Abstract
In this study, we correlated array-comparative genomic hybridization-defined abnormalities with survival in two different cohorts of patients treated with therapy based on high-dose melphalan with autologous stem-cell transplantation (64 from the Mayo Clinic and 67 from the University of Arkansas Medical School) and identified that several regions of genomic gains and losses were significantly associated with poorer survival. Three noncontiguous survival relevant regions covering 1p31-33 and two noncontiguous regions covering 20p12.3-12.1 were common between the two datasets. The prognostic relevance of these hotspots was validated in an independent cohort using fluorescent in situ hybridization, which showed that 1p31-32 loss is significantly associated with shorter survival (24.5 months versus 40 months, log-rank P-value=0.01), whereas 20p12 loss has a trend toward shorter survival (26.3 months versus 40 months, log-rank P-value=0.06). On multivariate analysis, 1p31-32 loss is an independent prognostic factor. On further analysis, the prognostic impact of 1p31-32 loss is due to shortening of post-relapse survival as there is no impact on complete response rates and progression-free survival.
Keywords: prognosis, array-comparative genomic hybridization, chromosome 1p
Introduction
Multiple myeloma is a post-germinal B-cell malignancy characterized by accumulation of clonal plasma cells in the bone marrow.1 Despite the introduction of novel agents into the treatment armamentarium, the disease is still incurable at present. From G-banding or spectral karyotyping analysis, it is known that myeloma cells are characterized by complex genetic aberrations, suggesting underlying genomic instability.2 Several recurrent genetic abnormalities have been identified and a number of these, including t(4;14), 17p13 deletion, 1q amplification, and hypodiploid karyotype, have been associated with shorter survival.3–14 However, earlier studies are hampered by some important limitations. Karyotypically defined survival-associated genetic aberrations are limited to 30% or less of patients that produced informative karyotype.15 In addition, the prognostic impact of karyotypic abnormalities may be due in large part to the proliferative nature of malignant cells that produce metaphases.16 Furthermore, the resolution of G-banding is low, and some abnormalities are karyotypically invisible. Fluorescent in situ hybridization (FISH)-based assay on interphase cells allows acquisition of useful information on larger number of patients, but is hampered by its obvious limitations as a discovery tool. Therefore, its application so far is limited to detecting recurrent abnormalities earlier defined by karyotyping or molecular methods.
Array-comparative genomic hybridization (aCGH) negates many of these limitations by having significantly higher resolution, not requiring metaphase cells, and allowing detection of genome-wide copy number changes. It, therefore, lends itself as a powerful discovery tool. Indeed, several earlier unappreciated genetic abnormalities have already been discovered in MM using this type of platform.17–20 In this study, we aim to define recurrent aCGH-defined genomic aberrations that are significantly associated with shorter survival and validated these using FISH in an independent cohort of patients.
Materials and methods
aCGH and gene expression profiling datasets
A Mayo Clinic dataset and the publicly available University of Arkansas Medical School (UAMS) dataset18 comprising 64 and 67 newly diagnosed multiple myeloma, respectively, were analyzed. The patients from Mayo Clinic were predominantly treated with dexamethasone-based induction therapy and single autologous stem-cell transplantation, whereas patients from the published dataset were treated with a stem-cell transplantation-based intensive sequential therapy called total therapy. For the Mayo Clinic dataset, aCGH hybridizations were performed using oligonucleotide CGH microarrays (Agilent Technologies, Santa Clara, CA, USA; Human Genome CGH Microarray 44B) that contained ~44 000 coding and noncoding human genome probes uniquely mapped to human genome. For the UAMS dataset, aCGH hybridizations were performed using oligonucleotide expression profiling microarrays (Agilent Technologies, Human 1A v2) that contained 22 500 probes overall among which 16 097 unique map positions were defined. For the Mayo Clinic dataset, gene expression profiling was performed using the Affymetrix U133A V2 chip (data available from GEO through accession number GSE6477), whereas for the UAMS dataset, gene expression was performed using the Affymetrix U133plus 2.0 chip (data available from GEO through accession number GSE2658).
Clinical validation dataset
We studied a cohort of 127 patients at the Mayo Clinic that had been treated with melphalan-based high-dose therapy. These patients are part of a transplant cohort that had already been used in earlier publications.14 In all cases studied, a research bone marrow sample had been obtained before high-dose therapy. All patients gave written informed consent for the collection and long-term storage of additional cells at the time of the bone marrow examination, including separate authorization for genetic studies and separate consent for review of their charts in accord with both federal regulations and Health Insurance Portability and Accountability Act guidelines. The consent was approved by the Institutional Review Board of the Mayo Clinic College of Medicine. Cytospin slides were made in all cases and the remaining sample was purified using CD138 magnetic beads when possible and stored in liquid nitrogen for future use. All patients in whom available samples were obtained were used in this analysis.
Detection of genomic aberration
For all datasets, log-ratios were treated as primary measure for copy number ratio between tumor and normal samples. To detect the regions of genomic aberration, we first constructed an artificial ‘reference’ chromosome of size 1000 for each sample and log-ratios of a sliding window (SW) of size 15 (~0.5Mb Windows) (25 for the UAMS dataset) was compared with the log-ratios of an artificial reference constructed for each sample. The ‘reference’ chromosome was constructed by recursively accepting 10 randomly sampled log-ratios from the autosomal regions whose P-value of Wilcoxon-signed-rank test against zero was larger than 0.05 until the size reach 1000. Then an SW of size 10 were constructed from the start of each chromosome and compared with the ‘reference’ chromosome by the Wilcoxon-rank sum test. The comparison for gains and losses were performed separately for each SW and this comparison procedure was repeated by advancing the SW for one probe each time until it reached the end of the chromosome. The P-values and their corresponding false discovery rates (q-values) for the copy number gain or loss were calculated independently for each SW (indicated as Pgain/Qgain or Ploss/Qloss) and assigned to the center position of the corresponding SW (Figure 1).
