Abstract
Although we have amassed extensive catalogues of signalling network components, our understanding of the spatiotemporal control of emergent network structures has lagged behind. Dynamic behaviour is starting to be explored throughout the genome, but analysis of spatial behaviours is still confined to individual proteins. The challenge is to reveal how cells integrate temporal and spatial information to determine specific biological functions. Key findings are the discovery of molecular signalling machines such as Ras nanoclusters, spatial activity gradients and flexible network circuitries that involve transcriptional feedback. They reveal design principles of spatiotemporal organization that are crucial for network function and cell fate decisions.
Signal transduction was once viewed as a collection of linear information transporting pipelines that related extracellular cues to specific genes. However, subsequent studies showed that different receptors often activate the same pathways and downstream effectors. Owing to pathway crosstalk, signals propagate through a tangled network of interconnecting proteins and cascades rather than through independent linear routes. Furthermore, findings from genome projects have revealed a new problem: there are fewer genes than biological processes. Hence, the concept that the specificity of biological processes is generated on the gene or even protein level erodes.
The idea of isolated pathways has given way to the concept of signalling networks, which allow a limited number of components to generate an exponentially larger number of outcomes owing to combinatorial interactions. Although we now can describe parts of these complex network topologies in detail, we still do not understand how they operate to generate biological specificity. It is like trying to plan a journey with an incomplete railway network map lacking train time schedules. Even the simple transport of a signal requires a network map, a time schedule and a notion of connectivities; that is, spatiotemporal coordination. However, biological networks not only transport, but also process and integrate signals. Crucial cell decisions, including whether to undergo proliferation, apoptosis and differentiation, are governed by the temporal dynamics and spatial distribution of key signalling effectors1–4. This realization provides a strong impetus to explore the emergent properties of signalling networks that are encoded by spatial and temporal dynamics.
This Review highlights current efforts towards understanding the spatial (three dimensional (3D)) and temporal (the fourth dimension) dynamics of signalling networks and discusses the challenges ahead. We summarize how temporal signalling dynamics control cell behaviour, and describe how the cellular localization of protein interactions and spatially distributed signalling processes add to the regulation of phenotypic responses. We conclude with a discussion on how spatial and temporal controls cooperate to choreograph the 4D dynamics of signalling networks that specify complex biological processes. Owing to space constraints we use selected examples to highlight these principles. This is an evolving field with sometimes competing hypotheses, which we discuss with a view of reconciling, when possible, and highlighting differences that may stimulate further research.
Temporal network dynamics and cell fate
The sharing of components between signalling pathways makes their function context dependent. How is context defined? A versatile way is through temporal specification. In the railway metaphor, all trains use common components and modules, but shifting the time when they run can determine which connections are enabled or disabled.
ERK signalling duration controls phenotypic responses
The first insights into how different cell surface receptors use shared pathways to generate specific cellular outcomes came from observations linking the duration of extracellular signal-regulated kinase (ERK) activation to cell fate decisions in rat pheochromocytoma PC-12 cells. In these experiments, transient ERK activation by epidermal growth factor (EGF) induced cell proliferation, whereas sustained ERK activation by nerve growth factor (NGF) induced differentiation1. Subsequent work extended the connection between ERK activation kinetics and cellular outcome to other cell types and conditions5. For instance, in human breast adenocarcinoma MCF-7 cells transient ERK activation by EGF induced proliferation, whereas sustained ERK activation by heregulin induced differentiation6. In squamous cell carcinoma SCC-12F cells sustained ERK activation by EGF and scatter factor/hepatocyte growth factor stimulated migration, whereas transient ERK activation by keratinocyte growth factor and insulin-like growth factor 1 stimulated proliferation7. These results show that temporal dynamics can specify distinct cell behaviours.
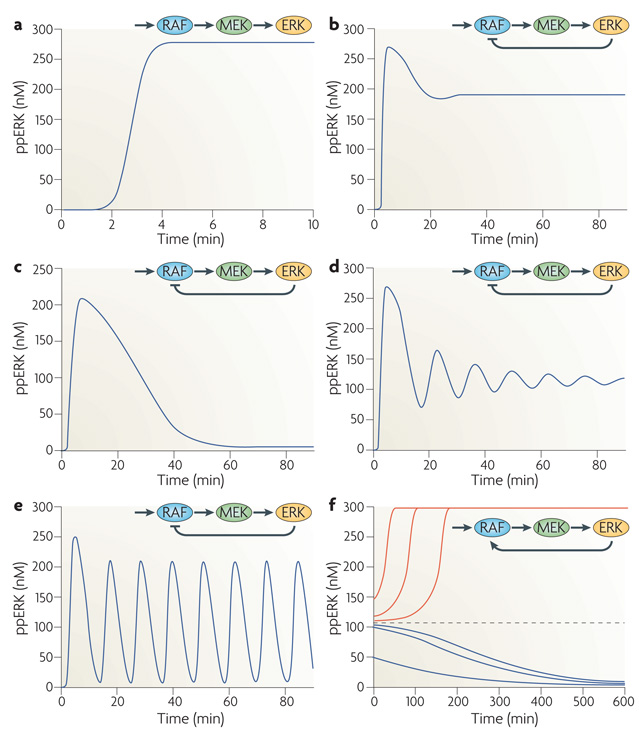
The ERK cascade comprises a three-tiered kinase module in which the first kinase phosphorylates and activates the second kinase, which phosphorylates the third kinase in a two-step, non-processive reaction. This theme is implemented by nature in several variations, commonly summarized as mitogen-activated protein kinase (MAPK) pathways. These currently include 15 MAPKs, the most prominent being ERK1 (also known as MAPK3), ERK2 (also known as MAPK1), Jun N-terminal kinase (JNK) and p38 MAPK pathways. Computational modelling shows that distinct modes of ERK spatiotemporal dynamics can emerge from different feedback wiring8–10. Depending on the feedback topologies and kinetic parameters, a MAPK cascade can display markedly different temporal responses to an identical constant stimulus: a monotone, sustained response (FIG. 1a); a transient, adaptive response including near-perfect adaptation (FIG. 1b,c); damped oscillations (FIG. 1d); sustained oscillations (FIG. 1e); and a switch-like, bistable response in which two stable steady states, Off and On, coexist (FIG. 1f). Different ERK temporal responses are also observed experimentally11–14. Although some details remain unclear, plausible mechanisms for the dynamic control of MAPK signalling in mammalian cells have been proposed. They include ERK-induced feedback phosphorylation of upstream kinases, for example RAF, the ability of which to activate MEK is impeded by ERK-mediated phosphorylation15, and their regulators, such as RAF kinase inhibitor protein (RKIP)16 and the Ras-activating guanine nucleotide exchange factor (GEF) Son of Sevenless (SOS)17. Note that the core Ras family of small GTPases has three members, HRAS, KRAS and NRAS, and unless specified we use Ras to refer to the family. In combination, these MAPK feedbacks can produce complex temporal activity patterns. In fact, oscillations in MAPK cascades were first theoretically predicted to occur as a result of negative feedback from ERK to SOS or RAF and of ultrasensitivity of the ERK responses to changes in the input8, and were later discovered experimentally12,14. Oscillations of ERK activity induced by negative feedback are enhanced by switch-like ERK activation caused by positive (double-negative) feedback arising from ERK-mediated inhibitory phosphorylation of RKIP16. Interestingly, both bistable and oscillatory MAPK dynamics can also arise from double phosphorylation of ERK and mutual sequestration when a kinase at an upper level (such as MAPK/ERK kinase (MEK)), forms a complex with a kinase (in this case ERK) at the next cascade level18,19.
Figure 1. Versatile MAPK dynamics.
Each panel schematically displays the RAF–MEK–ERK (RAF–MAPK/ERK kinase–extracellular signal-regulated kinase) cascade; the feedback from ERK to RAF, which is the initial mitogen-activated protein kinase (MAPK) activated by Ras-GTP, is indicated (when present). The different temporal responses of active, dually phosphorylated ERK (ppERK) to a constant Ras-GTP stimulus are obtained by changing the parameters of ERK-mediated feedback. A kinetic description of MAPK cascade reactions (rate equations) is given in Supplementary information S1 (table), in which parameter values are relative to the graph in part a. Parameters F and Kf describe the feedback regulation (F = 1, for no feedback, F < 1 for negative feedback, F > 1 for positive feedback; Kf equals ppERK concentration at which activation or inhibition is half-maximal), and indicates the maximal rate of a phosphatase reaction. a | No ERK–RAF feedback, F = 1. b | Negative ERK–RAF feedback, F=0.34, Kf = 25 nM. c | Negative ERK–RAF feedback, F=0.01, Kf = 1 nM, V7max = 0.175 nM s−1. d | Negative ERK–RAF feedback, F=0.27, Kf=25 nM. e | Negative ERK–RAF feedback, F=0.01, Kf=25 nM. f | Positive ERK–RAF feedback, F=5, Kf=100 nM, k1cat= 0.025 s−1, V11max =0.025 nMs−1. Depending on the initial conditions (pre-existing activity level of the cascade), ppERK either descends to the low activity state (blue curves) or approaches the high-affinity state (red curves); the dashed line indicates a threshold.
Inferring connections within signalling networks that underlie the observed complex dynamics is an emerging challenge in cell biology. It is not obvious how we can capture interactions between individual signalling nodes, as any activating or inhibitory stimulus applied to a particular node rapidly propagates through a network, causing widespread changes. One approach to untangle unknown network topologies, modular response analysis (MRA), groups many components into functional modules and infers their connections by measuring system-wide responses to systematic perturbations to all network modules20,21. Exploiting MRA, one study22 found that distinct temporal profiles of active ERK stimulated by EGF and NGF emerge from differential feedback wiring of the ERK cascade, with EGF eliciting negative feedback and NGF inducing positive feedback. Thus, networks are not hardwired, but can respond to different inputs by reconfiguring themselves. It will be interesting to study whether this plasticity just involves feedback loops or whether connections between network core structures are also subjected to dynamic remodelling.
ERK signalling induces transcriptional negative feedback
Phosphorylation of MAPKs is reversed by Ser/Thr phosphatases, Tyr phosphatases and dual specificity phosphatases (DUSPs). DUSPs are also known as MAPK phosphatases because they dephosphorylate ERK1, ERK2, JNKs and p38. Many DUSPs are immediate early genes (IEGs; that is, genes that are induced rapidly and do not require new protein synthesis for their transcription) induced by activated MAPKs23,24. ERK-mediated DUSP induction tightly controls ERK activity, and such transcriptional negative feedback is a common design principle of nearly all eukaryotic signalling pathways25,26. Differential induction and localization of DUSP isoforms in the cytoplasm and nucleus raises the intriguing possibility of different temporal dynamics for cytoplasmic and nuclear pools of ERK, and adds to the repertoire of signalling responses that determine cell fate decisions.
