrin matrices.80 MT2-MMP shows activity against laminin.70 MT2-MMP also activates MMP2 to the fully active form in a pathway that is TIMP-2-independent.81
Taken together, the data presented in the current study clearly demonstrate that increased gene expression of several ADAM and MMP protein family members (such as MMP1, MMP8, MMP15, or ADAM15) has occurred in melanoma tissue and was replicated in tissue cultures. The variable expression of MMP1, MMP2, and ADAM8 in the cultured melanoma cells highlights the heterogeneity of the metastatic melanoma samples, presumably indicating the presence of the different subtypes of melanoma in our sample pool. The expression of metalloproteinases can be regulated by microenvironmental effects,16,17,59,82,83 which may be recapitulated by the tumor tissue samples.
We found an overall tendency for several metalloproteinase targets to be as much as 100-fold up-regulated in culture, in fibroblasts, and in cultured melanoma cells. When establishing the in-house-derived cell lines, the tumor tissue is digested, and the cells are essentially set free of the extracellular structures. This may parallel the metastatic process by which tumor cells are breaking away from the primary tumor and forming the micrometastases or metastatic tumors. The success of the formation of the metastatic tumor depends on the survival of these cells. Metastasizing cells need to be able to invade the local tissue, break through the basal lamina, invade the capillary system, survive the harsh conditions of the circulation, extravasate, and proliferate at the distant site. Studies in animals show that typically far less than one in every 1000 malignant tumor cells that enter the bloodstream will survive to produce a tumor at a new site.84 To be able to potentially do all of this while adapting to the rapidly changing environment, elevated expression levels of numerous genes key in the metastatic process may be required. The primary cells in cell culture have to adapt to the drastically different environment of the tissue culture dish: attach and proliferate. Once these cells are established and capable of surviving the culturing and the passaging process, high copy numbers are no longer needed to produce the presumably elevated protein levels. This may reflect the decline in the gene expression levels of several MMPs from the first passage onward.
Alternatively, our study may simply reflect the artificial effects of cell culturing on metalloproteinase gene expression. In the culturing process, cells are transferred from a physiological tissue context to a monolayer culture. Structures, such as the ECM, are no longer present, leading to changes in the patterns of engagement of ligand–receptor interactions, which may modulate cell behaviors. In the tumor tissue, the stromal content might be as high as 90% (in the case of desmoplastic tumors, such as many carcinomas of the breast, stomach, and pancreas),85 and tumor–stroma interactions have been well documented to play a significant role in tumor development and progression.86 In cell culture, however, the population is homogenous; therefore, these heterotypic cell–cell interactions are not present. Lastly, some of these changes in expression patterns may reflect cells adapting to harshly different, new conditions. Environmental changes, such as pH or temperature imbalances, might impact changes in gene expression, morphology, and cell behavior. This reiterates the suggestion that experiments using cells that have been maintained in culture should be interpreted with great care.
Temporal Gene Expression of Metalloproteinases during Cell Culturing
Two in-house-derived metastatic melanoma cell lines (Mel80WM and Mel65WM) were used to investigate the effect of the culturing process on metalloproteinase gene expression compared with isogenic tissue. Mel80WM cells were used up until P7, and Mel65WM cells were investigated from P0 to P3.
The ADAM family members, ADAM9, ADAM12, and ADAM17, as well as four collagenolytic MMPs (MMP1, MMP2, MMP8, and MMP9) were targeted. For both cell lines, the three ADAMs showed less than one logarithmic unit difference between the highest and the lowest observed value (Fig. 2B and C). When comparing these results with those observed previously (Fig. 2A), ADAM12 and ADAM17 are consistent in that they are up-regulated in culture less than tenfold. ADAM9, however, showed more significant up-regulation previously in culture compared with the melanoma tissue samples (Fig. 2A) than what was observed in the Mel80WM or the Mel65WM cells.