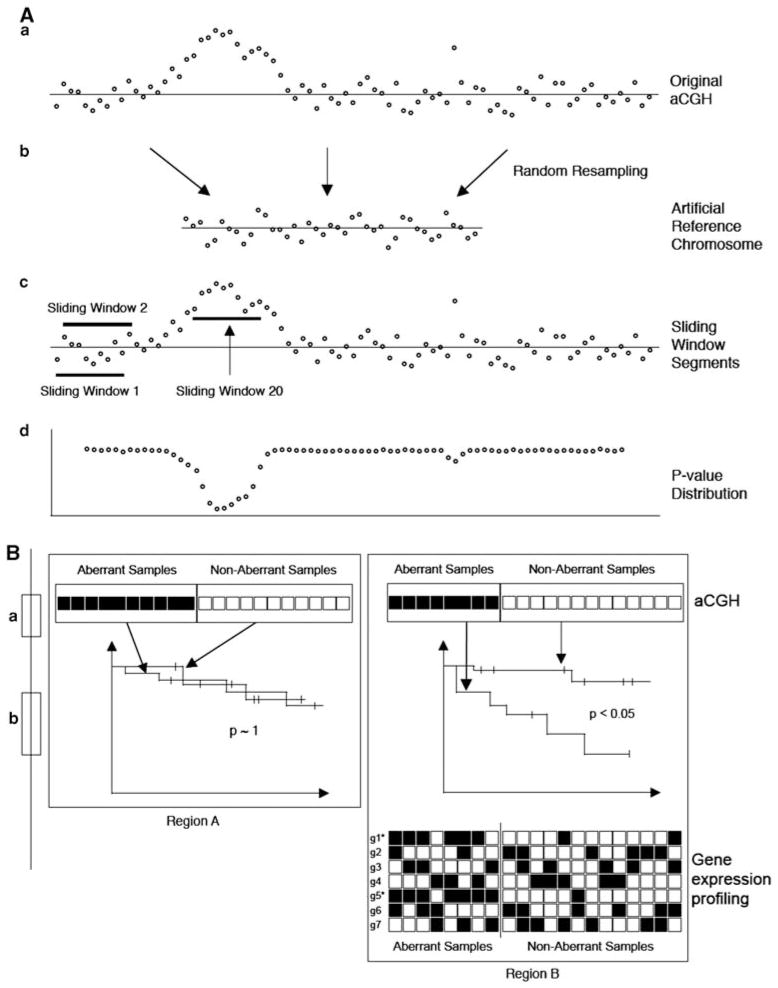
Figure 1.
The schematic diagram of the present approach. (A) Genomic aberration detection procedure and (B) the survival critical signature detection procedure are illustrated. (a) From the original aCGH, (b) a ‘reference’ chromosome is constructed and (c) a group of log-ratio values segmented by SWs are compared against those of the ‘reference’ chromosome, which subsequently result in (d) P-values. (B) Two chromosomal regions A and B are schematically compared here. aCGH data for each chromosomal regions are interrogated separately using the log-rank test to see whether survival difference is observed between samples that have genomic aberrations and those that do not. If statistically significant survival difference is observed (region B), expression profiles for genes in the chromosome region (g1,….,g7) are examined to see whether statistically significant association between copy number change and expression changes (gain and over-expression or loss and under-expression) can be detected using the Fisher’s exact test. Here, genes g1 and g5 show sufficient level of association and are marked*.
Detection of survival critical genomic aberrations
Survival difference was assessed between samples that have gain or loss of genomic regions as defined by the aforementioned method and those samples without using the method of Kaplan–Meier and the statistical significance was assessed by the log-rank test. The SW was identified as gained or lost if PGA<10−4 and QGA<10−3 (GA∈{gain, loss}). SW whose gains or losses led to poor prognosis with survival difference P-value ≤0.05 and happened to be ≥10% of all samples are considered survival critical and retained for further analysis. All calculations were performed using R system.
To detect the co-appearance of survival critical genomic aberrations (SCGAs) in different chromosomes, we performed unsupervised hierarchical clustering of a survival critical aberration matrix. For each chromosome, we first collected all SWs that were found to be survival critical and counted the number of SCGAs that each sample contained among the collected SWs. If a sample had SCGAs in more than half of all the collected SWs, it was given a +1 (gain) or −1 (loss) depending on the type of aberration for the chromosome. Otherwise, the sample was given a zero. The survival critical aberration matrix was constructed by collecting all the summary indicators across all chromosomes. In the hierarchical clustering, the distance between two vectors were measured by counting the number of elements that are the same, and complete linkage was used for linking two intermediate clusters.
Detection of survival critical gene expression
To prioritize genes that might be associated with poor survival, we measured the association between over- or under-expression of genes and the gain/loss of the same gene located within regions of gains/losses as detected by aCGH. This is measured for each gene within an SCGA and each sample. Specifically, we performed the Fisher’s exact test for each gene using the number of cases available in a 2×2 table. For the association between DNA gains and over-expression, the following categories will be reflected in the four cells: (1) over-expression on gained samples, (2) over-expression on not gained samples, (3) no over-expression on gained samples, and (4) no over-expression on not gained samples for the co-appearance of gains and over-expression. Analogous tests were performed for the co-appearance of losses and under-expression of genes.
MAS5.0-transformed gene expression data were median centered to the respective dataset. To identify over- and under-expression in the expression profiles, median normalized expression values of ≥1.5 were considered over-expressed and those ≤0.7 were considered under-expressed.
Fluorescent in situ hybridization
The cytoplasmic immunoglobulin light chain fluorescent in situ hybridization (cIg-FISH) technique that combines the interphase FISH strategy with fluorescent detection of cIg was used.21 For the detection of common chromosome abnormalities such as t(11;14)(q13;q32), t(4;14)(p16;q32), chromosome 13 deletion, 17p13 deletion, and 1q21 amplification, we used the same probes and conditions published earlier by us.4,14
To detect the survival critical locus detected from aCGH analysis, we used BAC RPCI11.C 960J16 whose insert contains PAK7 on 20p12 and BAC RPCI11.C 1141K22, which contains MSH4 on 1p31.1. To further ascertain the specificity of aCGH findings, we used BAC RPCI11.C 112C12, which contains PCSK2 on 20p11.2, and BAC CTD2542B21, which contains DAB1 on 1p32.2. We used CEP20 and fosmid G248P80487E4 located on 1p12 as a ploidy control.
Briefly, each BAC and fosmid was grown in selective media, and DNA was extracted and directly labeled with a Spectrum- Green or SpectrumOrange fluorophore by standard nick translation. PCR primers were designed to confirm that the insert was present and the labeled probe was then tested on normal metaphases to reaffirm localization of the probe and exclude cross-hybridization.