Transcriptional negative feedback generated by ERK-mediated DUSP expression can bring about ERK and DUSP oscillations that develop on a longer time-scale than oscillations arising from immediate negative feedback in the cytosol8,27. In haploid Saccharomyces cerevisiae, secreted pheromones activate MAPK signalling in cells of the opposite mating type, resulting in the formation of mating projections. A recent study shows that in these cells, oscillations of Fus3 MAPK activity depend on transcriptional induction of the MAPK phosphatase Msg5 and the negative regulator of pheromone receptor signalling, Sst2 (REF. 28). The Fus3 oscillations are observed on the same timescale (2–3 hours) as the periodic formation of additional mating projections28. Thus, oscillations in MAPK signalling activity have a physiological role in controlling gene expression on long timescales.
Discrete, digital outputs determine cell decisions
Cells in an organism are immersed in an ocean of growth factors and hormones. In addition, protein concentrations vary between individual cells29, adding intrinsic stochastic noise. How do cells discriminate between signal and noise? One possibility is that graded, analogue signals from receptors are converted into discrete, digital outputs, such as the all-or-none responses of signalling and gene expression cascades. Indeed, theoretical studies have shown that cell signalling circuits can act as analogue–digital converters, generating abrupt switches, multi-stable dynamics, excitable pulses and oscillations, and that these distinct outputs facilitate signal discrimination30–32 (BOX 1). Recent experimental work has shown that in different cell types and organisms, conversion of graded signals to digital outputs occurs at different levels. For example, in Xenopus laevis oocytes graded progesterone and sorbitol stimuli are converted into switch-like, all-or-none responses of the MOS–MEK–p42 MAPK33 and JNK cascades34 in the cytosol, whereas in Swiss 3T3 fibroblasts analogue ERK activation is converted into digital IEG responses in the nucleus2.
Box 1 | Temporal signalling dynamics.
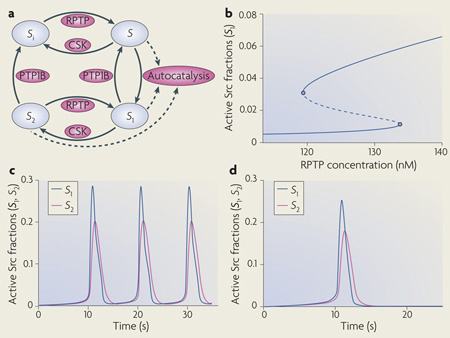
Understanding the temporal dynamics of signalling networks is facilitated by using kinetic schemes and ordinary differential equations (ODEs). Each signalling species is produced and consumed in particular reactions. The left-hand side of the ODE is the time derivative of a species concentration, and the right-hand side of the ODE is the algebraic sum of the reaction rates, which produce and consume that species. Because ODEs do not consider spatial dimension, this kinetic description implies a well-mixed, homogeneous reaction medium. This simplification facilitates the analysis of the effects of multiple inputs, feedback loops and pathway crosstalk on the dynamics of complex signalling networks106,107. Remarkably, already simple signalling motifs display intricate temporal dynamics18,27,30. For example, the basic activation–deactivation cycle of the Src Tyr kinase can show complex signalling dynamics that includes oscillations, toggle switches and excitable behaviour32. Src kinases can exist in four states (see the figure, part a). In the basal autoinhibited conformation (Si), Src is phosphorylated on the carboxy-terminal inhibitory Tyr and dephosphorylated on the activating Tyr in the catalytic domain. Both Tyr residues are dephosphorylated in the partially active form (S). In the first fully active conformation (S1) the inhibitory Tyr is dephosphorylated and the activating Tyr is phosphorylated, and in the second fully active form (S2) both Tyr are phosphorylated. A crucial non-linearity is brought about by intermolecular autophosphorylation of the activating Tyr (shown by dashed lines). Importantly, the complex Src dynamics do not require imposed external feedback loops and can occur at constant activities of Src inhibitors (such as C-terminal Src kinase (CSK)) and Src activators (such as protein Tyr phosphatase 1B (PTP1B)) and receptor-type protein Tyr phosphatases (RPTPs)). In different ranges of activities of these Src regulators, Src kinase activity can exhibit hysteresis (bistability) (see the figure, part b), oscillations (see the figure, part c), and excitable responses of active Src kinase fractions (see the figure, part d). Figure modified, with permission, from REF. 32 © National Academy of Sciences (2000).
Ultrasensitive and bistable responses (FIG. 1f) can be elicited by long-positive34 or short-positive feedback loops13,27. An example of long-positive feedback loops is RKIP inactivation by ERK16. Short-positive feedback loops arise when Ras-GTP (produced when SOS catalyses the exchange of GDP for GTP in the nucleotide binding pocket of Ras) binds to the SOS allosteric pocket. This causes a significant increase in the activity of this Ras-activating GEF, thereby stimulating further Ras activation35. Signal digitalization is also observed in the nucleus. In Swiss 3T3 fibroblasts EGF elicits transient ERK activity and negligible induction of the FOS IEG, whereas platelet-derived growth factor (PDGF) elicits sustained ERK activity and a substantial FOS response36. This is a result of cooperative expression and phosphorylation. EGF-induced ERK activity stimulates FOS transcription, but FOS is rapidly degraded. PDGF induces sustained activity of ERK and its substrate, ribosomal S6 kinase (RSK), which stabilize nascent FOS by phosphorylating it on multiple sites. Initial phosphorylation of FOS in the carboxyl terminus exposes an ERK docking site, resulting in further phosphorylation on Thr325 and Thr331 (REF. 36). We think that multi-site phosphorylation, and nested feed-forward stabilization loops from ERK and RSK to FOS, result in the observed switch-like expression response4,18,37. Thus, FOS serves as a binary cellular sensor of the duration and threshold intensity of ERK signalling36. All-or-none responses of ERK-induced IEG products generate distinct transcriptional programmes that lead to different cell phenotypes.
Spatiotemporal control of information
In addition to the temporal kinetic specification of network function, spatial control plays a major and complementary part. Our trains may leave at the same time but go to different destinations, or leave at different times heading for the same destination. Although the spatial component is less studied than the temporal one, its importance is highlighted by new findings.
Spatiotemporal organization controls network function
Recent systems biology work has elucidated how differences in the dynamic ERK interactome instigate cell fate decisions in PC12 cells3. Quantitative proteomics showed that ~20% of ERK interaction partners bind to ERK in a growth factor-regulated manner, and ~30% of these interactions are differentially regulated by EGF and NGF and affect differentiation. Interestingly, most of these interactions regulate spatial and temporal aspects of ERK signalling. Cytosolic interactions involve preformed complexes, from which ERK is released in response to stimulation by NGF and EGF. ERK nuclear interactions are induced in response to NGF, which stimulates the nuclear translocation of ERK (which is required for differentiation). In the nucleus ERK phosphorylates transcription factors and prompts the export of transcriptional inhibitors; interfering with these processes blocks differentiation. Thus, part of the NGF-specific transcriptional programme is initiated by the physical removal of inhibitors from the nucleus. In the cytosol, regulated ERK interactions include fine spatial compartmentalization processes. For example, NGF induces the sustained dissociation of the Ras GTPase-activating protein (RasGAP) NF1 from a Ras–ERK complex, prolonging Ras and ERK activation. NGF also induces the sustained release of ERK from the anchor protein PEA15, which sequesters ERK in the cytosol. Whenever tested, ERK interaction partners were also found to be ERK substrates, suggesting that ERK is anchored in the cytosol by its substrates. This ensures rapid ERK-mediated phosphorylation and localizes ERK signalling to different microcompartments that are determined by the expression and distribution of substrates.
It will be interesting to explore whether upstream ERK activators are also part of these preformed complexes and whether they reach them by random diffusion or active recruitment (BOX 2). The existence of binary and ternary scaffolds that bind different components of the Ras–RAF–mEK–ERK cascade suggests that both partially and fully scaffolded complexes occur. For instance, MAPK organizer 1 (MORG1) interacts with RAF, MEK and ERK, but also with MP1 (also known as MAPKSP1)38, which scaffolds MEK and ERK39. Thus, MORG1 can function as the scaffold of a scaffold, which may allow the modular assembly of a combinatorial variety of signalling complexes with distinct input–output functions. It is tempting to speculate that MORG1’s ability to regulate ERK activation only in response to selected stimuli is related to this super-scaffolding function. The participation of individual proteins in different signalling complexes will generate competing partitioning between the complexes and may accentuate the specification of functional outputs. This may serve as mechanism to coordinate signalling specificity, although this issue has not been explored.
Box 2 | Spatially distributed signalling.
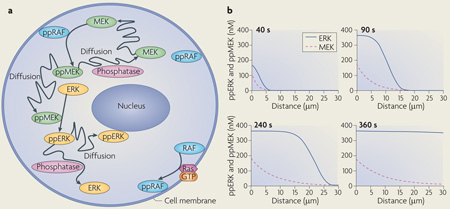
Ordinary differential equation-based models can account for spatial dynamics only by using coarse-grained compartmentalization; for example, by considering the cytoplasm and nucleus as two different, well-mixed compartments. More accurately, spatially distributed signalling processes are modelled using partial differential equations (PDEs), which contain derivatives with respect to time and space and are referred to as reaction–diffusion equations. Reaction–diffusion PDEs describe the concentration changes that are due to the production and consumption of a species in chemical reactions and its redistribution in space caused by diffusion and transport87,100. Solutions to reaction–diffusion equations are generally obtained by numerical integration, although for linear or saturated kinetics, stationary concentration profiles are obtained analytically88,108. In spatially distributed signalling networks, such as the β-adrenergic receptor pathway105 or mitogen-activated protein kinase (MAPK) cascades100, the initiating signals, cyclic AMP or RAF activity, respectively, are generated at the plasma membrane and subsequently spread into the cell through the sequential activation of downstream proteins (see the figure, part a, which illustrates the activation of successive kinases in the RAF–MEK–ERK (Raf–MAPK/ERK kinase–extracellular signal-regulated kinase) cascade). Importantly, the transmission of spatial information is controlled by feedback and feed-forward network motifs and cell shape104,109,110. For instance, bistability in a two-site ERK phosphorylation and dephosphorylation cycle18 was shown to generate a phosphoprotein wave that propagates from the surface into the cell interior100. The simulated time course of the propagation of MEK and ERK activation into the cytoplasm of a large cell, such as Xenopus laevis egg (cell radius 50 µm; nuclear radius 20 µm), is shown in the figure, part b. This wave propagation is facilitated by feedback inhibition of phosphatases owing to the generation of reactive oxygen species100. Positive feedback from dually phosphorylated ERK (ppERK) to cytoplasmic MEK further enhances the propagation span of the wave, making it possible to convey phosphorylation signals over exceedingly long distances. Such waves of protein phosphorylation and modification that travel with constant amplitude and velocity can transmit survival signalling along axons in developing neurons111,112. Figure in part b is modified, with permission, from Molecular Systems Biology REF.100 © 2006 Macmillan Publishers Ltd. All rights reserved.