MMP1, MMP2, MMP8, and MMP9 showed significant up-regulation in the early passages in both melanoma cell lines (Fig. 3B and C). MMP1 showed a spike in both cell lines; however, this was not as significant in the Mel80WM cells (3.3-fold) as in the Mel65WM cells (32-fold up-regulated). Although MMP1 continued to show varied expression levels in the Mel80WM cells, it stayed stable in the Mel65WM cells. In the MMP summaries (Fig. 3A), MMP1 showed variable expression (between 101 and 104 copies of mRNA) in the melanoma cells. One reason for this earlier result could have been the variable expression of the MMP1 gene as observed in Mel80WM. Depending on at what passage the examined cell line is, the copy number was variable compared with the tissue sample.
MMP2 showed slow, passage-wise up-regulation in Mel80WM culture, peaked at fivefold higher at P3, and then slowly down-regulated to over tenfold less than in tissue (Fig. 3B). In contrast, the expression of MMP2 was up-regulated 1000-fold at P2 in the Mel65WM cells (Fig. 3C), and it remained at that level for all subsequent passages. This MMP also showed variable expression in the summarized expressions (500–50,000 copies/5 ng RNA depending on the cell line; Fig. 3A).
The gene expression pattern of MMP8 mirrored that of MMP1 in the Mel80WM cells, showing variable copy numbers throughout the passages (Fig. 3B). In Mel65WM, the MMP8 copy numbers were at least tenfold higher than in the isogenic tissue sample (Fig. 3C). This MMP was up-regulated in the expression summaries (ten- to 100-fold depending on the cell line; Fig. 3A), which proved to be somewhat consistent with the passage-dependent observations.
MMP9 was consistently 100-fold up-regulated in both melanoma cell lines compared with the isogenic tissue samples (Fig. 3B and C). In the subsequent passages, the expression of MMP9 started decreasing steadily and disappeared by P4 (for Mel65WM) or P6 (for Mel80WM). The expression of MMP9 was also down-regulated in the summarized expression patterns (Fig. 3A).
It has been noted previously that long-term culturing of cancer cells may lead to phenotypic and genotypic changes.22 Commercially available, established cell lines are usually purchased at P50–P80 and above. When pairing the high number of doublings with the inherent genetic instability of immortal or transformed cells,21 the phenotypic and genotypic changes observed are not surprising. Qualitative and quantitative changes in MMP expression patterns were demonstrated previously in primary and passaged cultures of rabbit corneal fibroblasts.87 MMP9 secretion in HUVECs was shown to be passage-dependent.88 This secretion was restricted to very early passage cells, decreased rapidly as the cells were maintained in culture, and was not detected by the second passage.88 The present study, however, emphasizes the significance of radical changes in gene expression in primary cells and at the earliest passages.
SUMMARY
Despite the complexity of metalloproteinase regulation, five major levels of endogenous control can be recognized: gene transcription, mRNA stability, translation, proenzyme activation, and inhibition of enzymatic activity.89–92 Collectively, these mechanisms should confine degradative activity to those sites and situations where it is biologically necessary. However, tumor cells have developed multiple strategies to escape the checkpoints controlling metalloenzyme mRNA expression and proteolytic activity, acquiring new properties that lead to tumor growth and invasion. The present study has found a number of metalloproteinases whose expression differs in melanoma compared with normal tissue and cell lines.
Multiple, coordinated gene expression involving metalloproteinases may be critical for initiation of metastasis. For example, breast cancer metastasis to the lung uses epiregulin (a ligand for EGFR), COX2, MMP1, and MMP2.93 Also, the enzyme/inhibitor balance has been shown to favor the enzyme in breast cancer tumor tissue compared with adjacent, nontumor tissue,94 and thus, consideration of the levels of natural metalloproteinase inhibitors (such as TIMPs and reversion-inducing, cysteine-rich protein with Kazal motifs) is important. We have presently identified several metalloproteinases up-regulated in melanoma tissue, which can contribute to ECM degradation and facilitate tumor progression.
