Abstract
There is increasing interest in health interventions that incorporate genetic risk information. Although genetic feedback has been evaluated as an adjunct to smoking cessation interventions, its efficacy for reducing alcohol-related risks is unknown. The purpose of this study was to evaluate the feasibility, acceptability, and efficacy of a web-based alcohol intervention incorporating genetic feedback and risk information specific to ALDH2 genotype. The ALDH2*2 variant is associated with partial protection against alcohol dependence but confers significantly increased risk for alcohol-related cancers as a function of alcohol exposure. Two hundred Asian-American young adults were randomly assigned to receive web-based personalized genetic feedback or attention-control feedback. Genetic feedback included health risk information specific to alcohol-related cancer or alcohol dependence, depending on genotype. Outcomes included postintervention drinking behavior and theoretical correlates of behavior change. Genetic feedback and risk information resulted in significant reductions in 30-day drinking frequency and quantity among participants with the ALDH2*1/*2 genotype. Genetic feedback was rated highly by participants and also showed some effects on theoretical correlates of behavior change. Results provide initial evidence of the feasibility, acceptability, and brief efficacy of web-based genetic feedback for reducing alcohol-related health risks associated with ALDH2 genotype.
Keywords: Alcohol use, Aldehyde dehydrogenase, Intervention, Personalized feedback, Genetic feedback, Internet
The prospect of personalized medicine has generated substantial interest in behavioral interventions that incorporate genomic risk information [1, 2]. Given the complexities of characterizing the genetic basis of common, multifactorial diseases, genome-based risk prediction remains a challenging goal. While genome-wide association studies are identifying risk alleles for common disorders [3], it is evident that these variants will be large in number and characterized by small associations with disease outcomes [4–6], currently yielding minimal predictive power above and beyond clinical indicators [7–9]. Additional challenges include the translation of empirically derived risk estimates into individualized interventions and evaluating whether and how genetic risk information might promote behavior change [10–12].
To date, the incorporation of genetic feedback in substance use interventions is almost exclusive to studies of tobacco use. Two randomized trials evaluated the efficacy of genetic feedback for GSTM1, which encodes an enzyme involved in detoxifying environmental carcinogens and shows an association with lung cancer risk [13, 14]. In one study, the addition of GSTM1 feedback to a multicomponent intervention resulted in greater abstinence rates 6 months (but not 12 months) later [15]. Another found that GSTM1 feedback led to decreases in smoking and greater motivation to quit smoking compared to a control group [16]. In one nonrandomized study, participants who received GSTM1 feedback showed high utilization of smoking cessation services, but risk perception did not differ across higher- and lower-risk genotype groups [17]. Other studies have focused on CYP2D6, which also has functional significance for the metabolism of environmental toxins. In one trial, the addition of CYP2D6 feedback to a multicomponent intervention predicted perceived health risks and perceived benefits of smoking cessation, as well as subsequent quit attempts, but did not predict cessation [18, 19]. A study evaluating feedback about the L-myc EcoR1 polymorphism found no overall effect on smoking rates [20]. Finally, providing smokers with genotype-specific feedback about risk for alpha-1 antitrypsin deficiency, a genetic condition that increases risk for emphysema, was associated with greater cessation rates among those at higher genetic risk [21]. Overall, the use of genetic feedback in smoking interventions has yielded modest and inconsistent effects on cognitive outcomes (e.g., risk perception) and no evidence of consistent or sustained effects on behavior.
The efficacy of genetic feedback interventions for reducing alcohol-related health risks has not been studied. Heavy alcohol consumption and alcohol dependence are associated with disease burden globally [22]; in particular, the risk for upper aerodigestive tract cancers increases significantly with cumulative alcohol exposure [23, 24]. Upper aerodigestive tract cancers are attributed largely to exposure to acetaldehyde, a metabolic byproduct of ethanol and an established animal carcinogen [23, 24]. The primary pathway of alcohol metabolism includes oxidation of ethanol to acetaldehyde by alcohol dehydrogenase enzymes, followed by oxidation of acetaldehyde, which is catalyzed primarily by the mitochondrial aldehyde dehydrogenase (ALDH) enzyme [25]. Genetic variations are demonstrated to influence the catalytic properties of these enzymes, leading to differences in rates of acetaldehyde production or elimination [25].
Variations in alcohol metabolizing genes are demonstrated to moderate risk for alcohol-related cancers [24, 26, 27]. ALDH2, which encodes the mitochondrial aldehyde dehydrogenase enzyme, shows the strongest association with cancer risk. The ALDH2*2 allele, which is almost exclusive to individuals of northeast Asian descent, encodes a functionally inactive enzyme subunit that leads to impaired acetaldehyde metabolism. Individuals with ALDH2*2 show increased levels of blood acetaldehyde and increased physiological responses to alcohol (e.g., skin flushing, tachycardia) following alcohol consumption [28]. For individuals homozygous for ALDH2*2 (ALDH2*2/*2 genotype), mitochondrial aldehyde dehydrogenase is completely inactive. As a result, these individuals show strong physiological reactions to alcohol, low drinking rates, and virtually no risk for alcohol dependence [28, 29]. ALDH2*2 heterozygotes (ALDH2*1/*2 genotype) also show elevated blood acetaldehyde during alcohol consumption and have lower rates of alcohol use and dependence than those with the common ALDH2*1/*1 genotype. However, protection against alcohol dependence in heterozygotes is incomplete; a sizable proportion report moderate or heavy drinking and some develop alcohol dependence [29].
It is widely established that ALDH2*2 heterozygotes who drink alcohol are at significantly increased risk for alcohol-related cancers, in particular squamous cell esophageal cancer (e.g., [26, 30–32]); this association shows a dose–response pattern [26, 33, 34]. Odds ratios for esophageal cancer risk for ALDH2*1/*2 individuals (compared to ALDH2*1/*1) have been estimated to range from four to 13 across Japanese studies [26], but odds ratios as high as 30–95 have been reported among heavy drinkers [26, 30, 34]. One meta-analysis, which included seven studies, reported summary odds ratios of 7.07 (95% confidence interval 3.67–13.6) for heavy drinkers and 2.49 (95% confidence interval 1.29–4.79) for moderate drinkers with the ALDH2*1/*2 genotype [35]. Notably, there is molecular evidence to support that elevated cancer risk among heterozygotes is attributable to acetaldehyde exposure during alcohol consumption [33]. Because ALDH2*1/*2 individuals are estimated to number 540 million worldwide—comprising 8% of the global population—experts have called for large-scale prevention efforts in this group [33].