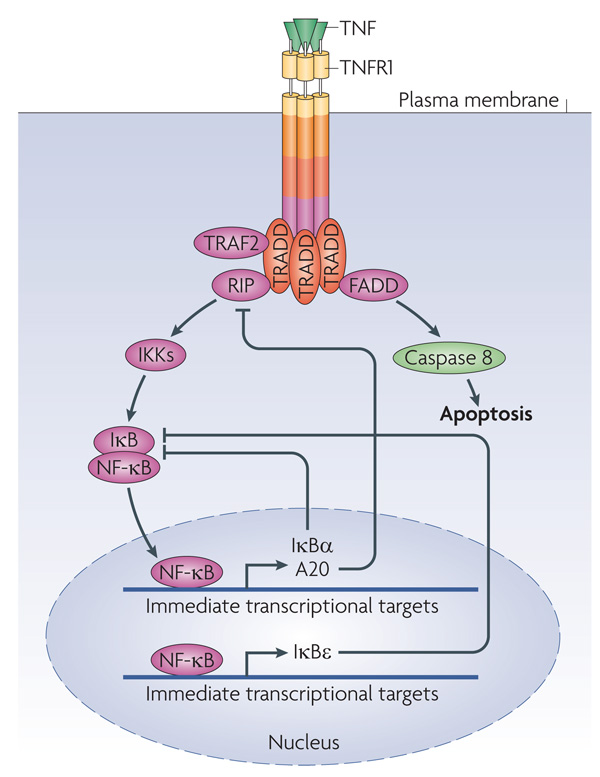
The output of nuclear factor-κB (NF-κB) is also regulated by spatiotemporal coordination (FIG. 2). NF-κB is inactive in the cytosol when tethered to inhibitor of NF-κB (IκB). IκB is phosphorylated by IκB kinase (IKK) and then degraded, resulting in the release and translocation of NF-κB to the nucleus, where it activates transcription, followed by NF-κB transport back to the cytoplasm40. The oscillation cycle of nucleocytoplasmic shuttling is finely tuned by several pathway inhibitors, including A20 and IκB isoforms, the expression of which is stimulated by NF-κB. IκB isoforms can bind NF-κB in the nucleus and force its export to the cytosol, where NF-κB is then retained. A20 blocks NF-κB activation by inducing the degradation of the adaptor receptor interacting protein (RIP), which mediates IKK activation41. Single cell analysis and modelling, using fluorescently labelled fusion proteins expressed at near physiological levels, show that different oscillation frequencies of nucleocytoplasmic NF-κB shuttling are determined by differentially timed stimulation pulses and the sequential induction of inhibitors42. There is a general caveat that exogenous expression may disturb the behaviour of signalling networks. However, recent results show that endogenous protein levels in individual cells vary extensively29, making artefacts from mild overexpression unlikely. A triple feedback model considering stochastic transcription of IKBA (the gene encoding IκBα) and A20, and delayed transcription of the gene encoding IκBε, predicts that IκBα and A20 cause oscillations of NF-κB shuttling, whereas IκBε increases response heterogeneity between cells42. This is due to the delayed transcriptional induction of IKBE, which adds stochastic noise42. The biological benefit of adding noise is not well understood, but can be related to desynchronizing oscillations in different cells. The cellular heterogeneity generated by noise could provide an advantage in which a cell population needs to react in a highly adaptive and selective way, such as in the immune response. Similar conclusions were derived from extensive stochastic simulations of the NF-κB pathway43. Importantly, the oscillation frequency modulates the specificity of gene expression42. These studies show that the spatial arrangement of signalling proteins is subject to dynamic regulation, and, vice versa, that spatial organization can specify kinetic activity profiles (FIG. 2).
Figure 2. Intrinsic transcriptional feedback inhibition of NF-κB.
In resting cells nuclear factor-κB (NF-κB) is inactive because inhibitor of NF-κB (IκB) retains it in the cytosol. On activation, for example by tumour necrosis factor (TNF), TNF receptor 1 (TNFR1) forms a complex with the adaptor protein TNFR-associated DEATH domain (TRADD), which recruits different proteins to initiate dual signalling pathways. TRADD recruits FAS-associated death domain (FADD) to promote apoptosis by stimulating caspase 8 activation. By contrast, TRADD recruits the adaptor receptor interacting protein (RIP) to counteract apoptosis by activating NF-κB. NF-κB activation is enabled as a result of the stimulus-induced degradation of IκBs following their phosphorylation by IκB kinases (IKKs), which releases NF-κB from its cytosolic anchor proteins so that it can translocate to the nucleus. However, nuclear NF-κB also induces the transcription of its own inhibitors. IκBs can bind to nuclear NF-κB and export it back to the cytosol, and A20 can interrupt receptor-mediated NF-κB activation by inducing the degradation of RIP.
Scaffolds: managers of spatiotemporal organization
The marriage of spatial and temporal orchestration is embodied in scaffolding proteins. Scaffolds are hallmarked by their ability to simultaneously bind two or more signalling proteins that typically have an enzyme–substrate relationship. The physical colocalization generates interesting properties, such as insulating signalling modules by physically tying them together, reducing reaction kinetics to zero order, enabling immediate feedback and anchoring protein complexes to distinct subcellular sites. Importantly, scaffolds allow the re-use of enzymes for different functions in a highly context-dependent manner, providing a simple solution to the dilemma of possessing fewer genes than processes. These properties make scaffolds ideally suited to operate as organizing principles in both synthetic and metabolic signalling networks. The first described examples of scaffolds were the A-kinase anchoring proteins (AKAPs), which have many diverse functions44. AKAPs (which have been extensively reviewed) often bind kinases, their phosphatases, upstream activators and downstream effectors. They assemble signalling platforms that orchestrate input–output relationships through physically coupled activation–deactivation cycles at highly localized sites in cells. The function of scaffolds in other systems, such as the MAPK pathway, is increasingly being appreciated and was also recently reviewed45–47. Here, we focus on open questions and try to delineate the design principles of how scaffolds contribute to spatiotemporal organization of signalling networks. To illustrate this, we use two examples (FIG. 3).
Figure 3. Scaffolds and spatial organization.
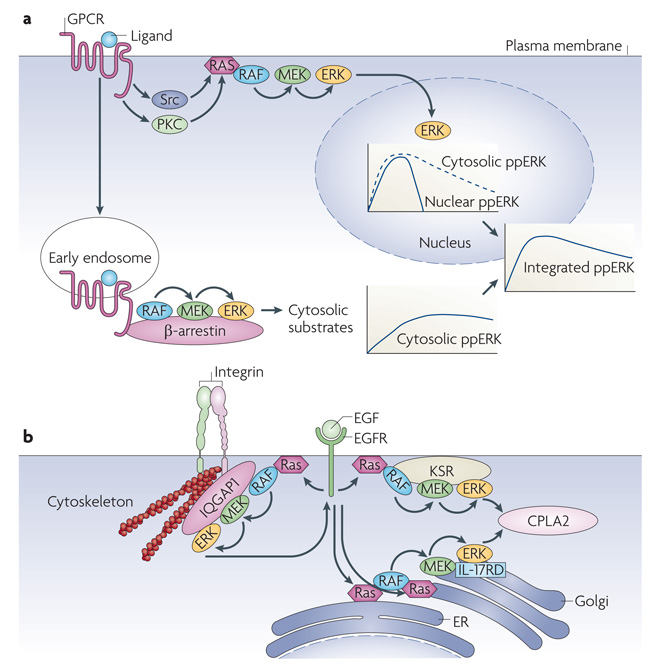
a | G-protein coupled receptors (GPCRs) activate extracellular signal-regulated kinase (ERK) through two spatially and temporally separated pathways. Mechanistic details are omitted for the sake of clarity and were reviewed previously113. Rapid ERK activation emanating from the plasma membrane through protein kinase C (PKC), Src, and receptor Tyr kinase stimulation is transient and β-arrestin independent, and allows ERK translocation to the nucleus. Sustained ERK activation is triggered from an endosomal β-arrestin-dependent RAF–MEK–ERK (RAF–MAPK/ERK kinase–extracellular signal-regulated kinase) module and restrains ERK signalling to the cytosol. The integrated dually phosphorylated ERK (ppERK) results from combining the nuclear and cytosolic ppERK levels. b | Ras activated at different subcellular compartments uses different scaffolding proteins to target ERK substrates. IQGAP1 (on the cytoskeleton) mediates negative ERK feedback phosphorylation of epidermal growth factor receptor (EGFR), and kinase suppressor of Ras (KSR) (at the plasma membrane) and interleukin-17 receptor D (IL-17RD; also known as SEF1) (in the Golgi) facilitate phosphorylation of cytoplasmic phospholipase A2 (CPLA2) by ERK activated at the plasma membrane or intracellular membranes, respectively. Ras activated at the endoplasmic reticulum (ER) might stimulate IL-17RD-bound ERK at the Golgi53. However, Ras can also be activated directly114 at the Golgi. In either case, ERK phosphorylation activates CPLA2 to generate arachidonic acid, which is a precursor to signalling molecules such as leukotrienes and prostaglandins.
The first is β-arrestin, a scaffold that coordinates the activation of multiple signalling pathways downstream of G-protein coupled receptors (GPCRs)48. GPCRs activate ERK through two spatially and temporally separated pathways. Rapid ERK activation emanating from the plasma membrane is transient, β-arrestin independent and allows ERK translocation to the nucleus. Sustained ERK activation is triggered by an endosomal RAF–MEK–ERK module that is assembled by β-arrestin scaffolding and restrains ERK signalling to the cytoplasm (FIG. 3a). Originally described as a protein that desensitizes signalling from GPCRs, β-arrestin has emerged as a multivalent scaffold protein that binds a large array of signalling molecules. Some of these molecules are downstream effectors that propagate signals into the cell, such as ERK, JNK3, p38 and AKT, whereas the others are enzymes that deactivate second messengers and inhibit GPCR-induced signalling, such as phosphodiesterases (which degrade cAMP) and diacylglycerol kinases (which degrade diacylglycerol)48–50. Typically, β-arrestin scaffolds whole signalling modules, such as RAF–MEK–ERK or the one formed by the JNK3 upstream kinases apoptosis signal-regulating kinase 1 (ASK1; also known as MEKK5) and MAPK kinase 4 (MKK4; also known as MAPKK4), and JNK3. This raises the question of how binding to the scaffold is regulated and how the correct specific assemblies are generated. The latter may be explained in part by the interaction sites that exist in individual components of the specific kinase cascades and lead to preformed modules51. A scaffold, in this case β-arrestin, would stabilize the preformed assemblies but also regulate the specificity, efficiency and amplitude of signal propagation52. However, β-arrestin still needs to pick up the appropriate module, and this may be regulated by spatial distribution and dynamics. For instance, as described below, the activation of the RAF–MEK–ERK cascade occurs at the plasma membrane in distinct Ras nanoclusters (see below) using kinase suppressor of Ras (KSR), a scaffold for RAF–MEK–ERK (FIG. 3b). GPCR-mediated ERK activation can be dissected into two phases: an early phase, which is β-arrestin independent and may correspond to nanocluster activation at the membrane, and a late phase, which extends ERK activity and is β-arrestin dependent. Interestingly, the early phase allows the nuclear translocation of ERK, whereas the late phase is triggered by endocytosed GPCRs and confines ERK signalling to the cytosol48 (FIG. 3a). This mechanism diversifies ERK function in GPCR signalling using spatial and temporal separation of activation, conceptually resembling signal splitters in electronic circuits. Thus, scaffolding can orchestrate signalling by defining the sequence of events in time and space.