Statistical analysis
Descriptive statistics were used to characterize the patients. The Fisher’s exact test was used to test for association between translocations and clinical characteristics for continuous variables. The Wilcoxon-rank sum test was used to test for differences between patient groups on the basis of their translocation status. The Fisher’s exact test was used to test differences among levels of categorical variables among patients with FISH abnormalities. Distributions for survival and progression- free survival were estimated with the Kaplan–Meier method. Log-rank testing was used to assess differences in survival for significance. Multivariable analysis was performed by the Cox method.
Results
Prevalence of genomic aberrations
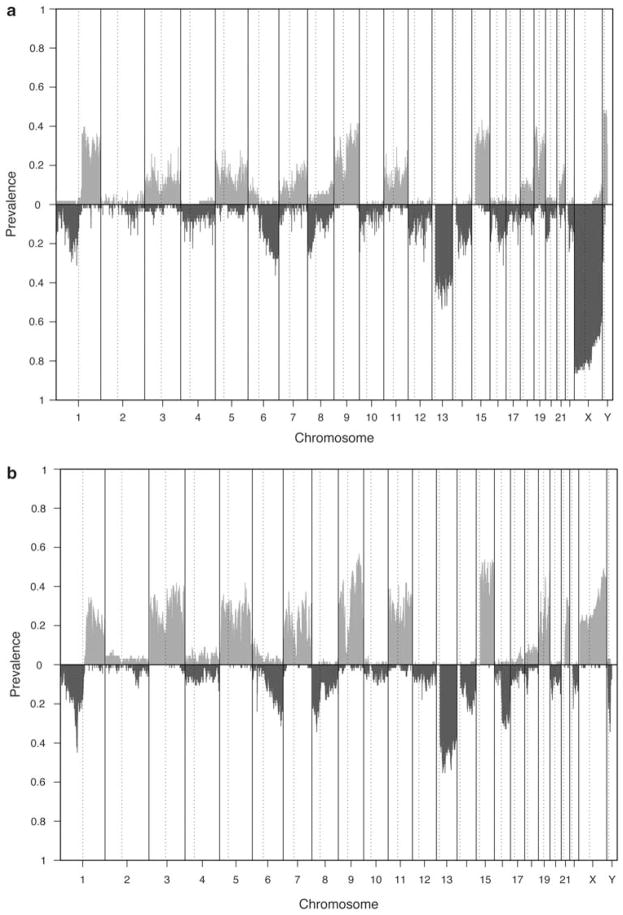
Although the resolution of the aCGH platforms used for the two datasets were different, the pattern of aberration were remarkably similar (Figure 2). The differences in the prevalence of aberrations on chromosome X and Y relates to the different use of control normal DNA in the two experiment protocol (control DNA from males in the Mayo dataset and control DNA from female in UAMS dataset). A number of aberrations in autosomal regions are common. Notably, gains in chromosomes 1q, 3, 5, 7, 9, 11, 15, 19, 21, and losses in chromosomes 1p, 6q, 8, 13q, 14q, 16q are prevalent and has been reported.18,19,22 The dominant gains in the odd-numbered chromosomes are due to the hyperdiploid MM subtypes, which are characterized by trisomies of chromosomes 3, 5, 7, 9, 11, 15, 19, 21.23 (Figure 2).
Figure 2.
Genomic aberration prevalence from (a) Mayo dataset and (b) UAMS dataset. Bars above x axis indicate prevalences for gains and below x axis for losses. Here, SWs whose PGA <10−4 and QGA<10−3 (GA ∈ {gain, loss}) are called aberrant.
Identification of survival critical regions and genes
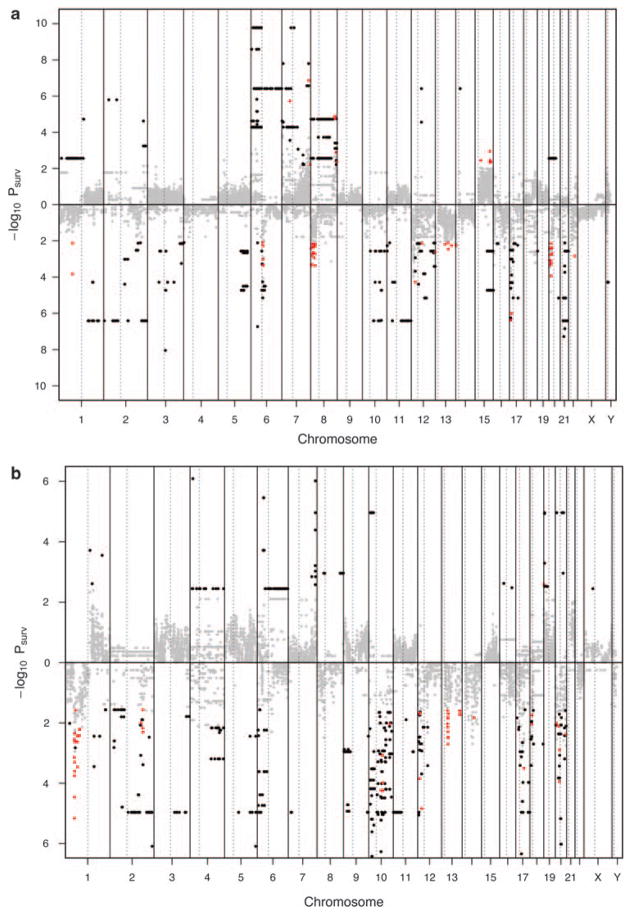
Clearly, not all the frequent aberrations are associated with poor prognosis (Figure 3). The number of SCGAs seems to be influenced to a certain extent by the resolution of the platform with the higher resolution Mayo Clinic dataset producing a much higher number of survival regions.
Figure 3.
Survival critical regions (SCR) from (a) Mayo dataset and (b) UAMS dataset. The log-rank test P-values for all SWs are shown with their chromosomal locations. Chromosomal locations in which the genomic aberrations led to poor survival with log-rank test P-value <0.05, corresponding q-value <0.05, and aberrations happened ≥10% of all samples are indicated in red. Chromosomal locations with log-rank test P-value <0.05, corresponding q-value <0.05, but aberrations happened < 10% of all samples are indicated in black.