One empirically supported approach for reducing alcohol-related risks is brief feedback and motivational enhancement interventions [36, 37]. Informed by motivational [38] and social psychological theories [39], these interventions typically incorporate personalized feedback about an individual’s drinking behavior relative to a given population or reference group. Theoretically, such information can enhance awareness of alcohol-related risks and highlight discrepancies between current behaviors and future goals [37, 39]. Whereas these interventions often provide normative feedback about the target behavior (alcohol use) in comparison to a reference group (e.g., college students), genetic feedback interventions provide individualized information about possible health risks based on one’s genotype relative to individuals with a different genotype [15–21]. While these two approaches appear compatible, personalized feedback interventions for alcohol use have not incorporated risk information specific to genetic variants. Additionally, whereas alcohol interventions have increasingly used web-based approaches to promote wide dissemination of personalized interventions [40, 41], few studies of genetic risk information have tested internet-delivered interventions [17].
The current study evaluated the feasibility, acceptability, and short-term efficacy of brief, web-based intervention incorporating personalized feedback and risk information specific to ALDH2 genotype. The primary aim was to examine whether genetic feedback and information about alcohol-related cancer risk would influence drinking behavior among individuals with the ALDH2*1/*2 genotype. However, a focus on ALDH2 allowed the additional goal of evaluating feedback about genetic risk for alcohol dependence. Specifically, meta-analyses show that the risk for alcohol dependence among ALDH2*1/*1 individuals is roughly 4.5 times higher than for ALDH2*1/*2 individuals and 8.3 times higher than for ALDH2*2/*2 individuals [29]. Thus, we evaluated genetic risk information specific to (a) alcohol-related cancers (targeting ALDH2*1/*2 individuals) and (b) alcohol dependence (targeting ALDH2*1/*1 individuals, given increased risk relative to ALDH2*1/*2 individuals). Changes in drinking behavior over a 30-day period following the intervention served as the primary outcome. Secondary outcomes included cognitive and motivational correlates of behavior change that were conceptually relevant based on theoretical considerations and prior empirical findings.
Method
Participants
The sample included 200 college students (46.5% male, mean age=20.2 years [SD=1.5]) of northeast Asian descent who participated in a prospective study of genetic and cognitive correlates of drinking behavior. Participants were initially recruited via phone and email based on university records and were eligible if they reported 100% Chinese, Korean, or Japanese heritage. On the whole, participants showed moderate drinking rates. Approximately 90% of the sample reported lifetime drinking; of these, 39% reported a recent heavy drinking episode (i.e., 4+ drinks for women or 5+ drinks for men in a single episode) and 17.5% met criteria for at-risk drinking based on a standardized measure. Detailed information on recruitment and drinking characteristics for this sample is available [42]. The racial composition of the sample was 57.5% Chinese, 33.5% Korean, and 9.0% Japanese. ALDH2 genotype distribution was 52.5% ALDH2*1/*1, 36.5% ALDH2*1/*2, and 11.0% ALDH2*2/*2.
Study Design
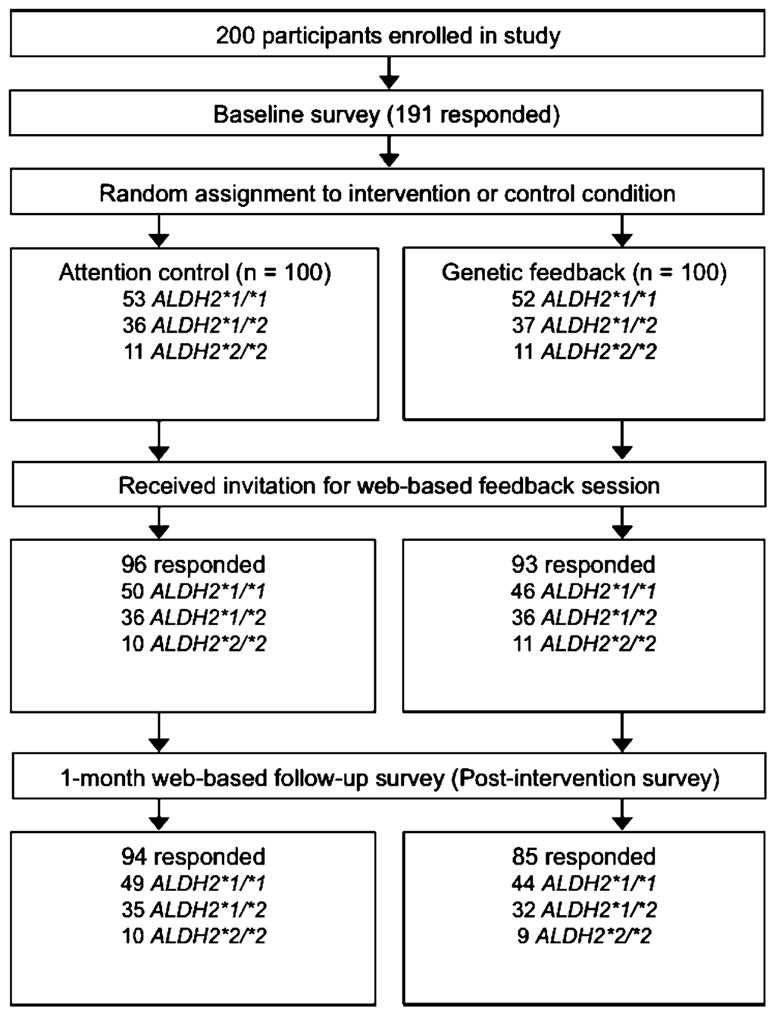
The study design is depicted in Fig. 1. As part of a larger prospective study, participants had completed an initial laboratory visit to provide informed consent and a blood sample for DNA analysis. Subsequently, participants completed two web-based assessments of drinking behavior; one occurred soon after the laboratory visit [42] and the other occurred 3 months thereafter. The latter assessment served as the “baseline” for the current study, as it was the assessment that directly preceded the intervention. Participants were randomly assigned to personalized genetic feedback (n=100) or attention-control feedback (n=100). Randomization was conducted within genotype group to ensure equal genotype distributions across conditions. Although investigators were not blind to participants’ condition, all personal contact with participants preceded randomization, minimizing chances of bias. Participants were unaware of their condition assignment but had been told that they would receive feedback about their ALDH2 genotype at some point during the study.
Fig. 1.
Study design
One month after randomization, participants received an email with a personalized identification number and an embedded link that directed them to their online feedback session. Web-based feedback and assessments were developed in DatStat Illume (DatStat., Inc., Seattle, WA, USA). Codes assigned to each participant based on genotype and treatment condition were preloaded into the web survey and linked to personal identification numbers to dictate which web-based feedback condition was presented. Participants completed a final (postintervention) assessment of drinking behavior 30 days after viewing their feedback. Upon completion of the study, individuals who had been assigned to attention-control feedback received an email with a link to view their genotype result and related risk information, ensuring that all participants had the chance to receive the information contained in the genetic feedback condition.