The second paradigm illustrates that the specificity of ERK substrate phosphorylation may be determined by the localization of upstream signals53,54. Again scaffolds may play prominent parts by directing ERK phosphorylation to distinct substrates in the cytosol or nucleus, depending on the subcellular structure on which Ras is activated. Ras activation is not confined to the plasma membrane and can also occur at intracellular membranes54. To phosphorylate cytosolic phospholipase A2 (CPLA2), ERK activated at the plasma membrane uses the KSR scaffold, whereas ERK activated at the endoplasmic reticulum (ER) employs interleukin-17 receptor D (IL-17RD; also known as SEF1). For feedback phosphorylation of the EGFR, ERK uses the IQGAP1 scaffold53 (FIG. 3b). So, depending on the input, differential scaffolding and subcellular targeting can allow a kinase module to signal to different effectors in parallel. We still know little about the molecular mechanisms, but new optical approaches will bridge this gap (BOX 3), and recent advances have started to clarify some of the membrane structures that organize signalling.
Box 3 | Microscopic technologies to analyse spatiotemporal organization of signalling networks.
Assembling signalling networks in time and space requires cellular imaging. Different methods provide high-resolution spatial and temporal information by spatially mapping molecules with respect to a defined cellular structure or another molecule. Each method has limitations, and complete visualization of a network requires multiple approaches.
2D imaging
Wide-field fluorescence imaging cannot provide three-dimensional (3D) resolution. This problem is partially mitigated by total internal reflection fluorescence microscopy, which limits imaging to a thin (~200 nm) slice of the cell that is adherent to the coverslip. Events occurring on or near the basal plasma membrane can be observed.
3D imaging
Three-dimensional imaging is achieved by confocal and two-photon microscopy; two-photon microscopy can access deep into samples. Both methods are diffraction limited. Higher resolution imaging is feasible with 4-Pi and stimulated emission depletion (STED) microscopy, which use non-linear de-excitation of fluorophores to bypass the resolution limit of diffraction. STED microscopy can visualize lipid rafts in live cells65.
FRET
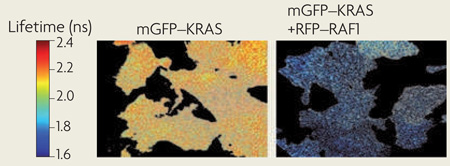
Methods using Förster resonant energy transfer (FRET) detect the proximity of molecules on length scales of 1–10 nm. Live cell FRET imaging can quantify and localize specific molecular interactions to cellular structures. Fluorescence lifetime imaging microscopy is predominantly used as a robust method to measure FRET. FRET between donor and acceptor fluorophores reduces the fluorescence lifetime of the donor, which can be used to generate a FRET image. For example, the lifetime of monomeric green fluorescent protein (mGFP) tagged to KRASGly12Val in BHK cells decreases when it is co-expressed with monomeric red fluorescent protein (mRFP) tagged to RAF1 owing to FRET between mGFP and mRFP after RAF1 is recruited to Ras nanoclusters (see the figure). Pixel-by-pixel fitting of two lifetimes and calibration using an mGFP–mRFP fusion protein allows the fraction of mGFP molecules undergoing FRET to be calculated (~30%). Fluorescence anisotropy microscopy, used to identify homo-FRET between identical florophores mapped the spatiotemporal dynamics of the nanoclustering of glycosylphosphatidylinositol-anchored protein66.
Single molecule and spectroscopic techniques
Single particle tracking and single fluorophore video tracking track the diffusion of single molecules with high spatial and temporal resolution. Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy (FCCS) measure fluorescent fluctuations in small confocal volumes. The data can be used to derive diffusion constants and, in the case of FCCS, infer and quantify molecular interactions without relying on FRET.
Image reproduced with permission, from Nature Cell Biology REF. 78 © 2007 Macmillan Publishers Ltd. All rights reserved.
An interesting new aspect emerged with the discovery that scaffolds can function as allosteric regulators of their client kinases. KSR can activate its client BRAF by side to side dimerization, presumably by an allosteric mechanism55. Finer mechanistic details were elaborated for the yeast scaffold Ste5, which functions in the pheromone mating pathway. Ste5 has two docking sites for its client kinase Fus3. The strong docking site stimulates Fus3 to phosphorylate and downregulate mating signalling through the Ste5 pathway56. The weak binding site assists the activation of Fus3 by allosterically improving its accessibility to phosphorylation by the upstream kinase Ste7, thereby enhancing pheromone signalling57. In addition, the localization of Ste5 is crucial for the quality of the signal output. Ste5 generates a graded output in the cell membrane (its natural location), whereas confining Ste5 to the cytosol enhances the inherent ultrasensitive activation of Fus3 (REF. 58).
Organelle apposition facilitates signal transfer
Just as scaffolds bring signalling molecules together to facilitate their interactions, the apposition of two organelles can create spatial highways for exchanging signalling molecules. For instance, the fact that mitochondria are close to the ER Ca2+-releasing channels overcomes the low affinities of mitochondrial Ca2+ transporters for calcium, allowing rapid Ca2+ accumulation in the mitochondrial matrix following the opening of ER Ca2+ stores59. Recently, the components of the macromolecular tether between the ER and mitochondria were identified as the integral ER membrane protein maintenance of mitochondrial morphology protein 1 (Mmm1) and the outer mitochondria membrane proteins mitochondrial distribution and morphology protein 10 (Mdm10), Mdm12 and Mdm34 (REF. 60). This large tether complex, termed ER–mitochondrion encounter structure (ERMES), contains multiple copies of these proteins. In addition, ERMES was shown to facilitate direct phospholipid exchange between the ER and mitochondria60.
Depletion of ER Ca2+ stores triggers the opening of store-operated Ca2+ channels in the plasma membrane, allowing extracellular Ca2+ to enter the cell. ER–plasma membrane junctions seem to be key regulators of Ca2+ influx, whereas the ER protein stromal interaction molecule 1 (STIM1) serves as a Ca2+ sensor61. STIM1 spanning the ER membrane aggregates into oligomeric complexes on Ca2+ store depletion and translocates to the plasma membrane to activate Ca2+ release-activated Ca2+ (CRAC) channels in non-excitable cells. Similarly to ER–mitochondrion contacts, an assembly of protein complexes that contain both ER and plasma membrane proteins is required for channelling signals at ER–plasma membrane junctions. The STIM oligomers directly bind the CRAC channel protein ORAI1 (also known as CRACM1), which opens the CRAC channel62. Subsequently, the smooth ER Ca2+ ATPase pump replenishes the ER Ca2+ stores.
Nano- and microscale signalling domains
The plasma membrane is a major platform for signal transduction. The architecture of the plasma membrane in turn can dictate network properties by sequestering signalling proteins in space and time. The plasma membrane is an asymmetric lipid bilayer, comprising > 7,000 lipid species. It is organized into ultrafine compartments by the engagement of transmembrane proteins with the submembrane cortical actin mesh. Classical diffusion occurs within individual compartments, but long-range diffusion across multiple compartments is impeded by compartment boundaries63. Inhomogeneities in the lipid bilayer also exist as a result of incomplete lipid mixing. These lipid assemblies include complexes of glycosphingolipid and cholesterol, known as lipid rafts64, which transiently exist on ~20 ms timescales and <20 nm length scales65. Lipid rafts can be stabilized by engagement with lipid-anchored proteins or cross linking66–68. As a result, the plasma membrane comprises a complex, non-random, dynamic array of lipids and protein–lipid complexes on many different length and timescales. The extent to which specific protein–lipid and lipid–lipid complexes are employed by cells has been debated64,67. However, there is clear evidence for protein–lipid and lipid-based sorting platforms functioning in endocytic and exocytic trafficking, membrane curvature formation, cell migration and polarity68.
Assembling protein–lipid nanodomains
Plasma membrane spatiotemporal dynamics can regulate signalling complexes that are permanently tethered, or transiently recruited to, the plasma membrane. One example is the formation of transient, nanoscale protein clusters (nanoclusters) that operate as temporary signalling platforms or reaction chambers. These clusters contain mixtures of kinases, phosphatases and other signalling proteins that are anchored directly to the membrane or by lipid-anchored regulatory proteins or subunits. Localization of proteins to such nanoscale domains can accelerate reaction rates of signalling events that require specific protein–protein or protein–lipid interactions69.
Fine control of input–output by signalling nanocircuits
Among the best-characterized membrane protein–lipid complexes are those formed by Ras GTPases. The three Ras isoforms, HRAS, KRAS and NRAS have a conserved guanine nucleotide-binding domain (G domain) but use different C-terminal anchors for membrane binding: HRAS and NRAS undergo farnesylation and acylation, whereas KRAS is only farnesylated and requires an adjacent polybasic domain for stable anchoring70. Ras proteins are arranged on the plasma membrane as a combination of nanoclusters and freely diffusing monomers71 (FIG. 4). A nanocluster comprises ~7 Ras proteins, has a radius of ~9 nm and an estimated lifetime of 0.5–1 s72,73 (FIG. 4a). The formation of highly dynamic nanoclusters involves a complex interplay between the Ras lipid anchor, plasma membrane elements, amino acids in the C-terminal Ras hypervariable region and its G domain, and ancillary scaffold proteins such as galectins. As a result, HRAS, KRAS and NRAS assemble into spatially distinct, non-mixed clusters, with further segregation between GTP-loaded and GDP-loaded proteins. For example, HRAS-GDP clusters are cholesterol and actin dependent, HRAS-GTP clusters are actin and cholesterol independent, and KRAS-GTP clusters are weakly actin dependent and cholesterol independent72. KRAS-GTP and HRAS-GTP clusters require specific scaffold proteins to promote assembly: these include galectin 1 or galectin 3, which are selectively recruited from the cytosol by HRAS-GTP and KRAS-GTP, respectively74–76. The scaffold stabilizes the G domain of a Ras-GTP monomer in a signalling-competent orientation with respect to the plasma membrane76,77.