The frequent gains observed in chromosomes 3, 5, 7, 9, 11, 15 were prognostically neutral, which is consistent with the known prognosis of H-MM.23,24 In the UAMS dataset, gain of several regions on 1q and loss of several regions on 1p are associated with shortened survival. In addition, 19p+, 13q−, 14q−, 18p−, and 20p− are also significantly associated with survival (Supplementary Table 1). For the Mayo Clinic dataset, gains of several regions on chromosome 5q, 7q, 8q, 15q, and loss of several regions on 1p, 8p, 12q, 13q, 16p, 16q, 17p, 20p, 22q are also significantly associated with survival (Supplementary Table 2). Only the following regions are common to the two dataset, including 1p33 gain, 1p31-32 loss, and 20p12 loss (two regions) (Table 1).
Table 1.
Survival hotspots common to Mayo and UAMS dataset Chr Cytoband Start End Gene
| Chr | Cytoband | Start | End | Gene |
|---|---|---|---|---|
| 1 | p33 | 48 536 864 | 50 284 012 | SPATA6, PPP1R8P, LOC644462, AGBL4, C1orf165 |
| 1 | p32.2-p32.1 | 57 095 535 | 58 723 069 | C8A, C8B, DAB1, LOC729423, LOC644928, LOC414357, OMA1 |
| 1 | p31.1 | 74 137 824 | 76 096 464 | LOC653733, LRRC44, FPGT, TNNI3K, C1orf173, CRYZ, C1orf171, LHX8, SLC44A5, LOC646417, LOC646425, ACADM, DLSTP, RABGGTB, SNORD45A, SNORD45B, MSH4 |
| 20 | p12.3-12.1 | 6 717 170 | 14 530 401 | pFUSIP1, HAO1, TXNDC13, PHKBP1, PLCB1, LOC728357, PLCB4, C20orf103, PAK7, ANKRD5, SNAP25, RPL23AP6, MKKS, C20orf94, JAG1, FAT1P1, HCG_2045828, RPS11P1, LOC441940, PGAM3P, LOC728573, LOC728450, BTBD3, PA2G4P2, C20orf38, C20orf82, LOC728600, TASP1, GAPDHP2, MRPS36P6, C20orf6, C20orf7, C20orf50, RPS3P1, SCYE1P, FLRT3, RNF11B |
| 20 | p12.1 | 14 772 372 | 17 558 841 | ENSAP, C20orf133, LOC613266, PPIAP17, LOC728628, C20orf23, RPLP0P1, RPL7AL3, SNRPB2, OTOR, PCSK2, BFSP1, RPS27AP2, DSTN, RRBP1 |
Abbreviation: UMAS, University of Arkansas Medical School.
We further assessed whether the genes contained within these common SCGAs had significantly lower mRNA expression in samples with and without the SCGAs. These are potentially the important genes deleted on these genomic regions that may impact survival (Table 1).
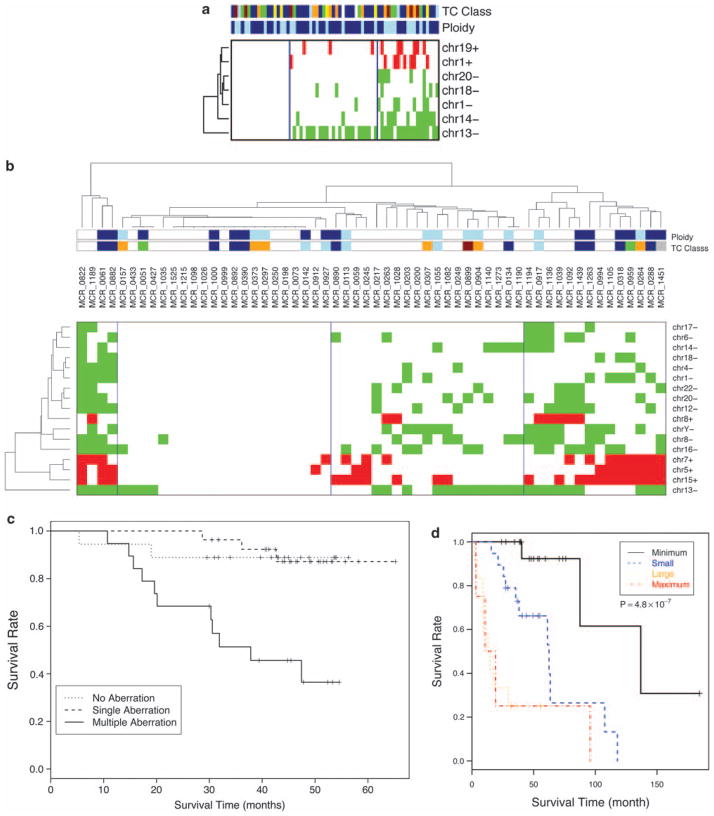
When these SCGAs are clustered, distinct patterns are seen in both datasets characterized by low, intermediate, or high number of these SCGAs. In both datasets, those patients with high number of SCGAs have significantly poorer survival. Of note, chromosome 13 deletion is seen across all the clusters from low to high SCGAs in both datasets. In fact, in some, it can be the only SCGA (Figure 4). This is consistent with it being one of the earliest SCGAs, whereas chromosome 1 abnormalities tend to occur in the setting of the cluster with the highest number of SCGAs, suggesting that it may be a late event.
Figure 4.
Clustering of samples according to the presence of survival critical regions in MM. Patients segregate into clusters based on the number of survival critical regions in both the (a) Mayo cohort and the (b) UAMS cohort. The clusters are not enriched for any ploidy or TC subtypes. The survival of patients is significant worse in tumors with the highest number of these survival critical regions in both the (c) Mayo and (d) UAMS cohort.
Validation of common survival critical region
We next validated several of these SCGAs using FISH in a separate dataset of 127 patients. For chromosome 1p, we used a probe against MSH4 on 1p31.1 and DAB1 on 1p32. Of the 127 patients, we had enough material for these assays in 122 patients. In these patients, the cases with loss of 1p31 overlapped with those with loss of 1p32 and so they were considered together as 1p31-32 loss. These abnormalities were detected in 19.7% of patients in our validation cohort. For 20p12.3-12.1, we used two probes targeting two genes (PAK7 and PCSK2) within this region. Material for FISH was available in 126 cases. In all cases, both genes were deleted together. The 20p12.3-12.1 loss was detected in 11.9% of patients. Loss of 1p31-32 and 20p12.3-12.1 occurred together in three patients.