Personalized Genetic Feedback
Web-based genetic feedback was designed to provide participants with their ALDH2 test result and genotype-specific risk information. The feedback session began with preliminary information designed to aid in feedback interpretation. This information conveyed that (a) alcohol use and alcohol use disorders are influenced both by genetic and environmental factors, (b) few genes have been consistently associated with alcohol-related behaviors, and (c) ALDH2 shows the strongest association with drinking behavior. This section also described the functional significance of ALDH2, the role of the aldehyde dehydrogenase enzyme in acetaldehyde elimination, the functional significance of ALDH2 variants (i.e., *1 and *2 alleles), and population differences in genotype frequencies. This section also described that ALDH2*2 is associated with increased levels of blood acetaldehyde during alcohol consumption and lower rates of alcohol dependence. Participants were prompted to proceed if they wanted to view their test result, at which point the result was presented (e.g., Your genotype is ALDH2*1/*2. This means that you have one copy of the *2 allele.).
Subsequent information varied based on genotype. ALDH2*1/*2 participants learned that their genotype is associated not only with a lower risk for alcohol dependence but also with significantly increased risk for alcohol-related cancers (i.e., five to 12 times greater risk for esophageal cancer versus ALDH2*1/*1 individuals, based on published studies available when the intervention was designed [31, 32, 34]). Feedback also explained that (a) increased cancer risk in ALDH2*1/*2 individuals is attributed to acetaldehyde exposure, (b) higher drinking levels are associated with increased risk, and (c) non-drinkers with this genotype do not show elevated risk.
Risk information for ALDH2*1/*1 participants was specific to alcohol dependence. These individuals learned that their genotype is associated with rates of alcohol dependence that are 4- to 5-fold higher than ALDH2*1/*2 individuals and 8- to 9-fold higher than ALDH2*2/*2 individuals [29]. Individuals with the ALDH2*2/*2 genotype did not receive information concerning increased risk for alcohol-related outcomes, as this genotype is associated with extremely low rates of alcohol use. These individuals received information that their genotype is associated with a significantly lower risk for alcohol dependence.
To enhance the validity of the information presented, the genetic feedback was framed in the context of empirical findings and was accompanied by hyperlinks that directed participants to relevant studies in the PubMed database. The genetic feedback concluded with an explicit statement that the information did not constitute a diagnosis or deterministic estimate of risk. Information also conveyed that the optimal method for reducing alcohol-related health risks is to moderate alcohol intake; this statement was accompanied by a brief list of generic strategies for moderating alcohol consumption (e.g., planning ahead; spacing drinks). Participants had the option of printing a summary of their feedback. All participants who viewed their feedback were also mailed a copy unless they opted out (n=33). Participants were encouraged to contact the study investigators with any questions. A genetic counselor was available to provide consultation to participants; no requests for information were received and no participants reported concerns.
Attention-Control Feedback
Participants assigned to the control group viewed a web-based attention-control feedback session. Feedback consisted of normative information about various college student behaviors collected from undergraduates in a prior survey (e.g., Of the students we surveyed, 73% played high school athletic sports). The purpose of using the attention-control feedback, as opposed to a wait-list control condition, was to minimize differences between experimental conditions by ensuring that control participants (a) received equivalent levels of contact with the study (e.g., email contact) and (b) participated in a web-based session.
Overview and Theoretical Basis of Primary and Secondary Outcome Variables
One limitation of research on behavior change following genetic risk information is the infrequent adoption of theory-based models [43]. Among studies that do invoke theoretical models, no one conceptual framework has demonstrated superiority, although specific constructs have emerged as important. These constructs include perceived susceptibility to health risks, emotional responses to risk information, and intentions for modifying behavior [43, 44]. Rather than aiming to comprehensively test any one theoretical model, the goal of the current study was to examine constructs that were conceptually relevant based on (a) prior studies evaluating genetic feedback and risk information and (b) theories of substance use behavior change.
The primary outcomes included three indices of alcohol use behavior at the baseline and postintervention assessments. Changes in drinking between time points served as the primary endpoints. Secondary outcomes included conceptually relevant cognitive, affective, and motivational variables. Risk perception, motivation, and intentions were assessed at baseline as well as immediately following the intervention (i.e., during the same web-based session at which participants logged on to view their feedback) to allow examination of the immediate impact of the feedback as influencing change in these variables. Fear arousal was assessed immediately following feedback presentation to examine differences in this outcome between genetic feedback and control conditions. Alcohol expectancies (measured at baseline and postintervention assessments) were included as a theoretically relevant variable that has been shown to correspond with changes in drinking [45]. Though not theoretically specific to behavior change, we also examined participant satisfaction with the feedback and interest in future genetic feedback (both assessed immediately following feedback, with the latter one assessed only in the genetic feedback condition) as measures of intervention acceptability.
Alcohol Use
Drinking quantity/frequency was assessed with the Daily Drinking Questionnaire, a validated measure of alcohol consumption [46]. Primary outcomes were 30-day frequency of alcohol use (assessed on a scale of 1 (“Not at all”) to 7 (“Once a day or more”)), maximum drinks consumed and typical number of drinks on weekend nights.
Risk Perception
Risk perception, a construct central to several theories of response to health risk information [43, 44], was measured in relation to alcohol-related health outcomes specifically. Because there was not a precedent for examining perceived risk for alcohol-related health outcomes in the context of genetic risk information, perceived risk was assessed with an item developed for the current study (“In your opinion, how likely is it that you will experience an alcohol-related health problem in the future?”), rated from 0 (completely unlikely) to 5 (completely likely).
Motivation
Motivation for reducing alcohol use was estimated using a change ruler used in prior work [47] based on motivational theories of behavior change [38]. Participants viewed a ruler with a analog scale numbered 0–10; lower numbers indicated precontemplation (0 = “I never think about my drinking”) and progressively higher numbers indicated commitment/action/maintenance (e.g., 5 = “I have decided to drink less,” 10 = “My drinking has changed. I now drink less than before”).
Behavioral Intentions
Behavioral intentions were assessed using three items modified from prior research on responses to genetic risk information [43]: “How likely are you to reduce your drinking in the next month?” (0 = very unlikely; 6 = very likely), “I plan to reduce my alcohol intake in the next month” (0 = very unlikely; 6 = very likely), and “I expect I will reduce my alcohol intake in the next month” (0 = very unlikely; 6 = very likely).
Fear Arousal
Given theoretical and empirical support for the role of affective response in genetic risk interpretation [43], participants reported on affective experiences in response to the feedback. We used items reported in prior research on genetic risk information [43] (i.e., tense, nervous, anxious, frightened, uncomfortable, worried), which were rated on a scale of 1 (not at all) to 7 (extremely). An exploratory principle component analysis with varimax rotation extracted one factor for these items; they were thus combined to form one scale (alpha=0.89).
Alcohol Expectancies
Alcohol expectancies (anticipated effects of alcohol consumption) were theoretically relevant to the current study in that expectancies consistently predict drinking behavior and are shown to change in accordance with changes in drinking [45]. Expectancies were assessed with the 38-item Comprehensive Effects of Alcohol measure [48]. Each expectancy item (e.g., “I would act sociable”) is rated on two components: the perceived likelihood of the outcome (1 = disagree, 4 = agree) and the subjective evaluation of the outcome (1 = bad, 5 = good). The measure includes global scales for positive and negative alcohol expectancies, resulting in four outcomes examined in the current study (i.e., positive/negative subscales and subjective evaluations (good/bad) of these expectancies).