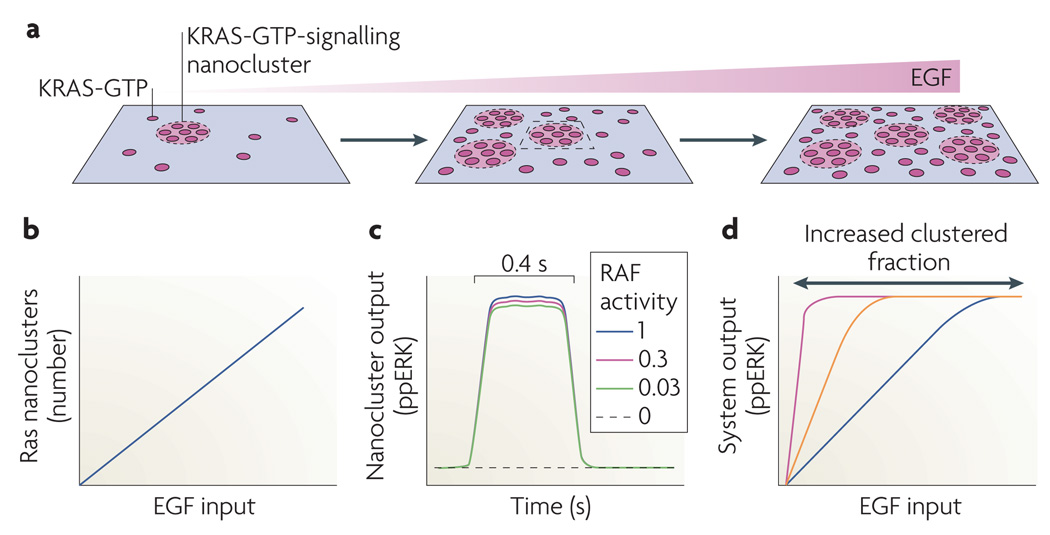
Figure 4. Ras nanoclusters digitize transmembrane signal transmission.
a | Activation of epidermal growth factor receptors (EGFRs) generates KRAS-GTP on the plasma membrane. A fixed proportion of these KRAS-GTP molecules (the clustered fraction) assemble into signalling nanoclusters. Each cluster has a radius of ~9 nm and contains ~7 KRAS-GTP molecules72. At higher EGF concentrations more nanoclusters form. b | Because KRAS-GTP levels are directly proportional to non-saturating EGF doses, and the KRAS-GTP clustered fraction remains constant as KRAS-GTP levels increase, the number of KRAS-GTP nanoclusters depends linearly on stimulating EGF concentration. c | After the recruitment of RAF and KSR–MEK–ERK (kinase suppressor of Ras–MAPK/ERK kinase–extracellular signal-regulated kinase) complexes from the cytosol, each nanocluster outputs a digital pulse of dually phosphorylated ERK (ppERK). The ppERK output is insensitive to RAF kinase input and is limited by disassembly of the nanocluster; a two-log range of relative RAF inputs is shown. d | As a result of b and c, the total system response to EGF, which is the aggregated digital outputs from all of the transiently active nanoclusters, is analogue (blue line). The gain of the response is increased if the KRAS clustered fraction increases from 40% to higher values (orange and purple lines).
The subset of Ras-GTP proteins found in nanoclusters is termed the clustered fraction (~40% in fibroblasts). Currently available data suggest that Ras-GTP nanoclusters might be the only sites of RAF recruitment and ERK activation on the plasma membrane; if Ras nanoclustering is abolished, ERK activation on the plasma membrane fails71,73,76,78. Conversely, increasing the KRAS-GTP clustered fraction enhances ERK signalling75,76. Ras-GTP nanoclusters recruit RAF and KSR–MEK–ERK complexes from the cytosol for activation. Because of the scaffolding, the ERK pathway module in a nanocluster generates the same activated dually phosphorylated ERK output for a wide range of RAF inputs and therefore operates as a low threshold switch78–80 (FIG. 4c). The generation of ERK is terminated by spontaneous disassembly of the nanocluster without requiring biochemical deactivation of the kinase cascade. This is a new use of spatiotemporal dynamics that eliminates the potential problem of hysteresis arising from dual processive ERK phosphorylation within the digital nanodomain81.
Spatiotemporal dynamics are also responsible for a cellular activated ERK response that is analogue with respect to EGF stimulation. As a result of non-equilibrium kinetics the Ras-GTP clustered fraction is approximately constant over a wide range of Ras-GTP levels. Consequently, there is a linear relationship between Ras-GTP levels and the number of nanoclusters generated on the plasma membrane72,78 (FIG. 4b). Thus, although each nanocluster delivers a brief, quantal activated ERK output (FIG. 4c), the total output of activated ERK from the plasma membrane is analogue (FIG. 4d). The Ras nanocluster circuitry therefore allows the plasma membrane to operate as an analogue–digital–analogue converter, digitizing the EGF analogue input signal for transmission across the plasma membrane by generating an appropriate number of Ras nanoclusters, and then regenerating the analogue EGF signal as a matched activated ERK output into the cytosol. The system operates with high fidelity, as expected of a digital signalling system78.
A similar analogue–digital–analogue converter operates through the glycosylphosphatidylinositol (GPI)-anchored protein CD59 nanocircuitry, which regulates immune cell activation82,83. Signalling is triggered by the formation of CD59 clusters. The Src family kinase LYN and the heterotrimeric G protein subunit Gαi are recruited to the clusters and activated, in turn activating phospholipase Cγ (PLCγ). The signal output from each cluster is a digital pulse of inositol-1,4,5-trisphosphate (InsP3); this is produced by the PLCγ-catalysed hydrolysis of phosphatidylinositol bisphosphate, which is released into the cytosol. As with Ras nanoclusters, the duration of the InsP3 pulse is limited by time and occurs on the same timescale as the output of activated ERK from a Ras nanocluster81–83. The overall system response is analogue because the InsP3 outputs from the individual clusters are summed in the cytosol and converted into an analogue Ca2+ signal following the activation of InsP3 receptors on intracellular calcium stores. The short lifetime of the Ras and CD59 clusters is crucial for high fidelity signal transmission because it allows for a high sampling rate of the analogue input signal. Indeed, computation shows that as the cluster lifetime increases, fidelity is lost and the system response becomes progressively digital, being determined by the biochemical kinetics and not plasma membrane spatiotemporal dynamics.
Given the similarity between the Ras and GPI-anchored nanocluster systems, it is tempting to speculate that this type of analogue–digital–analogue circuitry may represent a general mechanism for high fidelity signal transmission by lipid-anchored signalling proteins. Similarly to Ras, GPI-anchored proteins also show a fixed monomer to cluster distribution that violates simple mass action kinetics. Recent work has shown that this distribution is actively maintained and crucially dependent on cortical actin dynamics84. More broadly, the role of the unique architecture of the plasma membrane in supporting the assembly of analogue–digital–analogue converters brings into focus membrane spatiotemporal dynamics as a new regulator of signal transmission. Signal response is determined by the prevailing nanoclustered fractions of key signalling molecules. As the clustered fractions are dependent, among other things, on the state of the cortical actin cytoskeleton and the lipid content of the plasma membrane, these and other inputs, such as cell growth and metabolic state, migratory information and cell contact data, can be integrated to set the gain control of the analogue–digital–analogue converter (FIG. 4d).
Activation of the ERK cascade on the Golgi by HRAS or NRAS, or in the cytosol, depends on the input of RAF. Therefore, the activated ERK output from Ras nanoclusters if they operate on Golgi membranes is analogue, not digital79,80. Thus, subcellular localization crucially determines how the ERK module output is wired. These different system outputs from different compartments are biologically relevant. For example, in the developing mouse immune system, antigens that drive high strength activation of the Ras–ERK pathway from the plasma membrane lead to clonal deletion of T cells, whereas antigens that drive low strength activation from the Golgi result in clonal expansion85. So, spatial organization can directly determine the quality and biological effect of signalling output.
Chemical reactions and diffusion form spatial domains
Not long ago a cell was considered a bag of enzymes, in which biochemical reactions proceeded within the well-stirred, spatially uniform milieu of an enzymologist’s test tube. However, Alan Turing’s theoretical work showed that biochemical reactions can impose spatial order and break the symmetry of an initially homogeneous medium86. The fundamental novelty of Turing’s idea is that diffusion, which intuitively seems to be a process that eliminates spatial heterogeneity, can generate periodic spatial patterns in the initially uniform environment if the diffusion coefficients of interacting species are different. This laid the foundation of the physicochemical theory of morphogenesis87. If two morphogens, typically an activator and inhibitor, have different diffusivities, and the activator autocatalytically reproduces itself and stimulates its inhibitor, then the spatially uniform distribution can become unstable and drive the formation of heterogeneous spatial patterns. To prevent autocatalytic explosion, the inhibition process should be faster, and its diffusion coefficient should be larger than that of the activator. Spatial ranges of activation and inhibition are determined by the diffusion coefficient and the half-life of the species. Short-range, local activation and long-range inhibition govern periodic increases in the concentration of the activator and the spacing between repeating peaks. Similar spatial patterns occur when long-range inhibition is substituted by depletion of a substrate that is required to produce an activator and is consumed by activation87.
A different mechanism to generate positional information and spatial patterns exploits pre-existing heterogeneity in a cell88. Signal transduction proceeds through cycles of reversible covalent modification of target proteins, catalysed by an activator and deactivator, such as a kinase and phosphatase for a phosphorylated protein, or a GEF and GAP for a small GTPase. Owing to the presence of cellular structures, such as membranes, cytoplasm, organelles and chromosomes, opposing activator and deactivator enzymes are often spatially segregated. In other words, the intracellular environment for reactions and diffusion is initially inhomogeneous and does not resemble the uniform media traditionally considered for Turing-type models. For a protein that is phosphorylated by a membrane-bound kinase and dephosphorylated by a cytosolic phosphatase, a precipitous phosphorylation gradient with high concentrations of phosphorylated protein close to the membrane, and low concentrations in the cell interior, was predicted88. Provided that the phosphatase is far from saturation, the stationary phosphorylation profile decays almost exponentially with the distance from the membrane. The characteristic decay length (Lgrad) is determined by the diffusion coefficient (D) and the apparent first-order rate constant (k) of the phosphatase (deactivator) and does not depend on the kinase (activator) kinetics, . This simple spatial pattern is stable and occurs because of the pre-existing separation of opposing enzymes. Activity gradients of this type have been discovered experimentally for the small GTPase RAN89, the yeast MAPK Fus3 (REF. 90), protein Tyr phosphatase 1B91, aurora B kinase92 and the yeast protein kinase Pom1 (REF. 93).
If an active protein associates with other proteins to generate multi-protein complexes, or rapidly and reversibly binds to cytoskeleton elements, the apparent diffusion coefficient (D*) of this form becomes smaller than the diffusion coefficient (D) of a free inactive form94. Then, stable intracellular gradients of the total protein abundance arise from the spatial separation of activator and deactivator enzymes. These total protein gradients (Gradtotal) are less precipitous than gradients of the active form95(Grada): Gradtotal / Grada = (1−D*/D) < 1. Thus, stable stationary patterns of different signalling activities and protein abundances in diverse subcellular domains can arise from the spatial separation of opposing enzymes and diffusion.