The 1p31-32 loss had no significant correlation with other recurrent genetic aberrations such as 1q gain, t(4;14), t(11;14), 13 deletion, or 17p13 deletion, although the number of cases studied is small to draw any firm conclusions. On the other hand, 20p12.3-12.1 loss was significant associated with 17p13 deletion and a strong trend toward association with t(11;14) (Table 2).
Table 2.
Association of 1p loss and 20p loss with other recurrent genetic aberration
| Genetic aberrations | 1p 31-32 loss |
P-value | 20p12 loss |
P-value | ||
|---|---|---|---|---|---|---|
| Yes | No | Yes | No | |||
| t(11;14) | 0.74 | 0.05 | ||||
| Yes | 4 | 18 | 5 | 10 | ||
| No | 12 | 68 | 14 | 96 | ||
| t(4;14) | 0.52 | 0.38 | ||||
| Yes | 2 | 22 | 3 | 8 | ||
| No | 16 | 81 | 14 | 81 | ||
| 17p13 del | 1.00 | 0.001 | ||||
| Yes | 2 | 22 | 6 | 9 | ||
| No | 10 | 88 | 7 | 104 | ||
| 13 del | 0.49 | 0.27 | ||||
| Yes | 16 | 8 | 11 | 4 | ||
| No | 56 | 42 | 63 | 48 | ||
| 1q21 gain | 0.62 | 0.77 | ||||
| Yes | 8 | 16 | 5 | 10 | ||
| No | 27 | 71 | 33 | 78 | ||
There were no significant difference in complete response (CR) rates among patients with and without 1p31-32 loss and those with or without 20p12.3-12.1 loss. Among 24 patients with 1p31-32 loss, 7 (29%) had a complete remission whereas 31 of 98 (32%) patients without the deletion had a CR (P-value 0.8). Among the 15 patients with 20p12.3-12.1 loss, 5 (33%) achieved a CR, whereas 37 of 111 (33%) without the deletion achieved CR (P-value 1.0). Patients with either of these abnormalities remain in CR for shorter duration, with those having 1p31-32 loss remaining in CR for a median of 14.4 months compared with 32.2 months for those without, and those with 20p12.3-12.1 loss remaining in CR for a median of 19.9 months compared with 30 months for those without. However, these differences did not reach statistical significance, P-value 0.37 and 0.35, respectively, probably because of the few patients in CR (seven and five patients, respectively) with either abnormalities. Consistent with this, both 1p31-32 loss and 20p12.3-12.1 loss have no impact on progression-free survival. Progression-free survival of patients in the cohort with and without 1p31-32 loss is 12.8 months versus 16.3 months (log-rank P-value 0.28), whereas those with and without 20p12.3-12.1 loss is 10.4 months versus 16.9 months (log-rank P-value 0.1).
However, on univariate analysis, 1p31-32 loss was associated with shorter survival, with a median survival of 24.5 months compared with 40 months (log-rank P=0.01), validating our findings from the aCGH analysis. The 20p12.3-121 loss, on the other hand, showed a strong trend toward shorter survival, median survival 20.6 months versus 40 months (log-rank P=0.06), again consistent with the aCGH findings, although this did not reach statistical significance (Figure 5). The significantly shorter overall survival in patients with either of these genetic abnormalities is probably because of shorter postrelapse survival. Survival from relapse is significantly shorter for patients with 1p31-32 loss (12 months versus 19.3 months, log-rank P-value 0.01). Consistent with the overall survival results, survival from relapse is shorter for those with 20p12.3-12.1 loss, but this is not statistically significant (13.5 months versus 17.2 months, log-rank P-value 0.19). On multivariate analysis, including other genetic factors such as t(4;14), 1q gain, and 17p13 deletion, 1p loss remained an independent prognostic factor in addition to 17p13 loss and t(4;14) (Table 3). The 1p31-32 loss retained statistical significance (P-value 0.018) even when β-2 microglobulin was inserted into the model.
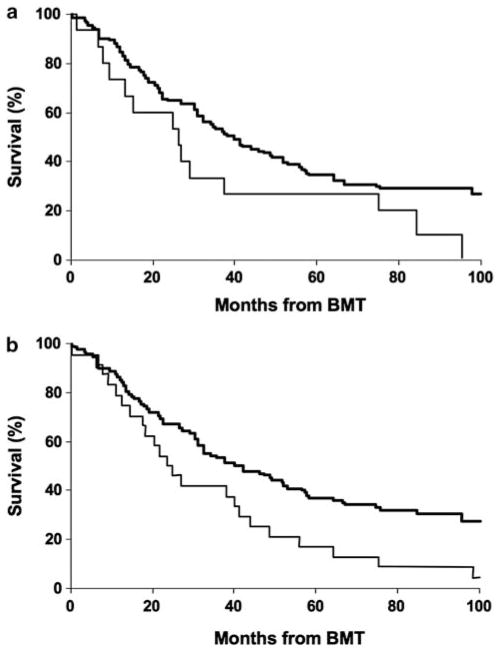
Figure 5.
Validation of these two SCR using FISH from for MSH4 (1p31.1) and PAK7 (20p12.3-12.1). In the Mayo bone marrow transplant (BMT) cohort, (a) patients with PAK7 deletion on 20p12.3-12.1 has a trend toward shorter survival (20.6 months versus 40 months, log-rank P-value=0.06), whereas those with (b) MSH4 deletion on 1p31.1 have a significantly shorter survival (24.5 months versus 40 months, log-rank P-value=0.01).
Table 3.
Cox proportional hazard regression model for overall survival (N=122)
| Variable | Hazard ratio (95% CI) | χ2P-value |
|---|---|---|
| 1p loss | 1.44 (1.12 to 1.82) | <0.010 |
| t(4;14) | 2.31 (1.23 to 4.07) | 0.01 |
| 17p13 del | 2.15 (1.03 to 4.02) | 0.04 |
| 1q21 gain | 1.08 (0.85 to 1.35) | 0.56 |
Discussion
In this study, we identified and validated two genomic regions of loss, 1p31-32 and 20p12.3-12.1, that are associated with shorter survival. In particular, 1p31-32 loss is an independent prognostic factor together with t(4;14) and 17p13 deletion in an independent cohort treated by high-dose therapy.