ACKNOWLEDGMENTS
The authors gratefully acknowledge support of this work by the National Institutes of Health (CA98799), the Robert A. Welch Foundation, the Texas Higher Education STAR Program (all to G.B.F.), and the National Institutes of Health, National Institute of Dental and Craniofacial Research (DE14318) COSTAR Program (to J.L.L.).
REFERENCES
- 1.Morton DL, Essner R, Kirkwood JM, Wollman RC. Malignant melanoma. In Kufe DW, Pollock RE, Weichselbaum RR, Bast RC, Gansler TS. (eds): Cancer Medicine, 6th ed Philadelpha, PA, USA: B.C. Decker, 2003 [Google Scholar]
- 2.Lucas R, McMichael T, Smith W, Armstrong B. Solar ultraviolet radiation: global burden of disease from solar ultraviolet radiation. World Health Organization: Environmental Burden of Disease Series, 2006 [Google Scholar]
- 3.Berwick M, Halpern A. Melanoma epidemiology. Curr Opin Oncol 1997;9:178–182 [DOI] [PubMed] [Google Scholar]
- 4.Clark WH, Elder DE, Jr., Guerry D, Epstein MN, Greene MH, van Horn M. A study of tumor progression: the precursor lesions of superficial spreading and nodular melanoma. Hum Pathol 1984;15:1147–1165 [DOI] [PubMed] [Google Scholar]
- 5.Bar-Eli M. Molecular mechanisms of melanoma metastasis. J Cell Physiol 1997;173:275–278 [DOI] [PubMed] [Google Scholar]
- 6.Welch DR, Goldberg SF. Molecular mechanisms controlling human melanoma progression and metastasis. Pathobiology 1997;65:311–330 [DOI] [PubMed] [Google Scholar]
- 7.Macgregor PF, Squire JA. Application of microarrays to the analysis of gene expression in cancer. Clin Chem 2002;48:1170–1177 [PubMed] [Google Scholar]
- 8.Rocks N, Paulissen G, El Hour M, et al. Emerging roles of ADAM and ADAMTS metalloproteinases in cancer. Biochimie 2008;90:369–379 [DOI] [PubMed] [Google Scholar]
- 9.Fridman R. Metalloproteinases and cancer. Cancer Metastasis Rev 2006;25:7–8 [DOI] [PubMed] [Google Scholar]
- 10.White JM. ADAMs: modulators of cell-cell and cell-matrix interactions. Curr Opin Cell Biol 2003;15:598–606 [DOI] [PubMed] [Google Scholar]
- 11.Schwettmann L, Tschesche H. Cloning and expression in Pichia pastoris of metalloprotease domain of ADAM 9 catalytically active against fibronectin. Protein Exp Purif 2001;21:65–70 [DOI] [PubMed] [Google Scholar]
- 12.Millichip MI, Dallas DJ, Wu E, Dale S, McKie N. The metallo-disintegrin ADAM10 (MADM) from bovine kidney has type IV collagenase activity in vitro. Biochem Biophys Res Commun 1998;245:594–598 [DOI] [PubMed] [Google Scholar]
- 13.Porter S, Clark IM, Kevorkian L, Edwards DR. The ADAMTS metalloproteinases. Biochem J 2005;386:15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Apte SS. A disintegrin-like and metalloprotease (reprolysin-type) with thrombospondin type 1 motif (ADAMTS) superfamily: functions and mechanisms. J Biol Chem 2009;284:31493–31497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roy R, Zhang B, Moses MA. Making the cut: protease-mediated regulation of angiogenesis. Exp Cell Res 2006;312:608–622 [DOI] [PubMed] [Google Scholar]
- 16.Gallagher PG, Bao Y, Prorock A, et al. Gene expression profiling reveals cross-talk between melanoma and fibroblasts: implications for host-tumor interactions in metastasis. Cancer Res 2005;65:4134–4146 [DOI] [PubMed] [Google Scholar]