Intricate concentration landscapes in single cells
Intricate landscapes of steady-state protein activities arise from different spatial localization of kinases and phosphatases and GEFs and GAPs on cell membranes, chromatin structures or in the cytoplasm. For many kinase cascades, such as MAPK cascades, the first level kinase is activated on the plasma membrane in response to external stimulation. When this occurs in a fully scaffolded environment, such as nanoclusters, the whole activation process is kinetically confined by spatial colocalization. However, when the cascade components are allowed to diffuse, either after the rapid disassembly of nanoclusters or alternative modes of activation in different compartments, the phosphorylation level and activity of the first kinase sharply decrease during a diffusion journey in the cytoplasm96. Because only the phosphorylated kinase fraction can stimulate a downstream kinase, a progressive reduction of the stimulation efficacy down the cascade occurs. This raises the question as to under what conditions signals emanating from the membrane robustly propagate into the cell interior. The assumption that both kinases and phosphatases are far from saturation, and that the apparent first-order rate constants do not change from layer to layer, allows us to formulate a simple criterion of signal propagation97. If the ratio of the phosphatase (deactivator) and kinase (activator) rate constants is much smaller than one, activation signals readily spread from the plasma membrane into the cell interior. The stationary activation profiles of successive kinases down the cascade display long, flat plateaus, which abruptly decay at the spatial locations following each other at almost constant space intervals. These intervals can be much larger than the characteristic decay length of the spatial activation profile for the initial kinase, spreading phosphorylation signals deep into a cell. If the ratio of the deactivator and activator rate constants is greater than one, the signal propagation stalls as the spatial profiles of successive activated proteins rapidly decay, confining the signal to the membrane97. More complex stable spatial patterns can arise from multisite phosphorylation of kinases in signalling cascades. For example, if a kinase at each level has two phosphorylation sites and the condition considered above for signal propagation is fulfilled, the activation profiles of dually phosphorylated forms at successive cascade levels display similar long, flat plateaus, whereas monophosphorylated kinases exhibit non-monotonous, transient concentration profiles. Their peaks are localized close to the places where the stationary fronts of dually phosphorylated kinases rapidly decay.
In GTPase cascades, an active GTPase can positively or negatively control GEFs or GAPs at many levels. For instance, a ‘ballet’ of small GTPases controls cytoskeletal dynamics, during which Cdc42 activates Rac and possibly Rho, whereas Rac and Rho inhibit each other98. Theoretical considerations showed that the spatial separation of GEFs and GAPs can lead to complex patterns of GTPase activities in a cell, in which activities can decrease, increase or exhibit peaks with an increase in the distance from the cellular structure (such as a membrane or chromosome) where the initial GEF is localized95. Such complex, non-monotonic, stable concentration profiles were recently reported for a chromosome-dependent RAN–importin-β cascade, coupled with a secondary phosphorylation network99. All these patterns are brought about by the spatial separation of opposing enzymes in activation–deactivation cycles of protein modification, rather than by the spatial symmetry breaking and instability of the spatially uniform distribution that occurs in the Turing mechanism.
Positive and negative feedback loops in protein cascades can bring dynamic instabilities in time and space27. For instance, travelling waves can occur in bistable MAPK cascades in which diffusion coefficients of components are assumed to be the same100 (BOX 2). Incorporation of the existing spatial heterogeneity brought about by cellular structures into Turing-type models seems to be promising to account for many intricate dynamic processes within single cells. For example, it was shown recently that nonlinear interactions between prototypic activator and inhibitor on the plasma membrane can lead to the emergence of Turing’s spatial patterns even for equal diffusion constants, provided that the exchange rates between the membrane and cytoplasm or decay rates in the cytoplasm are different for the activator and inhibitor101. Likewise, a Turing-type model was shown to account for the spontaneous initiation of cell polarization by the small GTPase Cdc42, its GEF Cdc24 and the effector protein bud emergence 1, which shuttle between the membrane and cytoplasm102. Alternative models of cell polarization based on bistable kinetics of the Cdc42–Rac–Rho network have also been proposed103. These discrepancies may reflect the infancy of our understanding, or simply the fact that nature has evolved more than one solution.
Conclusions
We have changed our perception of signalling pathways from linear pipelines to networks. We also have begun to rationalize how these network structures can determine the kinetics of distinct biochemical processes with high fidelity to translate them into specific biological responses. Along this way we have realized that specificity is generated by combinatorial assemblies and spatiotemporal dynamics rather than by a large number of genes with specific functions. We now face the challenge of explaining why evolution chose combinatorial assemblies over single pathway deterministic solutions. An obvious advantage of the former is that successful designs can be recycled and adapted for new purposes. Spatial and temporal separation can be a convenient means to specify signalling functions. This suggests that cell shape has an important role as it defines the spatial coordinates. Intriguing first glimpses were provided by work showing that cell shape controls the dynamics of localized biochemical activities104,105. This also highlights the need for new approaches, both conceptually and technologically, to move hand in hand for developing the insights and tools that allow us to survey the complex landscape of cell signalling.
Supplementary Material
Acknowledgments
We thank M. Tsyganov, J. Muñoz García, A. Kiyatkin and N. Kaimachnikov for discussions. This work was supported by Science Foundation Ireland under Grant No. 06/CE/B1129 and National Institutes of Health grants GM059570, GM066717. We apologize about not citing many pertinent contributions to the field because of space limitations.
Glossary
- Temporal dynamics
A quantitative description of how the system’s behaviour changes over time.
- Pheochromocytoma
An adrenal gland tumour that originates from cells derived from the neural crest.
- Adenocarcinoma
A cancer arising in glandular parts of epithelial tissues.
- Non-processive
Reaction mechanism in which the reactants dissociate after each partial reaction and have to encounter again for a new reaction. For instance, MEK phosphorylates ERK on two sites in a non-processive reaction that requires two distinct MEK–ERK interactions, in which only one site is phosphorylated per interaction event.
- Spatiotemporal dynamics
A description of how the system behaviour changes in space and time.
- Perfect adaptation
A term that, in control engineering, indicates the control strategy ensuring that the system output follows the desired course regardless of noise and variations in system parameters.
- Damped oscillations
Oscillations the amplitude of which decreases to zero while the system approaches a steady state.
- Sustained oscillations
Oscillations that continue indefinitely in time with constant amplitude and frequency.
- Guanine nucleotide exchange factor
A protein that catalyses the exchange of GDP for GTP for a GTP-binding protein.
- Ordinary differential equation
An equation in which differentiation occurs with respect to only a single independent variable, which is time for chemical kinetic equations.
- Analogue signal
A signal the quantity of which (for example, the concentration or activity (amplitude)) changes continuously in time and space, gradually increasing or decreasing.
- Digital output
A non-continuous signal that displays discrete levels, for instance zero or one.
- Interactome
The complete set of protein–protein interactions in a cell or organism. Interactome is often also used to designate the set of interaction partners of individual proteins.
- Partial differential equation
Contains partial derivatives with respect to two or more independent variables, which are time and the spatial coordinates for reaction–diffusion equations.
- GTPase-activating protein
A protein that facilitates the hydrolysis of GTP by a GTP-binding protein.
- Nanocluster
A transient, nanoscale array of plasma membrane proteins formed by lipid sorting and/or protein–protein interactions.
- 4-Pi
A laser scanning fluorescence microscope that uses two opposing objectives to improve axial spatial resolution.
- Stimulated emission depletion
A fluorescence microscopy technique that uses nonlinear de-excitation of fluorescent dyes to improve the spatial resolution of standard confocal microscopy.
- Farnesylation
A post-translational modification in which a farnesyl group (a hydrophobic group of three isoprene units) is conjugated to proteins, such as Ras GTPases, that contain a C-terminal CAAX motif. Farnesylation promotes attachment of the modified proteins to membranes.
- Hysteresis
A system that relates current inputs to different steady-state outputs, depending on the previous state of the system; that is, hysteresis provides a memory function to a system.
- Heterotrimeric G protein
A protein complex of three proteins (Gα, Gβ and Gγ). Gβ and Gγ form a tight complex, whereas Gα is part of the complex in its inactive, GDP-bound, form but dissociates in its active, GTP-bound, form. Both Gα and Gβγ can transmit downstream signals after activation.
- Phosphorylation gradient
A gradual change in the fraction of phosphorylated protein with distance
Footnotes
Competing interests statement
The authors declare no competing financial interests.
DATABASES
UniProtKB: http://www.uniprot.org ERK1 | ERK2 | Fus3 | HRAS | KRAS | NRAS | RAF | RAN | Ste5
FURTHER INFORMATION
Boris N. Kholodenko’s homepages: http://www.cellnetworks.org/nav.cgi?page=welcome; http://www.ucd.ie/sbi/peoplepartners/profiles/professorboriskholodenko
Nature pathway interaction database for Ras: http://pid.nci.nih.gov/search/intermediate_landing.shtml?molecule=RAS+family
Nature pathway interaction database for MEK: http://pid.nci.nih.gov/search/intermediate_landing.shtml?molecule=MEK1-2
Nature pathway interaction database for ERK: http://pid.nci.nih.gov/search/intermediate_landing.shtml?molecule=erk
SUPPLEMENTARY INFORMATION
See online article: S1 (table)
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1. Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. A conceptual breakthrough summarizing many experimental observations that different durations of ERK activity can result in different phenotypic responses.
- 2.Murphy LO, MacKeigan JP, Blenis J. A network of immediate early gene products propagates subtle differences in mitogen-activated protein kinase signal amplitude and duration. Mol. Cell. Biol. 2004;24:144–153. doi: 10.1128/MCB.24.1.144-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. von Kriegsheim A, et al. Cell fate decisions are specified by the dynamic ERK interactome. Nature Cell Biol. 2009;11:1458–1464. doi: 10.1038/ncb1994. Provides insight into a full set of protein–protein interactions involving ERK, and shows how ERK partners control ERK spatiotemporal dynamics and cell decisions.
- 4.Nakakuki T, et al. Ligand-specific c-Fos expression emerges from the spatiotemporal control of ErbB network dynamics. Cell. 2010 May 20; doi: 10.1016/j.cell.2010.03.054. (doi:10.16/j.cell.2010.03.054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meloche S, Pouyssegur J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene. 2007;26:3227–3239. doi: 10.1038/sj.onc.1210414. [DOI] [PubMed] [Google Scholar]
- 6.Nagashima T, et al. Quantitative transcriptional control of ErbB receptor signaling undergoes graded to biphasic response for cell differentiation. J. Biol. Chem. 2007;282:4045–4056. doi: 10.1074/jbc.M608653200. [DOI] [PubMed] [Google Scholar]
- 7.McCawley LJ, Li S, Wattenberg EV, Hudson LG. Sustained activation of the mitogen-activated protein kinase pathway. A mechanism underlying receptor tyrosine kinase specificity for matrix metalloproteinase-9 induction and cell migration. J. Biol. Chem. 1999;274:4347–4353. doi: 10.1074/jbc.274.7.4347. [DOI] [PubMed] [Google Scholar]
- 8. Kholodenko BN. Negative feedback and ultrasensitivity can bring about oscillations in the mitogen-activated protein kinase cascades. Eur. J. Biochem. 2000;267:1583–1588. doi: 10.1046/j.1432-1327.2000.01197.x. Predicted sustained MAPK oscillations that were later discovered experimentally (see references 12 and 14).