Feedback Interest/Satisfaction
Participants rated their interest/engagement in the feedback based on four items (informative, interesting, engaging, useful) assessed on a scale of 1 (not at all) to 7 (very). In addition, two items assessed participants’ interest in receiving future genetic information based on receiving feedback in this study. Items were phrased, “Based on learning about your ALDH2 genotype, are you more interested or less interested in having a genetic test in the future?” and “Based on this feedback, are you more interested or less interested in receiving feedback about health risks based on your genetic profile in the future?” Items were assessed on a scale of 1 (much less interested) to 7 (much more interested).
Feedback Comprehension
Genetic feedback participants were asked to recall (a) their genotype (ALDH2*1/*1, ALDH2*1/*2, or ALDH2*2/*2) and (b) whether the information they received indicated increased or decreased risk for alcohol-related cancers (for ALDH2*1/*2 participants) or alcohol dependence (for ALDH2*1/*1 participants).
Genotyping
Blood samples were analyzed at the Alcohol Research Center at Indiana University. DNA was isolated using the “HotSHOT” method [49] and TaqMan probes for allelic discrimination (Applied BioSystems, Foster City, CA, USA), as described previously [42]. Based on results using banked samples, single nucleotide polymorphism calling with this method was highly reliable (>99.9%). Any undetermined samples were repeated or sequenced until genotypes were obtained.
Data Analysis
Because genetic feedback content varied by genotype, Mann–Whitney nonparametric tests were conducted separately for each genotype group to detect significant intervention effects on the outcome variables. Main analyses involving the motivation, risk perception, and behavioral intentions scales made use of baseline to postintervention (i.e., self-report by participants immediately after receiving feedback) change scores as the main outcome of interest. Expectancy and alcohol use outcome variables made use of baseline to 1-month posttest change scores. Satisfaction and fear arousal were assessed immediately following the intervention and evaluated in between-group analyses. Effect sizes for Mann–Whitney statistics were calculated for all significant effects using the methods discussed by Newcombe [50, 51].
Results
Completion Rates and Preliminary Analyses
Ninety-three percent of genetic feedback participants and 96% of attention-control feedback participants viewed their feedback session. Among these groups, 94 and 85 individuals completed the 30-day postintervention survey, respectively. Response rates across all items assessed immediately following the feedback (i.e., risk perception, motivation, intentions, feedback satisfaction, and fear arousal) were 99–100%. Forty-three of 46 ALDH2*1/*1 participants, 34 of 36 ALDH2*1/*2 participants, and eight of 11 ALDH2*2/*2 participants correctly recalled their genotype. Forty-four of 46 ALDH2*1/*1 participants and 33 of 36 ALDH2*1/*2 participants correctly recalled information that their genotype was associated with relatively increased risk for alcohol dependence and alcohol-related cancers, respectively. Exploratory data analyses (e.g., histograms, descriptive statistics, and tests of normality) were conducted to examine outcome variables and thereby determine underlying distributions and detect outliers. Because no outcome variables were normally distributed, nonparametric analyses (i.e., Mann–Whitney tests) were applied. There were no significant baseline differences between genetic feedback and attention-control feedback groups on drinking variables (p’s>0.10). Descriptive statistics for outcome variables by genotype and intervention group are shown in Table 1.
Table 1.
Descriptive statistics for outcome variables by genotype and intervention group
| Variables | Baseline |
Posttest |
||||
|---|---|---|---|---|---|---|
| N | Mdn | M (SD) | N | Mdn | M (SD) | |
| Fear arousal | ||||||
| Intervention group | ||||||
| ALDH2*1/*1 | 46 | 1.63 | 2.10 (1.07) | |||
| Control group | ||||||
| ALDH2*1/*1 | 50 | 1.00 | 1.43 (0.79) | |||
| Intentions to reduce alcohol use | ||||||
| Intervention group | ||||||
| ALDH2*1/*1 | 44 | 1.00 | 1.72 (1.75) | 46 | 3.00 | 2.93 (1.63) |
| Control group | ||||||
| ALDH2*1/*1 | 46 | 2.17 | 2.16 (1.93) | 50 | 2.00 | 2.15 (1.80) |
| Risk perception | ||||||
| Intervention group | ||||||
| ALDH2*1/*1 | 45 | 0.00 | 0.53 (0.81) | 39 | 0.00 | 1.00 (1.45) |
| ALDH2*1/*2 | 36 | 0.00 | 0.33 (0.83) | 32 | 0.00 | 0.78 (1.21) |
| Control group | ||||||
| ALDH2*1/*1 | 48 | 0.00 | 0.77 (1.12) | 44 | 0.00 | 0.70 (1.05) |
| ALDH2*1/*2 | 36 | 0.00 | 0.33 (0.63) | 33 | 0.00 | 0.18 (0.46) |
| Expectancy outcomes | ||||||
| Evaluations of positive expectancies | ||||||
| Intervention group | ||||||
| ALDH2*1/*2 | 36 | 2.60 | 2.44 (0.77) | 32 | 2.55 | 2.38 (0.80) |
| Control group | ||||||
| ALDH2*1/*2 | 36 | 2.38 | 2.31 (0.69) | 33 | 2.47 | 2.40 (0.63) |
| Alcohol use outcomes | ||||||
| Peak quantity | ||||||
| Intervention group | ||||||
| ALDH2*1/*2 | 36 | 1.50 | 2.42 (2.90) | 32 | 0.50 | 1.53 (2.14) |
| Control group | ||||||
| ALDH2*1/*2 | 36 | 1.00 | 2.17 (3.38) | 35 | 1.00 | 2.03 (3.08) |
| Typical weekend quantity | ||||||
| Intervention group | ||||||
| ALDH2*1/*2 | 36 | 1.00 | 1.53 (1.80) | 32 | 0.00 | 1.06 (1.76) |
| Control group | ||||||
| ALDH2*1/*2 | 36 | 0.00 | 1.06 (1.87) | 35 | 0.00 | 1.17 (2.13) |
| Drinking frequency | ||||||
| Intervention group | ||||||
| ALDH2*1/*2 | 36 | 2.00 | 2.69 (3.51) | 32 | 1.00 | 2.06 (2.80) |
| Control group | ||||||
| ALDH2*1/*2 | 36 | 1.00 | 1.47 (2.12) | 33 | 1.00 | 1.52 (1.89) |
Note: Only significant group differences are depicted.