- 17.Zigrino P, Mauch C, Fox JW, Nischt R. ADAM-9 expression and regulation in human skin melanoma and melanoma cell lines. Int J Cancer 2005;116:853–859 [DOI] [PubMed] [Google Scholar]
- 18.Hoek KS. DNA microarray analyses of melanoma gene expression: a decade in the mines. Pigment Cell Res 2007;20:466–484 [DOI] [PubMed] [Google Scholar]
- 19.Masters JRW, Palson B. Cancer Cell Lines Part Ied. Dorchedt, The Netherlands: Kluwer Academic Publishers, 1999 [Google Scholar]
- 20.Tuschl G, Mueller SO. Effects of cell culture conditions on primary rat hepatocytes—cell morphology and differential gene expression. Toxicology 2006;218:205–215 [DOI] [PubMed] [Google Scholar]
- 21.Masramon L, Vendrell E, Tarafa G, et al. Genetic instability and divergence of clonal populations in colon cancer cells in vitro. J Cell Sci 2006;119:1477–1182 [DOI] [PubMed] [Google Scholar]
- 22.Van den Oord JJ, Sarasin A, Winnepenninckx V, Spatz A. Expression profiling of melanoma cell lines: in search of a progression-related molecular signature. Future Oncol 2007;3:609–611 [DOI] [PubMed] [Google Scholar]
- 23.Hsu M-Y, Elder DE, Herlyn M. Melanoma: the Wistar melanoma (WM) cell lines. In Masters JRW, Palsson B. (eds): Human Cell Culture, vol I, Cancer Cell Lines Part 1 Dordrecht, The Netherlands: Kluwer Academic Publishers, 1999;259–274 [Google Scholar]
- 24.Giricz O, Lauer-Fields JL, Fields GB. The normalization of gene expression data in melanoma: investigating the use of glyceraldehyde 3-phosphate dehydrogenase and 18S ribosomal RNA as internal reference genes for quantitative real-time PCR. Anal Biochem 2008;380:137–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marshall OJ. PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics 2004;20:2471–2472 [DOI] [PubMed] [Google Scholar]
- 26.Cal S, Obaya AJ, Llamazares M, Garabaya C, Quesada V, López-Otín C. Cloning, expression analysis, and structural characterization of seven novel human ADAMTSs, a family of metalloproteinases with disintegrin and thrombospondin-1 domains. Gene 2002;283:49–62 [DOI] [PubMed] [Google Scholar]
- 27.Puxeddu I, Pang YY, Harvey A, et al. The soluble form of a disintegrin and metalloprotease 33 promotes angiogenesis: implications for airway remodeling in asthma. J Allergy Clin Immunol 2008;121:1400–1406 [DOI] [PubMed] [Google Scholar]
- 28.Rosendahl MS, Ko SC, Long DL, et al. Identification and characterization of a pro-tumor necrosis factor-α-processing enzyme from the ADAM family of zinc metalloproteases. J Biol Chem 1997;272:24588–24593 [DOI] [PubMed] [Google Scholar]
- 29.Lunn CA, Fan X, Dalie B, et al. Purification of ADAM 10 from bovine spleen as a TNFα convertase. FEBS Lett 1997;400:333–335 [DOI] [PubMed] [Google Scholar]
- 30.Sanderson MP, Erickson SN, Gough PJ, et al. ADAM10 mediates ectodomain shedding of the betacellulin precursor activated by p-aminophenylmercuric acetate and extracellular calcium influx. J Biol Chem 2005;280:1826–1837 [DOI] [PubMed] [Google Scholar]
- 31.Moss ML, Bomar M, Liu Q, et al. The ADAM10 prodomain is a specific inhibitor of ADAM10 proteolytic activity and inhibits cellular shedding events. J Biol Chem 2007;282:35712–35721 [DOI] [PubMed] [Google Scholar]