- 9.Brightman FA, Fell DA. Differential feedback regulation of the MAPK cascade underlies the quantitative differences in EGF and NGF signalling in PC12 cells. FEBS Lett. 2000;482:169–174. doi: 10.1016/s0014-5793(00)02037-8. [DOI] [PubMed] [Google Scholar]
- 10. Kiyatkin A, et al. Scaffolding protein Grb2-associated binder 1 sustains epidermal growth factor-induced mitogenic and survival signaling by multiple positive feedback loops. J. Biol. Chem. 2006;281:19925–19938. doi: 10.1074/jbc.M600482200. A working model of combinatorially complex interactions of multidomain proteins that control phosphoinositide 3-kinase and ERK pathway crosstalk.
- 11.Birtwistle MR, et al. Ligand-dependent responses of the ErbB signaling network: experimental and modeling analyses. Mol. Syst. Biol. 2007;3:144. doi: 10.1038/msb4100188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakayama K, Satoh T, Igari A, Kageyama R, Nishida E. FGF induces oscillations of Hes1 expression and Ras/ERK activation. Curr. Biol. 2008;18:R332–R334. doi: 10.1016/j.cub.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Das J, et al. Digital signaling and hysteresis characterize Ras activation in lymphoid cells. Cell. 2009;136:337–351. doi: 10.1016/j.cell.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shankaran H, et al. Rapid and sustained nuclear-cytoplasmic ERK oscillations induced by epidermal growth factor. Mol. Syst. Biol. 2009;5:332. doi: 10.1038/msb.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dougherty MK, et al. Regulation of Raf-1 by direct feedback phosphorylation. Mol. Cell. 2005;17:215–224. doi: 10.1016/j.molcel.2004.11.055. [DOI] [PubMed] [Google Scholar]
- 16.Shin SY, et al. Positive- and negative-feedback regulations coordinate the dynamic behavior of the Ras-Raf-MEK-ERK signal transduction pathway. J. Cell Sci. 2009;122:425–435. doi: 10.1242/jcs.036319. [DOI] [PubMed] [Google Scholar]
- 17.Douville E, Downward J. EGF induced SOS phosphorylation in PC12 cells involves P90 RSK-2. Oncogene. 1997;15:373–383. doi: 10.1038/sj.onc.1201214. [DOI] [PubMed] [Google Scholar]
- 18.Markevich NI, Hoek JB, Kholodenko BN. Signaling switches and bistability arising from multisite phosphorylation in protein kinase cascades. J. Cell Biol. 2004;164:353–359. doi: 10.1083/jcb.200308060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiao L, Nachbar RB, Kevrekidis IG, Shvartsman SY. Bistability and oscillations in the Huang-Ferrell model of MAPK signaling. PLoS Comput. Biol. 2007;3:1819–1826. doi: 10.1371/journal.pcbi.0030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kholodenko BN. Untangling the signalling wires. Nature Cell Biol. 2007;9:247–249. doi: 10.1038/ncb0307-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kholodenko BN, et al. Untangling the wires: a strategy to trace functional interactions in signaling and gene networks. Proc. Natl Acad. Sci. USA. 2002;99:12841–12846. doi: 10.1073/pnas.192442699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Santos SD, Verveer PJ, Bastiaens PI. Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nature Cell Biol. 2007;9:324–330. doi: 10.1038/ncb1543. Direct experimental determination of context-dependent and time-varying topology of dynamic connections between MAPK cascade components.
- 23.Brondello JM, Brunet A, Pouyssegur J, McKenzie FR. The dual specificity mitogen-activated protein kinase phosphatase-1 and -2 are induced by the p42/p44MAPK cascade. J. Biol. Chem. 1997;272:1368–1376. doi: 10.1074/jbc.272.2.1368. [DOI] [PubMed] [Google Scholar]
- 24.Patterson KI, Brummer T, O’Brien PM, Daly RJ. Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem. J. 2009;418:475–489. doi: 10.1042/bj20082234. [DOI] [PubMed] [Google Scholar]
- 25.Amit I, et al. A module of negative feedback regulators defines growth factor signaling. Nature Genet. 2007;39:503–512. doi: 10.1038/ng1987. [DOI] [PubMed] [Google Scholar]
- 26.Legewie S, Herzel H, Westerhoff HV, Bluthgen N. Recurrent design patterns in the feedback regulation of the mammalian signalling network. Mol. Syst. Biol. 2008;4:190. doi: 10.1038/msb.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kholodenko BN. Cell-signalling dynamics in time and space. Nature Rev. Mol. Cell Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hilioti Z, et al. Oscillatory phosphorylation of yeast Fus3 MAP kinase controls periodic gene expression and morphogenesis. Curr. Biol. 2008;18:1700–1706. doi: 10.1016/j.cub.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sigal A, et al. Variability and memory of protein levels in human cells. Nature. 2006;444:643–646. doi: 10.1038/nature05316. [DOI] [PubMed] [Google Scholar]
- 30.Tyson JJ, Chen KC, Novak B. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr. Opin. Cell Biol. 2003;15:221–231. doi: 10.1016/s0955-0674(03)00017-6. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Hao N, Dohlman HG, Elston TC. Bistability, stochasticity, and oscillations in the mitogen-activated protein kinase cascade. Biophys. J. 2006;90:1961–1978. doi: 10.1529/biophysj.105.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaimachnikov NP, Kholodenko BN. Toggle switches, pulses and oscillations are intrinsic properties of the Src activation/deactivation cycle. FEBS J. 2009;276:4102–4118. doi: 10.1111/j.1742-4658.2009.07117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrell JE, Jr, Machleder EM. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–898. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- 34.Bagowski CP, Ferrell JE., Jr Bistability in the JNK cascade. Curr. Biol. 2001;11:1176–1182. doi: 10.1016/s0960-9822(01)00330-x. [DOI] [PubMed] [Google Scholar]
- 35. Freedman TS, et al. A Ras-induced conformational switch in the Ras activator Son of sevenless. Proc. Natl Acad. Sci. USA. 2006;103:16692–16697. doi: 10.1073/pnas.0608127103. Direct experimental evidence of Ras–SOS positive feedback.
- 36. Murphy LO, Smith S, Chen RH, Fingar DC, Blenis J. Molecular interpretation of ERK signal duration by immediate early gene products. Nature Cell Biol. 2002;4:556–564. doi: 10.1038/ncb822. Demonstrates how a short and prolonged duration of ERK signalling can be sensed at the level of IEGs.
- 37.Mangan S, Zaslaver A, Alon U. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J. Mol. Biol. 2003;334:197–204. doi: 10.1016/j.jmb.2003.09.049. [DOI] [PubMed] [Google Scholar]
- 38.Vomastek T, et al. Modular construction of a signaling scaffold: MORG1 interacts with components of the ERK cascade and links ERK signaling to specific agonists. Proc. Natl Acad. Sci. USA. 2004;101:6981–6986. doi: 10.1073/pnas.0305894101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teis D, Wunderlich W, Huber L. A. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev. Cell. 2002;3:803–814. doi: 10.1016/s1534-5807(02)00364-7. [DOI] [PubMed] [Google Scholar]
- 40.Vallabhapurapu S, Karin M. Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 41.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF- κB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 42. Ashall L, et al. Pulsatile stimulation determines timing and specificity of NF-κB-dependent transcription. Science. 2009;324:242–246. doi: 10.1126/science.1164860. Provides a combined experimental and mathematical analysis of the nucleo-cytoplasmic shuttling cycles of NF-κB and how they relate to specifying gene expression.
- 43.Kim D, Kolch W, Cho KH. Multiple roles of the NF-κB signaling pathway regulated by coupled negative feedback circuits. FASEB J. 2009;23:2796–2802. doi: 10.1096/fj.09-130369. [DOI] [PubMed] [Google Scholar]
- 44.Beene DL, Scott JD. A-kinase anchoring proteins take shape. Curr. Opin. Cell Biol. 2007;19:192–198. doi: 10.1016/j.ceb.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nature Rev. Mol. Cell Biol. 2005;6:827–837. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- 46.McKay MM, Morrison DK. Integrating signals from RTKs to ERK/MAPK. Oncogene. 2007;26:3113–3121. doi: 10.1038/sj.onc.1210394. [DOI] [PubMed] [Google Scholar]
- 47.Shaw AS, Filbert EL. Scaffold proteins and immune-cell signalling. Nature Rev. Immunol. 2009;9:47–56. doi: 10.1038/nri2473. [DOI] [PubMed] [Google Scholar]
- 48.DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. β-arrestins and cell signaling. Annu. Rev. Physiol. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- 49.Perry SJ, et al. Targeting of cyclic AMP degradation to β2-adrenergic receptors by β-arrestins. Science. 2002;298:834–836. doi: 10.1126/science.1074683. [DOI] [PubMed] [Google Scholar]
- 50.Nelson CD, et al. Targeting of diacylglycerol degradation to M1 muscarinic receptors by β-arrestins. Science. 2007;315:663–666. doi: 10.1126/science.1134562. [DOI] [PubMed] [Google Scholar]
- 51.Tanoue T, Nishida E. Molecular recognitions in the MAP kinase cascades. Cell Signal. 2003;15:455–462. doi: 10.1016/s0898-6568(02)00112-2. [DOI] [PubMed] [Google Scholar]
- 52. Levchenko A, Bruck J, Sternberg PW. Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc. Natl Acad. Sci. USA. 2000;97:5818–5823. doi: 10.1073/pnas.97.11.5818. A kinetic model showing that scaffold organization of a kinase cascade markedly changes the input–output relationships.
- 53. Casar B, et al. Ras subcellular localization defines extracellular signal-regulated kinase 1 and 2 substrate specificity through distinct utilization of scaffold proteins. Mol. Cell Biol. 2009;29:1338–1353. doi: 10.1128/MCB.01359-08. Provides insight in how Ras signalling from different membrane compartments uses different scaffold proteins for the ERK pathway to selectively target downstream ERK substrates.
- 54. Chiu VK, et al. Ras signalling on the endoplasmic reticulum and the Golgi. Nature Cell Biol. 2002;4:343–350. doi: 10.1038/ncb783. Shows that Ras signalling can emanate from different subcellular membrane compartments and activate different downstream pathways.