Primary Alcohol Use Outcomes
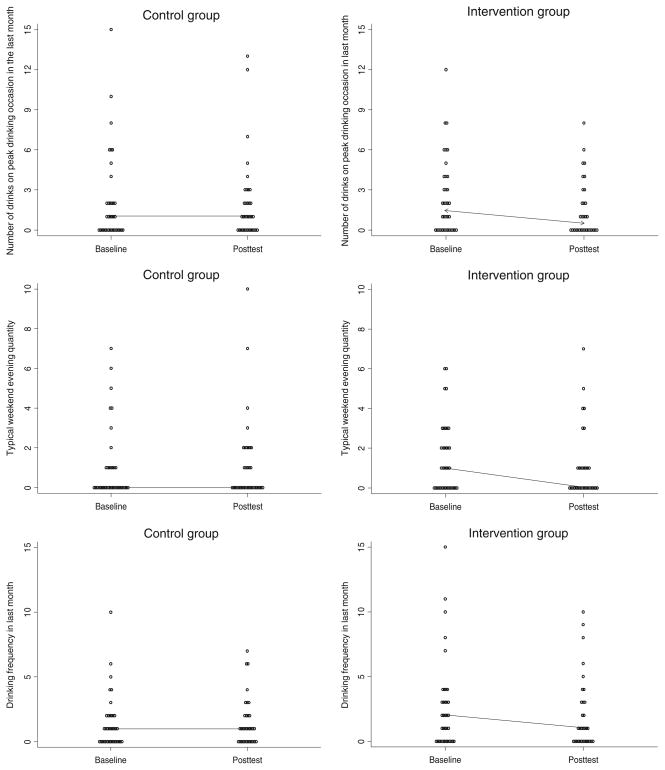
Results indicated significant intervention effects on drinking outcomes for ALDH2*1/*2 participants (Table 2). As shown in Fig. 2, ALDH2*1/*2 participants who received genetic feedback showed significantly greater baseline to posttest decreases in drinking quantity and frequency than attention-control feedback participants. No significant intervention effects were found for other groups (p’s>0.25).
Table 2.
Intervention group differences on outcome variables by genotype
| Variables | Number | U | z | p | θ | θ CI |
|---|---|---|---|---|---|---|
| Participant satisfaction | ||||||
| Informative | ||||||
| ALDH2*1/*1 | 96 | 259 | −6.79 | <0.001 | 0.89 | 0.80–0.94 |
| ALDH2*1/*2 | 72 | 228 | −4.82 | <0.001 | 0.82 | 0.70–0.90 |
| ALDH2*2/*2 | 21 | 22.5 | −2.35 | 0.019 | 0.80 | 0.54–0.92 |
| Interesting | ||||||
| ALDH2*1/*1 | 96 | 273 | −6.66 | <0.001 | 0.88 | 0.79–0.93 |
| ALDH2*1/*2 | 72 | 300 | −4.00 | <0.001 | 0.77 | 0.64–0.86 |
| ALDH2*2/*2 | 21 | 17 | −2.74 | 0.006 | 0.85 | 0.60–0.95 |
| Engaging | ||||||
| ALDH2*1/*1 | 96 | 262 | −6.63 | <0.001 | 0.89 | 0.80–0.94 |
| ALDH2*1/*2 | 72 | 298 | −4.00 | <0.001 | 0.77 | 0.64–0.86 |
| ALDH2*2/*2 | 21 | 13.5 | −2.97 | 0.003 | 0.88 | 0.64–0.96 |
| Useful | ||||||
| ALDH2*1/*1 | 96 | 131 | −7.58 | <0.001 | 0.94 | 0.87–0.98 |
| ALDH2*1/*2 | 72 | 226 | −4.81 | <0.001 | 0.83 | 0.71–0.99 |
| ALDH2*2/*2 | 21 | 10.5 | −3.19 | 0.001 | 0.90 | 0.67–0.98 |
| Fear arousal | ||||||
| ALDH2*1/*1 | 96 | 699 | −3.72 | <0.001 | 0.70 | 0.58–0.79 |
| Intentions to reduce drinking | ||||||
| ALDH2*1/*1 | 90 | 563 | −3.70 | <0.001 | 0.72 | 0.61–0.81 |
| Risk perception | ||||||
| ALDH2*1/*1 | 93 | 820.5 | −2.22 | 0.026 | 0.62 | 0.50–0.72 |
| ALDH2*1/*2 | 72 | 395 | −3.38 | <0.001 | 0.70 | 0.56–0.80 |
| Alcohol expectancies | ||||||
| Evaluation of positive expectancies | ||||||
| ALDH2*1/*2 | 65 | 340.5 | 2.50 | 0.013 | 0.68 | 0.54–0.79 |
| Alcohol use outcomes | ||||||
| Peak quantity | ||||||
| ALDH2*1/*2 | 67 | 374 | 2.43 | 0.015 | 0.67 | 0.53–0.78 |
| Typical weekend quantity | ||||||
| ALDH2*1/*2 | 67 | 374 | 2.59 | 0.010 | 0.67 | 0.53–0.78 |
| Drinking frequency | ||||||
| ALDH2*1/*2 | 65 | 360.5 | 2.26 | 0.024 | 0.66 | 0.52–0.77 |
Only statistically significant tests are depicted. θ =0.5 indicates that the two compared distributions match perfectly. As the θ moves toward 1.0, the distributions are less and less similar. The confidence intervals (θ CI) indicate whether the effect size is reliably different from 0.5
Fig. 2.
Baseline to postintervention (30-day) changes in drinking outcomes for ALDH2*1/*2 participants (lines indicate median changes)
Secondary Outcomes
Within the ALDH2*1/*1 group, genetic feedback participants showed significantly greater increases in risk perception and intentions to change drinking behavior, as well as greater fear arousal, compared to attention-control feedback participants (Tables 1 and 2). Within the ALDH2*1/*2 group, genetic feedback predicted greater baseline to posttest increases in risk perception compared to attention-control feedback. Among ALDH2*1/*2 participants, the genetic feedback group reported significant reductions in their evaluations of “positive” alcohol expectancies following the intervention compared to those receiving attention-control feedback. That is, positive alcohol expectancies were rated as being less desirable after the intervention compared to before the intervention in this group. No other significant intervention effects on secondary outcomes were observed (p’s>0.05).
Participant Ratings of Web-Based Feedback
Regardless of genotype, participants rated the genetic feedback as significantly more informative, interesting, engaging, and useful than attention-control feedback (Table 2). Ratings of the genetic feedback on these items averaged 5.8 on the 1–7 scale, indicating good acceptability. These participants also reported high interest in receiving a genetic test (M=5.87, SD=1.13) and genetic risk information (M=6.03, SD=1.16) in the future based on receiving genetic feedback in this study.
Discussion
This study evaluated a brief, web-based intervention incorporating personalized genetic feedback and risk information specific to ALDH2 genotype. Participants with the ALDH2*1/*2 genotype who received personalized genetic feedback and risk information reported significant reductions in 30-day drinking frequency, quantity, and peak consumption compared to those assigned to a control condition. Genetic feedback also predicted significant increases in risk perception (for ALDH2*1/*1 and ALDH2*1/*2 individuals) and intentions to reduce drinking (for ALDH2*1/*1 individuals). Ratings of the genetic feedback indicated high interest and engagement, as well as high interest in receiving future genetic risk information. Overall, the results provide initial support for the feasibility and acceptability of a web-based genetic feedback intervention targeting substance use behavior in college students, as well the potential efficacy of this approach for reducing health risks associated with ALDH2*2.