- 32.Iba K, Albrechtsen R, Gilpin BJ, Loechel F, Wewer UM. Cysteine-rich domain of human ADAM 12 (Meltrin α) supports tumor cell adhesion. Am J Pathol 1999;154:1489–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iba K, Albrechtsen R, Gilpin B, et al. The cysteine-rich domain of human ADAM 12 supports cell adhesion through syndecans and triggers signaling events that lead to β1 integrin-dependent cell spreading. J Cell Biol 2000;149:1143–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy R, Wewer UM, Zurakowski D, Pories SE, Moses MA. ADAM12 cleaves extracellular matrix proteins and correlates with cancer status and stage. J Biol Chem 2004;279:51323–51330 [DOI] [PubMed] [Google Scholar]
- 35.Mochizuki S, Okada Y. ADAMs in cancer cell proliferation and progression. Cancer Sci 2007;98:621–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lendeckel U, Kohl J, Arndt M, Carl-McGrath S, Donat H, Röcken C. Increased expression of ADAM family members in human breast cancer and breast cancer cell lines. J Cancer Res Clin Oncol 2004;131:41–48 [DOI] [PubMed] [Google Scholar]
- 37.Carl-McGrath S, Lendeckel U, Ebert M, Roessner A, Röcken C. The disintegrin-metalloproteinases ADAM9, ADAM12, and ADAM15 are upregulated in gastric cancer. Int J Oncol 2005;26:17–24 [PubMed] [Google Scholar]
- 38.Trochon-Joseph V, Martel-Renoir D, Mir LM, et al. Evidence of antiangiogenic and antimetastatic activities of the recombinant disintegrin domain of metargidin. Cancer Res 2004;64:2062–2069 [DOI] [PubMed] [Google Scholar]
- 39.Black RA, Rauch CT, Kozlosky, et al. A metalloproteinase disintegrin that releases tumor-necrosis factor-α from cells. Nature 1997;385:729–733 [DOI] [PubMed] [Google Scholar]
- 40.Kenny PA. TACE: a new target in epidermal growth factor receptor dependent tumors. Differentiation 2007;75:800–808 [DOI] [PubMed] [Google Scholar]
- 41.Holbro T, Hynes NE. ErbB receptors: directing key signaling networks throughout life. Annu Rev Pharmacol Toxicol 2004;44:195–217 [DOI] [PubMed] [Google Scholar]
- 42.Sahin U, Weskamp G, Kelly K, et al. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J Cell Biol 2004;164:769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicholson RI, Gee JMW, Harper ME. EGFR and cancer prognosis. Eur J Cancer 2001;37:9–15 [DOI] [PubMed] [Google Scholar]
- 44.Giambernardi TA, Sakaguchi AY, Gluhak J, et al. Neutrophil collagenase (MMP-8) is expressed during early development in neural crest cells as well as in adult melanoma cells. Matrix Biol 2001;20:577–587 [DOI] [PubMed] [Google Scholar]
- 45.Palavalli LH, Prickett TD, Wunderlich JR, et al. Analysis of the matrix metalloproteinase family reveals that MMP8 is often mutated in melanoma. Nat Genetics 2009;241:518–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zigrino P, Kuhn I, Bäuerle T, et al. Stromal expression of MMP-13 is required for melanoma invasion and metastasis. J Invest Dermatol 2009;129:2686–2693 [DOI] [PubMed] [Google Scholar]
- 47.Väisänen AH, Kallioinen M, Turpeenniemi-Hujanen T. Comparison of the prognostic value of matrix metalloproteinases 2 and 9 in cutaneous melanoma. Hum Pathol 2008;29:377–385 [DOI] [PubMed] [Google Scholar]
- 48.Nikkola J, Vihinen P, Vlaykova T, Hahka-Kemppinen M, Kähäri V-M, Pyrhönen S. High expression levels of collagenase-1 and stromelysin-1 correlate with shorter disease-free survival in human metastatic melanoma. Int J Cancer 2002;97:432–438 [DOI] [PubMed] [Google Scholar]