Although 1p loss has been earlier identified as a relatively common genetic aberration in myeloma and is associated with shorter survival, these were all studies based on conventional karyotype.25–27 A recent study in 87 patients by cIg-FISH using a 1p21 probe showed that 1p21 deletion is present in 20% of patients tested and is an independent prognostic factor.28 In our study, using the high resolution conferred by the aCGH platform, we were able to identify several independent prognostically relevant regions in 1p31-32. This is not necessarily the minimally deleted or most commonly deleted region on chromosome 1p in MM. Our strategy did not identify 1p21, which in the aforementioned study was selected based on minimal area of deletion from an earlier studies. This further reinforces the concept that some very common regions of genetic aberration may be bystanders in the tumorigenic process, whereas others may have a more significant function in driving the process. Furthermore, not all lesions with an important functional role in tumor biology will be important for prognosis. For example, lesions that establish the malignant clone or lead to expansion may be prognostically neutral, whereas those that lead to aggressive characteristics such as proliferation or drug resistance may be more important for survival. From our analysis, it seems that there is an additive effect of the presence of survival relevant genomic aberration with tumors that have more of them having shorter survival. It is attractive to speculate that the genes, pathways, and functions affected by the individual aberrations may have synergistic effect to the advantage of the tumor cells while detrimental to the host.
The 1p31-32 loss is not associated with any known important genetic aberrations in MM such as t(4;14), 1q gain, 13 deletion, or 17p13 deletion, and it has independent prognostic impact on overall survival, but not progression-free survival. This was mainly because of significantly shorter response duration and post-relapse survival. This may suggest relatively shorter survival with salvage treatment. However, information regarding salvage therapies for only a small proportion of the Mayo cohort was known. Therefore, a significant difference in the type of salvage treatment between those with and without 1p31-32 loss cannot be excluded as the main reason for this difference in post-relapse survival, although this is less likely because the prognostic relevance of 1p31-32 loss is consistently observed in three independent cohorts. On the other hand, our earlier study on 1q21 gain shows an association with high-risk genetic factors, such as t(4;14),4 and on multivariate analysis, loses its prognostic significance. We also identified another locus on 20p that has a strong trend toward an association with shorter survival. However, it is strongly associated with 17p13 deletion, which may account for most of its prognostic impact.
In an attempt to identify critical genes located on these survival relevant loci, which may be integrally involved in mediating the short survival, we examined gene expression data in the same tumors from which the aCGH data was derived. In particular, we wanted to identify genes within these loci that had significantly reduced gene expression in those tumors with deletion of the loci. The assumption was that these genes will be more specifically targeted by the genetic aberrations and not involved in other process or pathways that may be activated by other mechanisms that may be important for survival. Therefore, our strategy is not unexpected to identify genes that may be deregulated by other mechanism in addition to DNA copy changes. This is also consistent with the fact that we did not identify CDKN2C (p18), which is located on 1p32 and has been implicated to have a function in disease pathogenesis29,30 and whose gene expression has also been shown to correlate with survival.30 However, it is also clear from these studies that there is poor correlation between CDKN2C expression and genomic loss of CDKN2C,29,30 suggesting that the p18 pathways may be affected by a number of mechanisms. Another example is that the genes we identified in the 1p31-32 locus did not contain the genes located on 1p that were part of a 70-gene high-risk signature developed by the University of Arkansas group.31 The 70-gene signature was enriched with genes located on chromosome 1p and 1q, but it is likely that the high-risk signature is a composite of the prognostic impact of 1q gain, 1p loss, and proliferation. Indeed, many of the genes in the 70-gene are involve in the cell cycle and used in proliferation signatures in other malignancies. Furthermore, these genes were not concentrated on certain regions of 1p. This highlights again that the genes on 1p that form part of the high-risk gene signatures maybe deregulated by trans-mechanisms and hence will not be identified by our strategy. This is also consistent with our earlier finding that over-expression of CSK1B has different prognostic impact compared with gain of 1q21 that contain the CKS1B locus.4
Of the potentially important genes with correlated reduced expression on 1p31-32, DAB1 has been recently implicated as a potential tumor suppressor gene on a large genomic region that is a fragile site commonly deleted in cancers.32 MSH4 is a member of the MutH homolog family that has been implicated in DNA instability in cancer, as it is involved in DNA repair.33 On 20p12.3-12.1, PLCB1 deletion has been implicated in progression from myelodysplastic syndrome to acute myeloid leukemia.34 There are other genes such as RABGGTB on 1p31 and JAG1 on 20p12.1, which have increased expression in diffuse large B-cell lymphoma and acute myeloid leukemia, respectively.35,36 In fact, JAG1 over-expression is associated with poorer prognosis in breast cancer.37 However, these genes are unlikely to be relevant in our setting, as these genetic loci are deleted and the genes under-expressed. The exact function that these candidates have in myeloma biology will have to be further investigated and these works are currently ongoing.
One limitation of our study is the resolution of the platforms used. Higher resolution platforms are currently available. Despite this, the potential of using an unbiased whole genome platform, such as aCGH, to identify prognostically relevant genomic aberrations was shown. It is likely that with larger datasets and higher resolution platform, smaller regions or additional regions associated with survival may be identified. However, this still does not detract from the validity of this study.
Another important caveat is the relatively small number of patients in these cohorts. However, our study highlights the potential prognostic relevance of 1p loss, which should be systematically studied by FISH in the context of large clinical trials to confirm its prognostic relevance, especially in relation to better established genetic-risk factors such as t(4;14), t(14;16), 17p13 deletion, and even 1q21 gain.
In conclusion, we identified and independently validated novel and focal regions of genomic losses that are associated with poorer survival in myeloma. In addition, we identified genes within these regions that may be important in mediating the poor outcome. It is likely that different genomic information, DNA gains or loss, gene expression profiles, epigenetic changes such as miRNA expression and DNA methylation, and even clinical prognostic factors will provide relevant prognostic impact and integration of these factors into multivariate models will provide the most powerful prognostic system.