Previous studies incorporating genetic feedback as an adjunct to smoking interventions have focused primarily on adults who were motivated for cessation. In contrast, this study targeted nontreatment-seeking young adults who generally reported little motivation to reduce their drinking, perhaps reflective of modest drinking rates in this sample. In this context, evidence for significant intervention effects among ALDH2*1/*2 individuals is notable for at least two reasons. First, given that even moderate drinking is associated with elevated cancer risk in this group, interventions for moderate drinkers could have a significant impact on overall disease burden in this subgroup [33]. Second, the results are consistent with the notion that personalized genetic feedback might be efficacious for addressing complex behaviors even when individuals are precontemplative of change. These results provide a basis for examining the efficacy of more comprehensive primary interventions addressing alcohol use among young adults with the ALDH2*1/*2 genotype [33]. To the extent that such interventions might influence drinking trajectories, primary interventions could play a significant role in reducing cancer risk attributable to cumulative alcohol exposure.
Our focus on ALDH2 allowed the secondary goal of evaluating health risk information specific to genetic risk for alcohol dependence. In contrast to the risk information delivered to ALDH2*1/*2 participants, providing ALDH2*1/*1 individuals with information about increased risk for alcohol dependence did not influence drinking. Because genetic feedback precludes random assignment to feedback conditions, the two feedback groups could not be compared directly. However, evaluating these two types of feedback in same study allows for some speculation about differential intervention effects. One possibility is that genetic risk information specific to a medical diagnosis is more salient than risk information for a substance use disorder. For example, consistent with existing theory on responses to genetic risk information [44, 52], to the extent that a cancer diagnosis is perceived as (a) more severe or (b) having a stronger genetic basis compared to alcohol dependence, cancer-related risk information would be perceived as more salient. Perceived disease severity and perceived genetic contribution to disease are therefore two constructs of theoretical relevance for future studies [44, 52]. An alternative explanation for the differential intervention effects is that preexisting differences across genotype groups could influence intervention response. For instance, because ALDH2*1/*2 individuals show heightened physiological responses to alcohol, genetic information could have been more salient for this group in that they could explicitly relate genetic feedback information to past drinking experiences. Differences in drinking history across genotypes could also contribute to differential intervention response.
Whereas associations of individual genetic variants with substance use behavior are usually small, focusing on ALDH2 allowed for the communication of relatively large effect sizes (i.e., odds ratios) with respect to alcohol-related health outcomes. Theoretically, the ability to explain increasing variance in genetic risk should increase the efficacy of genetic feedback interventions [53]. However, simulation studies suggest that explaining even moderate variance in genetic risk for complex disorders will require the consideration of vast numbers of susceptibility alleles [5]. Although such studies are underway, accurate prediction of genetic risk for complex diseases using genome-wide panels is not yet a reality [4]. In the interim, studies of genetic feedback that focus on established risk markers, as was the goal in this study, can be informative from a proof-of-concept standpoint. It is also noteworthy that genetic feedback appears capable of promoting health protective behaviors irrespective of whether participants have the higher-or lower-risk genotype [17, 54] or whether preventative measures exist for the disorder in question [55]. Thus, one possibility is that simply providing personalized genetic feedback could enhance intervention uptake or efficacy, irrespective of the specificity of test results.
Limitations of this study include a moderately sized sample, a relatively short follow-up period, and a focus on a specific population. Additionally, given the preliminary nature of this study and its goals to establish intervention feasibility and acceptability, we did not aim to test a comprehensive theoretical model or to conduct formal meditational analyses in evaluating intervention effects. Future studies could address additional theoretical constructs relevant for health risk perception (for example, self-efficacy, coping appraisals, perceived disease severity [43, 44, 52]) as mediators or moderators of intervention efficacy. Notably, experimental analog studies have proven useful for evaluating effects of genetic risk information on psychological and motivational responses in the context of established health behavior theories [43, 53] and would be useful for refining interventions such as the one described here. Another limitation concerns our assessment of risk perception, which relied on a single-item measure developed for the current study. Ideally, future studies could develop improved risk perception measures with established psychometric properties. Using self-report measures of drinking behavior is also a potential limitation, as the veracity of participants’ reports cannot be confirmed.
The current findings offer initial evidence for the feasibility, acceptability, and short-term efficacy of brief interventions for reducing alcohol-related health risks among individuals with ALDH2*2. Whereas large-scale interventions in primary care settings have recently been recommended for this purpose [33], the current results suggest that web-based approaches, which have the capacity for broad impact, could also be feasible and effective as a delivery method for primary interventions. Based on the established efficacy of traditional brief motivational enhancement interventions for alcohol use [36–37], one direction for future research would be to integrate genetic risk information with personalized normative feedback specific to individual drinking patterns. Additionally, the feasibility of a web-based genetic feedback approach for targeting alcohol use in this study of college students suggests that similar methods could be useful for addressing other health behaviors in young adult populations.
Acknowledgments
This research was supported by National Institute on Alcohol Abuse and Alcoholism (NIAAA) grants F31AA016440 and K02AA00269 and a Small Grant Award from the University of Washington Alcohol and Drug Abuse Institute. Genotyping services were provided by the Genomics and Molecular Biology Core of the Alcohol Research Center Indiana, which is funded by NIAAA grant P60AA07611-20. The authors thank Melanie Pepin, MS, CGC, for her assistance with this study.