- 49.Blackburn JS, Rhodes CH, Coon CI, Brinckerhoff CE. RNA interference inhibition by matrix metalloproteinase-1 prevents melanoma metastasis by reducing tumor collagenase activity and angiogenesis. Cancer Res 2007;67:10849–10858 [DOI] [PubMed] [Google Scholar]
- 50.Iida J, McCarthy JB. Expression of collagenase-1 (MMP-1) promotes melanoma growth through the generation of active transforming growth factor-β. Melanoma Res 2007;17:205–213 [DOI] [PubMed] [Google Scholar]
- 51.Kondratiev S, Gnepp DR, Yakirevich E, et al. Expression and prognostic role of MMP2, MMP9, MMP13, and MMP14 matrix metalloproteinases in sinonasal and oral malignant melanomas. Human Pathol 2008;39:337–343 [DOI] [PubMed] [Google Scholar]
- 52.Benbow U, Brinckerhoff CE. The AP-1 site and MMP gene regulation: what is all the fuss about? Matrix Biol 1997;15:519–526 [DOI] [PubMed] [Google Scholar]
- 53.Borden P, Heller RA. Transcriptional control of matrix metalloproteinases and the tissue inhibitors of matrix metalloproteinases. Crit Rev Eukaryot Gene Expr 1997;7:159–178 [DOI] [PubMed] [Google Scholar]
- 54.Westermarck J, Kahari V-M. Regulation of matrix metalloproteinase expression in tumor invasion. FASEB J 1999;13:781–792 [PubMed] [Google Scholar]
- 55.Koblinski JE, Ahram M, Sloane BF. Unraveling the role of proteases in cancer. Clin Chim Acta 2000;291:113–135 [DOI] [PubMed] [Google Scholar]
- 56.Nelson AR, Fingleton B, Rothenberg ML, Matrisian LM. Matrix metalloproteinases: biologic activity and clinical implications. J Clin Oncol 2000;18:1135–1149 [DOI] [PubMed] [Google Scholar]
- 57.Väisänen A, Tuominen H, Kallioinen M, Turpeenniemi-Hujanen T. Matrix metalloproteinase-2 (72 kDa type IV collagenase) expression occurs in the early stage of human melanocytic tumor progression and may have prognostic value. J Pathol 1996:180:283–289 [DOI] [PubMed] [Google Scholar]
- 58.Väisänen A, Kallioinen M, Taskinen PJ, Turpeenniemi-Hujanen T. Prognostic value of MMP-2 immunoreactive protein (72 kD type IV collagenase) in primary skin melanoma. J Pathol 1998;186:51–58 [DOI] [PubMed] [Google Scholar]
- 59.Bodey B, Bodey JB, Siegel SE, Kaiser HF. Matrix metalloproteinase expression in malignant melanomas: tumor-extracellular matrix interactions in invasion and metastasis. In Vivo 2001;15:57–64 [PubMed] [Google Scholar]
- 60.Kurschat P, Wickenhauser C, Groth W, Krieg T, Mauch C. Identification of activated matrix metalloproteinase-2 (MMP-2) as the main gelatinolytic enzyme in malignant melanoma by in situ zymography. J Pathol 2002;197:179–187 [DOI] [PubMed] [Google Scholar]
- 61.Hofmann UB, Westphal JR, van Muijen GNP, Ruiter DJ. Matrix metalloproteinases in human melanoma. J Invest Dermatol 2000;115:337–344 [DOI] [PubMed] [Google Scholar]
- 62.Pereira AM, Strasberg-Rieber M, Rieber M. Invasion-associated MMP-2 and MMP-9 are up-regulated intracellularly in concert with apoptosis linked to melanoma cell detachment. Clin Exp Metastasis 2005;22:285–295 [DOI] [PubMed] [Google Scholar]
- 63.Levi E, Fridman R, Miao H-Q, Ma Y-S, Yayon A, Vlodavsky I. Matrix metalloproteinase 2 releases active soluble ectodomain of fibroblast growth factor receptor 1. Proc Natl Acad Sci USA 1996;93:7069–7074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer 2002;2:161–174 [DOI] [PubMed] [Google Scholar]