Supplementary Material
Acknowledgments
RF is a Clinical Investigator of the Damon Runyon Cancer Research Fund and is a Clinical Investigator at the Mayo Clinic. WJC is supported by NMRC Clinician Scientist Investigator award. JJK is supported by the Gene and Mary Lou Kurtz Fellowship in Multiple Myeloma Research. RF is also supported in part by the Donaldson Charitable fund Trust, grants R01 CA83724-01, SPORE P50, CA100707-01, and P01 CA62242 from the National Cancer Institute. Dr Fonseca is also supported by the grant CA015083 from the National Cancer Institute.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Author’s Contribution
Wee J Chng performed experimental analysis and wrote the manuscript. Tae-Hoon Chung performed the data analysis. Scott Van Wier performed FISH experiments. Jonathan J Keats performed aCGH experiments. Angela Baker performed aCGH experiments. P Leif Bergsagel designed the study and approved the manuscript. Morie A Gertz contributed the validation dataset and performed the analysis. John Carpten designed the study and approved the manuscript. Rafael Fonseca designed the study and wrote the manuscript.
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
References
- 1.Kyle RA, Rajkumar SV. Multiple myeloma. Blood. 2008;111:2962–2972. doi: 10.1182/blood-2007-10-078022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fonseca R, Barlogie B, Bataille R, Bastard C, Bergsagel PL, Chesi M, et al. Genetics and cytogenetics of multiple myeloma: a workshop report. Cancer Res. 2004;64:1546–1558. doi: 10.1158/0008-5472.can-03-2876. [DOI] [PubMed] [Google Scholar]
- 3.Fonseca R, Blood E, Rue M, Harrington D, Oken MM, Kyle RA, et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood. 2003;101:4569–4575. doi: 10.1182/blood-2002-10-3017. [DOI] [PubMed] [Google Scholar]
- 4.Fonseca R, Van Wier SA, Chng WJ, Ketterling R, Lacy MQ, Dispenzieri A, et al. Prognostic value of chromosome 1q21 gain by fluorescent in situ hybridization and increase CKS1B expression in myeloma. Leukemia. 2006;20:2034–2040. doi: 10.1038/sj.leu.2404403. [DOI] [PubMed] [Google Scholar]
- 5.Avet-Loiseau H, Attal M, Moreau P, Charbonnel C, Garban F, Hulin C, et al. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myelome. Blood. 2007;109:3489–3495. doi: 10.1182/blood-2006-08-040410. [DOI] [PubMed] [Google Scholar]
- 6.Chang H, Qi C, Yi QL, Reece D, Stewart AK. p53 gene deletion detected by fluorescence in situ hybridization is an adverse prognostic factor for patients with multiple myeloma following autologous stem cell transplantation. Blood. 2005;105:358–360. doi: 10.1182/blood-2004-04-1363. [DOI] [PubMed] [Google Scholar]
- 7.Chang H, Qi X, Trieu Y, Xu W, Reader JC, Ning Y, et al. Multiple myeloma patients with CKS1B gene amplification have a shorter progression-free survival post-autologous stem cell transplantation. Br J Haematol. 2006;135:486–491. doi: 10.1111/j.1365-2141.2006.06325.x. [DOI] [PubMed] [Google Scholar]
- 8.Chang H, Sloan S, Li D, Zhuang L, Yi QL, Chen CI, et al. The t(4;14) is associated with poor prognosis in myeloma patients undergoing autologous stem cell transplant. Br J Haematol. 2004;125:64–68. doi: 10.1111/j.1365-2141.2004.04867.x. [DOI] [PubMed] [Google Scholar]
- 9.Fassas AB, Spencer T, Sawyer J, Zangari M, Lee CK, Anaissie E, et al. Both hypodiploidy and deletion of chromosome 13 independently confer poor prognosis in multiple myeloma. Br J Haematol. 2002;118:1041–1047. doi: 10.1046/j.1365-2141.2002.03757.x. [DOI] [PubMed] [Google Scholar]
- 10.Shaughnessy J, Jacobson J, Sawyer J, McCoy J, Fassas A, Zhan F, et al. Continuous absence of metaphase-defined cytogenetic abnormalities, especially of chromosome 13 and hypodiploidy, ensures long-term survival in multiple myeloma treated with total therapy I: interpretation in the context of global gene expression. Blood. 2003;101:3849–3856. doi: 10.1182/blood-2002-09-2873. [DOI] [PubMed] [Google Scholar]
- 11.Smadja NV, Bastard C, Brigaudeau C, Leroux D, Fruchart C. Hypodiploidy is a major prognostic factor in multiple myeloma. Blood. 2001;98:2229–2238. doi: 10.1182/blood.v98.7.2229. [DOI] [PubMed] [Google Scholar]
- 12.Keats JJ, Reiman T, Maxwell CA, Taylor BJ, Larratt LM, Mant MJ, et al. In multiple myeloma, t(4;14)(p16;q32) is an adverse prognostic factor irrespective of FGFR3 expression. Blood. 2003;101:1520–1529. doi: 10.1182/blood-2002-06-1675. [DOI] [PubMed] [Google Scholar]
- 13.Zhan F, Sawyer J, Gupta S, Huang Y, Anaissie E, Xu H, et al. Elevated expression of CKS1B at 1q21 is highly correlated with short survival in myeloma. Blood. 2004;104:77a. [Google Scholar]
- 14.Gertz MA, Lacy MQ, Dispenzieri A, Greipp PR, Litzow MR, Henderson KJ, et al. Clinical implications of t(11;14)(q13;q32), t(4;14)(p16.3;q32), and -17p13 in myeloma patients treated with high-dose therapy. Blood. 2005;106:2837–2840. doi: 10.1182/blood-2005-04-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai JL, Zandecki M, Mary JY, Bernardi F, Izydorczyk V, Flactif M, et al. Improved cytogenetics in multiple myeloma: a study of 151 patients including 117 patients at diagnosis. Blood. 1995;85:2490–2497. [PubMed] [Google Scholar]
- 16.Rajkumar SV, Fonseca R, Dewald GW, Therneau TM, Lacy MQ, Kyle RA, et al. Cytogenetic abnormalities correlate with the plasma cell labeling index and extent of bone marrow involvement in myeloma. Cancer Genet Cytogenet. 1999;113:73–77. doi: 10.1016/s0165-4608(99)00009-6. [DOI] [PubMed] [Google Scholar]