- 55.Rajakulendran T, Sahmi M, Lefrancois M, Sicheri F, Therrien M. A dimerization-dependent mechanism drives RAF catalytic activation. Nature. 2009;461:542–545. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 56.Bhattacharyya RP, et al. The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science. 2006;311:822–826. doi: 10.1126/science.1120941. [DOI] [PubMed] [Google Scholar]
- 57.Good M, Tang G, Singleton J, Remenyi A, Lim WA. The Ste5 scaffold directs mating signaling by catalytically unlocking the Fus3 MAP kinase for activation. Cell. 2009;136:1085–1097. doi: 10.1016/j.cell.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takahashi S, Pryciak PM. Membrane localization of scaffold proteins promotes graded signaling in the yeast MAP kinase cascade. Curr. Biol. 2008;18:1184–1191. doi: 10.1016/j.cub.2008.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rizzuto R, et al. Ca2+ transfer from the ER to mitochondria: when, how and why. Biochim. Biophys. Acta. 2009;1787:1342–1351. doi: 10.1016/j.bbabio.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kornmann B, et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roos J, et al. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J. Cell Biol. 2005;169:435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park CY, et al. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 2009;136:876–890. doi: 10.1016/j.cell.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kusumi A, et al. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 2005;34:351–378. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- 64.Simons K, Toomre D. Lipid rafts and signal transduction. Nature Rev. Mol. Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 65.Eggeling C, et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 66.Sharma P, et al. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell. 2004;116:577–589. doi: 10.1016/s0092-8674(04)00167-9. [DOI] [PubMed] [Google Scholar]
- 67.Hancock JF. Lipid rafts: contentious only from simplistic standpoints. Nature Rev. Mol. Cell Biol. 2006;7:456–462. doi: 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hanzal-Bayer MF, Hancock JF. Lipid rafts and membrane traffic. FEBS Lett. 2007;581:2098–2104. doi: 10.1016/j.febslet.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 69.Nicolau DV, Jr, Burrage K, Parton RG, Hancock JF. Identifying optimal lipid raft characteristics required to promote nanoscale protein-protein interactions on the plasma membrane. Mol. Cell Biol. 2006;26:313–323. doi: 10.1128/MCB.26.1.313-323.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hancock JF, Paterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- 71.Hancock JF, Parton RG. Ras plasma membrane signalling platforms. Biochem. J. 2005;389:1–11. doi: 10.1042/BJ20050231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Plowman SJ, Muncke C, Parton RG, Hancock JF. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc. Natl Acad. Sci. USA. 2005;102:15500–15505. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Murakoshi H, et al. Single-molecule imaging analysis of Ras activation in living cells. Proc. Natl Acad. Sci. USA. 2004;101:7317–7322. doi: 10.1073/pnas.0401354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Belanis L, Plowman SJ, Rotblat B, Hancock JF, Kloog Y. Galectin-1 is a novel structural component and a major regulator of h-ras nanoclusters. Mol. Biol. Cell. 2008;19:1404–1414. doi: 10.1091/mbc.E07-10-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shalom-Feuerstein R, et al. K-ras nanoclustering is subverted by overexpression of the scaffold protein galectin-3. Cancer Res. 2008;68:6608–6616. doi: 10.1158/0008-5472.CAN-08-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Plowman SJ, Ariotti N, Goodall A, Parton RG, Hancock JF. Electrostatic interactions positively regulate K-Ras nanocluster formation and function. Mol. Cell. Biol. 2008;28:4377–4385. doi: 10.1128/MCB.00050-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abankwa D, Gorfe AG, Inder K, Hancock JF. Ras membrane orientation and nanodomain localization generate isoform diversity. Proc. Natl Acad. Sci. USA. 2010;107:1130–1135. doi: 10.1073/pnas.0903907107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tian T, et al. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nature Cell Biol. 2007;9:905–914. doi: 10.1038/ncb1615. [DOI] [PubMed] [Google Scholar]
- 79. Harding A, Tian T, Westbury E, Frische E, Hancock JF. Subcellular localization determines MAP kinase signal output. Curr. Biol. 2005;15:869–873. doi: 10.1016/j.cub.2005.04.020. Shows that in mammalian cells the MAPK cascade can operate as a switch with different sensitivity to the input signals from the plasma membrane and cytoplasm.
- 80.Inder K, et al. Activation of the MAPK module from different spatial locations generates distinct system outputs. Mol. Biol. Cell. 2008;19:4776–4784. doi: 10.1091/mbc.E08-04-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harding AS, Hancock JF. Using plasma membrane nanoclusters to build better signaling circuits. Trends Cell Biol. 2008;18:364–371. doi: 10.1016/j.tcb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Suzuki KG, Fujiwara TK, Edidin M, Kusumi A. Dynamic recruitment of phospholipase Cγ at transiently immobilized GPI-anchored receptor clusters induces IP3-Ca2+ signaling: single-molecule tracking study 2. J. Cell Biol. 2007;177:731–742. doi: 10.1083/jcb.200609175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Suzuki KG, et al. GPI-anchored receptor clusters transiently recruit Lyn and Gα for temporary cluster immobilization and Lyn activation: single-molecule tracking study 1. J. Cell Biol. 2007;177:717–730. doi: 10.1083/jcb.200609174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goswami D, et al. Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell. 2008;135:1085–1097. doi: 10.1016/j.cell.2008.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Daniels MA, et al. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 2006;444:724–729. doi: 10.1038/nature05269. [DOI] [PubMed] [Google Scholar]
- 86. Turing AM. The chemical basis of morphogenesis. Phil. Trans. R. Soc. Lond. B Biol. Sci. 1952;237:37–72. Shows that diffusion can destabilize spatially uniform steady-state distribution, resulting in heterogeneous spatial concentration patterns.
- 87.Gierer A. Generation of biological patterns and form: some physical, mathematical, and logical aspects. Prog. Biophys. Mol. Biol. 1981;37:1–47. doi: 10.1016/0079-6107(82)90019-0. [DOI] [PubMed] [Google Scholar]
- 88. Brown GC, Kholodenko BN. Spatial gradients of cellular phospho-proteins. FEBS Lett. 1999;457:452–454. doi: 10.1016/s0014-5793(99)01058-3. Shows that the spatial separation of opposing enzymes in a protein modification cycle brings about protein activity gradients and non-uniform spatial profiles.
- 89.Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- 90.Maeder CI, et al. Spatial regulation of Fus3 MAP kinase activity through a reaction-diffusion mechanism in yeast pheromone signalling. Nature Cell Biol. 2007;9:1319–1326. doi: 10.1038/ncb1652. [DOI] [PubMed] [Google Scholar]
- 91.Yudushkin IA, et al. Live-cell imaging of enzyme-substrate interaction reveals spatial regulation of PTP1B. Science. 2007;315:115–119. doi: 10.1126/science.1134966. [DOI] [PubMed] [Google Scholar]
- 92.Fuller BG, et al. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 2008;453:1132–1136. doi: 10.1038/nature06923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moseley JB, Mayeux A, Paoletti A, Nurse P. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature. 2009;459:857–860. doi: 10.1038/nature08074. [DOI] [PubMed] [Google Scholar]
- 94.Kholodenko BN. Spatially distributed cell signalling. FEBS Lett. 2009;583:4006–4012. doi: 10.1016/j.febslet.2009.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stelling J, Kholodenko BN. Signaling cascades as cellular devices for spatial computations. J. Math. Biol. 2008;58:35–55. doi: 10.1007/s00285-008-0162-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kholodenko BN. MAP kinase cascade signaling and endocytic trafficking: a marriage of convenience? Trends Cell Biol. 2002;12:173–177. doi: 10.1016/s0962-8924(02)02251-1. Shows theoretically that the propagation of phosphorylation signals solely by diffusion can be terminated by cytoplasmic phosphatases. The study suggests that in large cells, motor-driven trafficking of endosomes and scaffolds carrying phosphorylated kinases or assembled signalling complexes is required for signal transduction.
- 97.Munoz-Garcia J, Neufeld Z, Kholodenko BN. Positional information generated by spatially distributed signaling cascades. PLoS Comput. Biol. 2009;5:e1000330. doi: 10.1371/journal.pcbi.1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol. Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 99.Athale CA, et al. Regulation of microtubule dynamics by reaction cascades around chromosomes. Science. 2008;322:1243–1247. doi: 10.1126/science.1161820. [DOI] [PubMed] [Google Scholar]
- 100. Markevich NI, Tsyganov MA, Hoek JB, Kholodenko BN. Long-range signaling by phosphoprotein waves arising from bistability in protein kinase cascades. Mol. Syst. Biol. 2006;2:61. doi: 10.1038/msb4100108. Shows the possibility of waves of protein phosphorylation travelling through the cytoplasm or long neuron axons.
- 101.Levine H, Rappel WJ. Membrane-bound Turing patterns. Phys. Rev. E Stat. Nonlin Soft Matter Phys. 2005;72 doi: 10.1103/PhysRevE.72.061912. 061912. [DOI] [PubMed] [Google Scholar]
- 102.Goryachev AB, Pokhilko AV. Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Lett. 2008;582:1437–1443. doi: 10.1016/j.febslet.2008.03.029. [DOI] [PubMed] [Google Scholar]
- 103.Mori Y, Jilkine A, Edelstein-Keshet L. Wave-pinning and cell polarity from a bistable reaction-diffusion system. Biophys. J. 2008;94:3684–3697. doi: 10.1529/biophysj.107.120824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Meyers J, Craig J, Odde DJ. Potential for control of signaling pathways via cell size and shape. Curr. Biol. 2006;16:1685–1693. doi: 10.1016/j.cub.2006.07.056. [DOI] [PubMed] [Google Scholar]
- 105.Neves SR, et al. Cell shape and negative links in regulatory motifs together control spatial information flow in signaling networks. Cell. 2008;133:666–680. doi: 10.1016/j.cell.2008.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Borisov N, et al. Systems-level interactions between insulin-EGF networks amplify mitogenic signaling. Mol. Syst. Biol. 2009;5:256. doi: 10.1038/msb.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang CC, Cirit M, Haugh JM. PI3K-dependent cross-talk interactions converge with Ras as quantifiable inputs integrated by Erk. Mol. Syst. Biol. 2009;5:246. doi: 10.1038/msb.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Berezhkovskii AM, Coppey M, Shvartsman SY. Signaling gradients in cascades of two-state reaction-diffusion systems. Proc. Natl Acad. Sci. USA. 2009;106:1087–1092. doi: 10.1073/pnas.0811807106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kholodenko BN, Kolch W. Giving space to cell signaling. Cell. 2008;133:566–567. doi: 10.1016/j.cell.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 110.Neves SR, Iyengar R. Models of spatially restricted biochemical reaction systems. J. Biol. Chem. 2009;284:5445–5449. doi: 10.1074/jbc.R800058200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kholodenko BN. Four-dimensional organization of protein kinase signaling cascades: the roles of diffusion, endocytosis and molecular motors. J. Exp. Biol. 2003;206:2073–2082. doi: 10.1242/jeb.00298. [DOI] [PubMed] [Google Scholar]
- 112.Rishal I, Fainzilber M. Retrograde signaling in axonal regeneration. Exp. Neurol. 2009;223:5–10. doi: 10.1016/j.expneurol.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 113.Luttrell LM. Composition and function of G protein-coupled receptor signalsomes controlling mitogen-activated protein kinase activity. J. Mol. Neurosci. 2005;26:253–264. doi: 10.1385/JMN:26:2-3:253. [DOI] [PubMed] [Google Scholar]
- 114.Fehrenbacher N, Bar-Sagi D, Philips M. Ras/MAPK signaling from endomembranes. Mol. Oncol. 2009;3:297–307. doi: 10.1016/j.molonc.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.