- 65.Dean RA, Overall CM. Proteomics discovery of metalloproteinase substrates in the cellular context by iTRAQTM labeling reveals a diverse MMP-2 substrate degradome. Mol Cell Proteomics 2007;6:611–623 [DOI] [PubMed] [Google Scholar]
- 66.Okada A, Tomasetto C, Lutz Y, Bellocq J-P, Rio M-C, Basset P. Expression of matrix metalloproteinases during rat skin wound healing: evidence that membrane type-1 matrix metalloproteinase is a stromal activator of pro-gelatinase A. J Cell Biol 1997;137:67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pulyaeva H, Bueno J, Polette M, et al. MT1-MMP correlates with MMP-2 activation potential seen after epithelial to mesenchymal transition in human breast carcinoma cells. Clin Exp Metastasis 1997;15:111–120 [DOI] [PubMed] [Google Scholar]
- 68.Gilles C, Polette M, Piette J, et al. High level of MT-MMP expression is associated with invasiveness of cervical cancer cells. Int J Cancer 1996;65:209–213 [DOI] [PubMed] [Google Scholar]
- 69.Zhang W, Matrisian LM, Holmbeck K, Vick CC, Rosenthal EL. Fibroblast-derived MT1-MMP promotes tumor progression in vitro and in vivo. BMC Cancer 2006;6:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.D'Ortho M-P, Will H, Atkinson S, et al. Membrane-type matrix metalloproteinases 1 and 2 exhibit broad-spectrum proteolytic capacities comparable to many matrix metalloproteinases. Eur J Biochem 1997;250:751–757 [DOI] [PubMed] [Google Scholar]
- 71.Butler GS, Dean RA, Tam EM, Overall CM. Pharmacoproteomics of a metalloproteinase hydroxamate inhibitor in breast cancer cells: dynamics of a membrane type 1 matrix metalloproteinase-mediated membrane protein shedding. Mol Cell Biol 2008;28:4896–4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sato H, Takino T, Okada Y, et al. A matrix metalloproteinase expressed on the surface of invasive tumor cells. Nature 1994;370:61–65 [DOI] [PubMed] [Google Scholar]
- 73.Knäuper V, Will H, Lopez-Otin C, et al. Cellular mechanisms for human procollagenase-3 (MMP-13) activation: evidence that MT1-MMP (MMP-14) and gelatinase A (MMP-2) are able to generate active enzyme. J Biol Chem 1996;271:17124–17131 [DOI] [PubMed] [Google Scholar]
- 74.Nakada M, Nakamura H, Ikeda E, et al. Expression and tissue localization of membrane-type 1, 2, and 3 matrix metalloproteinases in human astrocytic tumors. Am J Pathol 1999;154:417–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Davidson B, Goldberg I, Berner A, et al. Expression of membrane-type 1, 2, and 3 matrix metalloproteinases messenger RNA in ovarian carcinoma cells in serous effusions. Am J Clin Pathol 2001;115:517–524 [DOI] [PubMed] [Google Scholar]
- 76.Kitagawa Y, Kunimi K, Uchibayashi T, Sato H, Namiki M. Expression of messenger RNAs for membrane-type 1, 2, and 3 matrix metalloproteinases in human renal cell carcinomas. J Urol 1999;162:905–909 [DOI] [PubMed] [Google Scholar]
- 77.Ueno H, Nakamura H, Inoue M, et al. Expression and tissue localization of membrane-types 1, 2, and 3 matrix metalloproteinases in human invasive breast carcinomas. Cancer Res 1997;57:2055–2060 [PubMed] [Google Scholar]
- 78.Lafleur MA, Drew AF, de Sousa EL, et al. Upregulation of matrix metalloproteinases (MMPs) in breast cancer xenografts: a major induction of stromal MMP-13. Int J Cancer 2005;114:544–554 [DOI] [PubMed] [Google Scholar]