Contributor Information
Christian S. Hendershot, Email: chender@unm.edu, The Mind Research Network, Albuquerque, NM, USA. Center on Alcoholism, Substance Abuse and Addictions (CASAA), University of New Mexico, Albuquerque, NM, USA
Jacqueline M. Otto, Department of Psychology, University of Washington, Seattle, WA, USA
Susan E. Collins, Department of Psychiatry and Behavioral Sciences, University of Washington, Seattle, WA, USA
Tiebing Liang, Indiana University School of Medicine, Indianapolis, IN, USA
Tamara L. Wall, Department of Psychiatry, University of California, San Diego, La Jolla, CA, USA. Psychology Service, Veterans Affairs San Diego Healthcare System, San Diego, CA, USA. Veterans Medical Research Foundation, San Diego, CA, USA
References
- 1.Bell J. Predicting disease using genomics. Nature. 2004;429:453–456. doi: 10.1038/nature02624. [DOI] [PubMed] [Google Scholar]
- 2.Guttmacher AE, Collins FS. Realizing the promise of genomics in biomedical research. JAMA. 2005;294:1399–1402. doi: 10.1001/jama.294.11.1399. [DOI] [PubMed] [Google Scholar]
- 3.The Wellcome Trust Case Control Consortium. Genome-wide association study of 14, 000 cases of seven common diseases and 3, 000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kraft P, Hunter DJ. Genetic risk prediction: Are we there yet? N Engl J Med. 2009;360:1701–1703. doi: 10.1056/NEJMp0810107. [DOI] [PubMed] [Google Scholar]
- 5.Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007;17:1520–1528. doi: 10.1101/gr.6665407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk of complex disease. Curr Opin Genet Dev. 2008;18:257–263. doi: 10.1016/j.gde.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Lango H, Palmer CNA, Morris AD, et al. Assessing the combined impact of 18 common genetic variants of modest effect sizes on type 2 diabetes risk. Diabetes. 2008;57:3129–3135. doi: 10.2337/db08-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyssenko V, Jonsson A, Almgren P, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008;359:2220–2232. doi: 10.1056/NEJMoa0801869. [DOI] [PubMed] [Google Scholar]
- 9.van Hoek M, Dehghan A, Wittentan JCM, et al. Predicting type 2 diabetes based on polymorphisms from genome-wide association studies: A population-dased study. Diabetes. 2008;57:3122–3128. doi: 10.2337/db08-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henrikson NB, Bowen D, Burke W. Does genomic risk information motivate people to change their behavior? Genome Med. 2009;1:37. doi: 10.1186/gm37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lerman C, Croyle RT, Tercyak KP, Hamann H. Genetic testing: Psychological aspects and implications. J Consult Clin Psychol. 2002;70:784–797. doi: 10.1037//0022-006x.70.3.784. [DOI] [PubMed] [Google Scholar]
- 12.Janssens AC, van Duijn CM. Genome-based prediction of common diseases: Methodological considerations for future research. Genome Med. 2009;1:20. doi: 10.1186/gm20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benhamou S, Lee WJ, Alexandrie AK, et al. Meta- and pooled analyses of the effects of glutathione S-transferase M1 polymorphisms and smoking on lung cancer risk. Carcinogenesis. 2002;23:1343–1350. doi: 10.1093/carcin/23.8.1343. [DOI] [PubMed] [Google Scholar]
- 14.Carlsten C, Sagoo GS, Frodsham AJ, Burke W, Higgins JPT. Glutathione S-transferase M1 (GSTM1) polymorphisms and lung cancer: A literature-based systematic HuGE review and meta-analysis. Am J Epidemiol. 2008;167:759–774. doi: 10.1093/aje/kwm383. [DOI] [PubMed] [Google Scholar]
- 15.McBride CM, Bepler G, Lipkus IM, et al. Incorporating genetic susceptibility feedback into a smoking cessation program for African-American smokers with low income. Cancer Epidemiol Biomarkers Prev. 2002;11:521–528. [PubMed] [Google Scholar]
- 16.Sanderson SC, Humphries SE, Hubbart C, Hughes E, Jarvis MJ, Wardle J. Psychological and behavioural impact of genetic testing smokers for lung cancer risk: A phase II exploratory trial. J Health Psychol. 2008;13:481–494. doi: 10.1177/1359105308088519. [DOI] [PubMed] [Google Scholar]
- 17.Sanderson SC, O’Neill SC, White DB, et al. Responses to online GSTM1 genetic test results among smokers related to patients with lung cancer: A pilot study. Cancer Epidemiol Biomarkers Prev. 2009;18:1953–1961. doi: 10.1158/1055-9965.EPI-08-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lerman C, Gold K, Audrain J, et al. Incorporating bio-markers of exposure and genetic susceptibility into smoking cessation treatment: Effects on smoking-related cognitions, emotions, and behavior change. Health Psychol. 1997;16:87–99. doi: 10.1037//0278-6133.16.1.87. [DOI] [PubMed] [Google Scholar]
- 19.Audrain J, Boyd NR, Roth J, Main D, Caporaso NE, Lerman C. Genetic susceptibility testing in smoking-cessation treatment: One-year outcomes of a randomized trial. Addict Behav. 1997;22:741–751. doi: 10.1016/s0306-4603(97)00060-9. [DOI] [PubMed] [Google Scholar]
- 20.Ito H, Matsuo K, Wakai K, et al. An intervention study of smoking cessation with feedback on genetic cancer susceptibility in Japan. Prev Med. 2006;42:102–108. doi: 10.1016/j.ypmed.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 21.Carpenter MJ, Strange C, Jones Y, et al. Does genetic testing result in behavioral health change? Changes in smoking behavior following testing for alpha-1 antitrypsin deficiency. Ann Behav Med. 2007;33:22–28. doi: 10.1207/s15324796abm3301_3. [DOI] [PubMed] [Google Scholar]
- 22.Rehm J, Mathers C, Popova S, Thavorncharoensap M, Teerawattananon Y, Patra J. Global burden of disease and injury and economic cost attributable to alcohol use and alcohol-use disorders. Lancet. 2009;373:2223–2233. doi: 10.1016/S0140-6736(09)60746-7. [DOI] [PubMed] [Google Scholar]
- 23.Baan R, Straif K, Grosse Y, et al. Carcinogenicity of alcoholic beverages. Lancet Oncol. 2007;8:292–293. doi: 10.1016/s1470-2045(07)70099-2. [DOI] [PubMed] [Google Scholar]
- 24.Lachenmeier DW, Kanteres F, Rehm J. Carcinogenicity of acetaldehyde in alcoholic beverages: Risk assessment outside ethanol metabolism. Addiction. 2009;104:533–550. doi: 10.1111/j.1360-0443.2009.02516.x. [DOI] [PubMed] [Google Scholar]
- 25.Edenberg HJ. The genetics of alcohol metabolism—role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30:5–13. [PMC free article] [PubMed] [Google Scholar]
- 26.Chen YJ, Chen C, Wu DC, et al. Interactive effects of lifetime alcohol consumption and alcohol and aldehyde dehydrogenase polymorphisms on esophageal cancer risks. Int J Cancer. 2006;119:2827–2831. doi: 10.1002/ijc.22199. [DOI] [PubMed] [Google Scholar]
- 27.Hiyama T, Yoshihara M, Tanaka S, Chayama K. Genetic polymorphisms and esophageal cancer risk. Int J Cancer. 2007;121:1643–1658. doi: 10.1002/ijc.23044. [DOI] [PubMed] [Google Scholar]