- 17.Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrasco DR, Tonon G, Huang Y, Zhang Y, Sinha R, Feng B, et al. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell. 2006;9:313–325. doi: 10.1016/j.ccr.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 19.Largo C, Saez B, Alvarez S, Suela J, Ferreira B, Blesa D, et al. Multiple myeloma primary cells show a highly rearranged unbalanced genome with amplifications and homozygous deletions irrespective of the presence of immunoglobulin-related chromosome translocations. Haematologica. 2007;92:795–802. doi: 10.3324/haematol.11052. [DOI] [PubMed] [Google Scholar]
- 20.Largo C, Alvarez S, Saez B, Blesa D, Martin-Subero JI, Gonzalez-Garcia I, et al. Identification of overexpressed genes in frequently gained/amplified chromosome regions in multiple myeloma. Haematologica. 2006;91:184–191. [PubMed] [Google Scholar]
- 21.Ahmann GJ, Jalal SM, Juneau AL, Christensen ER, Hanson CA, Dewald GW, et al. A novel three-color, clone-specific fluorescence in situ hybridization procedure for monoclonal gammopathies. Cancer Genet Cytogenet. 1998;101:7–11. doi: 10.1016/s0165-4608(97)00058-7. [DOI] [PubMed] [Google Scholar]
- 22.Walker BA, Leone PE, Jenner MW, Li C, Gonzalez D, Johnson DC, et al. Integration of global SNP-based mapping and expression arrays reveals key regions, mechanisms, and genes important in the pathogenesis of multiple myeloma. Blood. 2006;108:1733–1743. doi: 10.1182/blood-2006-02-005496. [DOI] [PubMed] [Google Scholar]
- 23.Smadja NV, Fruchart C, Isnard F, Louvet C, Dutel JL, Cheron N, et al. Chromosomal analysis in multiple myeloma: cytogenetic evidence of two different diseases. Leukemia. 1998;12:960–969. doi: 10.1038/sj.leu.2401041. [DOI] [PubMed] [Google Scholar]
- 24.Chng WJ, Santana-Davila R, Van Wier SA, Ahmann GJ, Jalal SM, Bergsagel PL, et al. Prognostic factors for hyperdiploid-myeloma: effects of chromosome 13 deletions and IgH translocations. Leukemia. 2006;20:807–813. doi: 10.1038/sj.leu.2404172. [DOI] [PubMed] [Google Scholar]
- 25.Qazilbash MH, Saliba RM, Ahmed B, Parikh G, Mendoza F, Ashraf N, et al. Deletion of the short arm of chromosome 1 (del 1p) is a strong predictor of poor outcome in myeloma patients undergoing an autotransplant. Biol Blood Marrow Transplant. 2007;13:1066–1072. doi: 10.1016/j.bbmt.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 26.Wu KL, Beverloo B, Lokhorst HM, Segeren CM, van der Holt B, Steijaert MM, et al. Abnormalities of chromosome 1p/q are highly associated with chromosome 13/13q deletions and are an adverse prognostic factor for the outcome of high-dose chemotherapy in patients with multiple myeloma. Br J Haematol. 2007;136:615–623. doi: 10.1111/j.1365-2141.2006.06481.x. [DOI] [PubMed] [Google Scholar]
- 27.Debes-Marun CS, Dewald GW, Bryant S, Picken E, Santana-Davila R, Gonzalez-Paz N, et al. Chromosome abnormalities clustering and its implications for pathogenesis and prognosis in myeloma. Leukemia. 2003;17:427–436. doi: 10.1038/sj.leu.2402797. [DOI] [PubMed] [Google Scholar]
- 28.Chang H, Ning Y, Qi X, Yeung J, Xu W. Chromosome 1p21 deletion is a novel prognostic marker in patients with multiple myeloma. Br J Haematol. 2007;139:51–54. doi: 10.1111/j.1365-2141.2007.06750.x. [DOI] [PubMed] [Google Scholar]
- 29.Dib A, Peterson TR, Raducha-Grace L, Zingone A, Zhan F, Hanamura I, et al. Paradoxical expression of INK4c in proliferative multiple myeloma tumors: bi-allelic deletion vs increased expression. Cell Div. 2006;1:23. doi: 10.1186/1747-1028-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leone PE, Walker BA, Jenner MW, Chiecchio L, Dagrada G, Protheroe RK, et al. Deletions of CDKN2C in multiple myeloma: biological and clinical implications. Clin Cancer Res. 2008;14:6033–6041. doi: 10.1158/1078-0432.CCR-08-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaughnessy JD, Jr, Zhan F, Burington BE, Huang Y, Colla S, Hanamura I, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2006;109:2276–2284. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- 32.McAvoy S, Zhu Y, Perez DS, James CD, Smith DI. Disabled-1 is a large common fragile site gene, inactivated in multiple cancers. Genes Chromosomes Cancer. 2008;47:165–174. doi: 10.1002/gcc.20519. [DOI] [PubMed] [Google Scholar]
- 33.Her C, Zhao N, Wu X, Tompkins JD. MutS homologues hMSH4 and hMSH5: diverse functional implications in humans. Front Biosci. 2007;12:905–911. doi: 10.2741/2112. [DOI] [PubMed] [Google Scholar]
- 34.Lo Vasco VR, Calabrese G, Manzoli L, Palka G, Spadano A, Morizio E, et al. Inositide-specific phospholipase c beta1 gene deletion in the progression of myelodysplastic syndrome to acute myeloid leukemia. Leukemia. 2004;18:1122–1126. doi: 10.1038/sj.leu.2403368. [DOI] [PubMed] [Google Scholar]
- 35.Stirewalt DL, Meshinchi S, Kopecky KJ, Fan W, Pogosova-Agadjanyan EL, Engel JH, et al. Identification of genes with abnormal expression changes in acute myeloid leukemia. Genes Chromosomes Cancer. 2008;47:8–20. doi: 10.1002/gcc.20500. [DOI] [PubMed] [Google Scholar]
- 36.Linderoth J, Eden P, Ehinger M, Valcich J, Jerkeman M, Bendahl PO, et al. Genes associated with the tumour microenvironment are differentially expressed in cured versus primary chemotherapy-refractory diffuse large B-cell lymphoma. Br J Haematol. 2008;141:423–432. doi: 10.1111/j.1365-2141.2008.07037.x. [DOI] [PubMed] [Google Scholar]
- 37.Dickson BC, Mulligan AM, Zhang H, Lockwood G, O’Malley FP, Egan SE, et al. High-level JAG1 mRNA and protein predict poor outcome in breast cancer. Mod Pathol. 2007;20:685–693. doi: 10.1038/modpathol.3800785. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.