- 79.Ohnishi Y, Tajima S, Ishibashi A. Coordinate expression of membrane type-matrix metalloproteinases-2 and 3 (MT2-MMP and MT3-MMP) and matrix metalloproteinase-2 (MMP-2) in primary and metastatic melanoma cells. Eur J Dermatol 2001;11:420–423 [PubMed] [Google Scholar]
- 80.Hotary KB, Yana I, Sabeh F, et al. Matrix metalloproteinases regulate fibrin-invasive activity via MT1-MMP-dependent and independent processes. J Exp Med 2002;195:295–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morrison CJ, Butler GS, Bigg HF, Roberts CR, Soloway PD, Overall CM. Cellular activation of MMP-2 (gelatinase A) by MT2-MMP occurs via a TIMP-2-independent pathway. J Biol Chem 2001;276:47402–47410 [DOI] [PubMed] [Google Scholar]
- 82.Benbow U, Schoenermark MP, Mitchell TI, et al. A novel host/tumor cell interaction activates matrix metalloproteinase 1 and mediates invasion through type I collagen. J Biol Chem 1999;274:25371–25378 [DOI] [PubMed] [Google Scholar]
- 83.Li L, Dragulev B, Zigrino P, Mauch C, Fox JW. The invasive potential of human melanoma cell lines correlates with their ability to alter fibroblast gene expression in vitro and the stromal microenvironment in vivo. Int J Cancer 2009;125:1796–1804 [DOI] [PubMed] [Google Scholar]
- 84.Alberts B. Cells in Their Social Context Molecular Biology of the Cell, 5th ed New York, NY, USA: Garland Publishing, 2007 [Google Scholar]
- 85.Connolly JL, Schnitt SJ, Wang HH, Dvorak AM, Dvorak HF. Principles of cancer pathology. In Kufe DW, Pollock RE, Weichselbaum RR, Bast RC, Jr, Gansler TS. (eds): Cancer Medicine, 6th ed Philadelpha, PA, USA: B.C. Decker, 2003 [Google Scholar]
- 86.Mueller MM, Fusenig NE. Tumor-stroma interactions directing phenotype and progression of epithelial skin tumor cells. Differentiation 2002;70:486–497 [DOI] [PubMed] [Google Scholar]
- 87.Fini ME, Girard MT. The pattern of metalloproteinase expression by corneal fibroblasts is altered by passage in cell culture. J Cell Sci 1990;97:373–383 [DOI] [PubMed] [Google Scholar]
- 88.Arkell J, Jackson CJ. Constitutive secretion of MMP9 by early-passage cultured human endothelial cells. Cell Biochem Function 2003;21:381–386 [DOI] [PubMed] [Google Scholar]
- 89.Reunanen N, Li S-P, Ahonene M, Foschi M, Han J, Kähäri V-M. Activation of p38a mitogen-activated protein kinase enhances collagenase-1 (MMP-1) and stromelysin-1 (MMP-3) expresssion by mRNA stabilization. J Biol Chem 2002;277:32360–32368 [DOI] [PubMed] [Google Scholar]
- 90.Reuben PM, Cheung HS. Regulation of matrix metalloproteinase (MMP) gene expression by protein kinases. Front Biosci 2006;11:1199–1215 [DOI] [PubMed] [Google Scholar]
- 91.Ra HJ, Parks WC. Control of matrix metalloproteinase catalytic activity. Matrix Biol 2007;26:587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Murphy G, Nagase H. Progress in matrix metalloproteinase research. Mol Aspects Med 2008;29:290–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gupta GP, Nguyen DX, Chiang AC, et al. Mediators of vascular remodeling co-opted for sequential steps in lung metastasis. Nature 2007;446:765–770 [DOI] [PubMed] [Google Scholar]
- 94.Figueira RC, Gomes LR, Neto JS, Silva FC, Silva ID, Sogayar MC. Correlation between MMPs and their inhibitors in breast cancer tumor tissue specimens and in cell lines with different metastatic potential. BMC Cancer 2009;9:20. [DOI] [PMC free article] [PubMed] [Google Scholar]