- 28.Peng GS, Yin SJ. Effect of the allelic variants of aldehyde dehydrogenase ALDH2*2 and alcohol dehydrogenase ADH1B*2 on blood acetaldehyde concentrations. Hum Genomics. 2009;3:121–127. doi: 10.1186/1479-7364-3-2-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luczak SE, Glatt SJ, Wall TL. Meta-analyses of ALDH2 and ADH1B with alcohol dependence in Asians. Psychol Bull. 2006;132:607–621. doi: 10.1037/0033-2909.132.4.607. [DOI] [PubMed] [Google Scholar]
- 30.Yang CX, Matsuo K, Ito H, et al. Esophageal cancer risk by ALDH2 and ADH2 polymorphisms and alcohol consumption: Exploration of gene–environment and gene–gene interactions. Asian Pac J Cancer Prev. 2005;6:256–262. [PubMed] [Google Scholar]
- 31.Yokoyama A, Muramatsu T, Ohmori T, et al. Alcohol-related cancers and aldehyde dehydrogenase-2 in Japanese alcoholics. Carcinogenesis. 1998;19:1383–1387. doi: 10.1093/carcin/19.8.1383. [DOI] [PubMed] [Google Scholar]
- 32.Yokoyama A, Muramatsu T, Omori T, et al. Alcohol and aldehyde dehydrogenase gene polymorphisms influence susceptibility to esophageal cancer in Japanese alcoholics. Alcohol Clin Exp Res. 1999;23:1705–1710. [PubMed] [Google Scholar]
- 33.Brooks PJ, Enoch MA, Goldman D, Li TK, Yokoyama A. The alcohol flushing response: An unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med. 2009;6(3):e1000050. doi: 10.1371/journal.pmed.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yokoyama T, Yokoyama A, Kato H, et al. Alcohol flushing, alcohol and aldehyde dehydrogenase genotypes, and risk for esophageal squamous cell carcinoma in Japanese men. Cancer Epidemiol Biomarkers Prev. 2003;12:1227–1233. [PubMed] [Google Scholar]
- 35.Lewis SJ, Smith GD. Alcohol, ALDH2, and esophageal cancer: A meta-analysis which illustrates the potentials and limitations of a Mendelian randomization approach. Cancer Epidemiol Biomarkers Prev. 2005;14:1967–1971. doi: 10.1158/1055-9965.EPI-05-0196. [DOI] [PubMed] [Google Scholar]
- 36.Collins SE, Carey KB, Sliwinski MJ. Mailed personalized normative feedback as a brief intervention for at-risk college drinkers. J Stud Alcohol. 2002;63:559–567. doi: 10.15288/jsa.2002.63.559. [DOI] [PubMed] [Google Scholar]
- 37.Larimer ME, Lee CM, Kilmer JR, et al. Personalized mailed feedback for college drinking prevention: A randomized clinical trial. J Consult Clin Psychol. 2007;75:285–293. doi: 10.1037/0022-006X.75.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller WR, Rollnick S. Motivational interviewing: Preparing people for change. New York: Guilford; 2002. [Google Scholar]
- 39.Lewis MA, Neighbors C. Social norms approaches using descriptive drinking norms education: A review of the research on personalized normative feedback. J Am Coll Health. 2006;54:213–218. doi: 10.3200/JACH.54.4.213-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cunningham JA, Wild TC, Cordingley J, van Mierlo T, Humphreys K. A randomized controlled trial of an internet-based intervention for alcohol abusers. Addiction. 2009;104:2023–2032. doi: 10.1111/j.1360-0443.2009.02726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neighbors C, Lee CM, Lewis MA, Fossos N, Walter T. Internet-based personalized feedback to reduce 21st-birthday drinking: A randomized controlled trial of an event-specific prevention intervention. J Consult Clin Psychol. 2009;77:51–63. doi: 10.1037/a0014386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hendershot CS, Collins SE, George WH, et al. Associations of ALDH2 and ADH1B genotypes with alcohol-related phenotypes in Asian young adults. Alcohol Clin Exp Res. 2009;33:839–847. doi: 10.1111/j.1530-0277.2009.00903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wright AJ, French DP, Weinman J, Marteau TM. Can genetic risk information enhance motivation for smoking cessation? An analogue study. Health Psychol. 2006;25:740–752. doi: 10.1037/0278-6133.25.6.740. [DOI] [PubMed] [Google Scholar]
- 44.Gooding HC, Organista K, Burack J, Biesecker BB. Genetic susceptibility testing from a stress and coping perspective. Soc Sci Med. 2006;62:1880–1890. doi: 10.1016/j.socscimed.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 45.Jones BT, Corbin W, Fromme K. A review of expectancy theory and alcohol consumption. Addiction. 2001;96:57–72. doi: 10.1046/j.1360-0443.2001.961575.x. [DOI] [PubMed] [Google Scholar]
- 46.Collins RL, Parks GA, Marlatt GA. Social determinants of alcohol-consumption: The effects of social-interaction and model status on the self-administration of alcohol. J Consult Clin Psychol. 1985;53:189–200. doi: 10.1037//0022-006x.53.2.189. [DOI] [PubMed] [Google Scholar]
- 47.LaBrie JW, Quinlan T, Schiffman JE, Earleywine ME. Performance of alcohol and safer sex change rulers compared with readiness to change questionnaires. Psychol Addict Behav. 2005;19:112–115. doi: 10.1037/0893-164X.19.1.112. [DOI] [PubMed] [Google Scholar]
- 48.Fromme K, Stroot EA, Kaplan D. Comprehensive effects of alcohol: Development and psychometric assessment of a new expectancy questionnaire. Psychol Assess. 1993;5:19–26. [Google Scholar]
- 49.Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT) Biotechniques. 2000;29:52. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- 50.Newcombe RG. Confidence intervals for an effect size measure based on the Mann–Whitney statistic. Part 1: General issues and tail-area-based methods. Stat Med. 2005;25:543–557. doi: 10.1002/sim.2323. [DOI] [PubMed] [Google Scholar]
- 51.Newcombe RG. Confidence intervals for an effect size measure based on the Mann–Whitney statistic. Part 2: Asymptotic methods and evaluation. Stat Med. 2005;25:559–573. doi: 10.1002/sim.2324. [DOI] [PubMed] [Google Scholar]
- 52.Marteau TM, Weinman J. Self-regulation and the behavioural response to DNA risk information: A theoretical analysis and framework for future research. Soc Sci Med. 2006;62:1360–1368. doi: 10.1016/j.socscimed.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 53.Cameron LD, Sherman KA, Marteau TM, Brown PM. Impact of genetic risk information and type of disease on perceived risk, anticipated affect, and expected consequences of genetic tests. Health Psychol. 2009;28:307–316. doi: 10.1037/a0013947. [DOI] [PubMed] [Google Scholar]
- 54.McBride CM, Alford SH, Reid RJ, Larson EB, Baxevanis AD, Brody LC. Characteristics of users of online personalized genomic risk assessments: Implications for physician–patient interactions. Genet Med. 2009;11:582–587. doi: 10.1097/GIM.0b013e3181b22c3a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chao S, Roberts JS, Marteau TM, Silliman R, Cupples LA, Green RC. Health behavior changes after genetic risk assessment for Alzheimer disease: The REVEAL Study. Alzheimer Dis Assoc Disord. 2008;22:94–97. doi: 10.1097/WAD.0b013e31815a9dcc. [DOI] [PMC free article] [PubMed] [Google Scholar]