Abstract
p53 suppresses tumor development by responding to unauthorized cell proliferation, growth factor or nutrient deprivation, and DNA damage. Distinct pathways have been identified that cause p53 activation, including ARF-dependent response to oncogene activation, ribosomal protein-mediated response to abnormal rRNA synthesis, and ATM-dependent response to DNA damage. Elucidating the mechanisms of these signaling events are critical for understanding tumor suppression by p53 and development of novel cancer therapeutics. More than a decade of research has established the ATM kinase as a key molecule that activates p53 after DNA damage. Our recent study revealed that ATM phosphorylation of MDM2 is likely to be the key step in causing p53 stabilization. Upon activation by ionizing irradiation, ATM phosphorylates MDM2 on multiple sites near its RING domain. These modifications inhibit the ability of MDM2 to poly-ubiquitinate p53, thus leading to its stabilization. MDM2 phosphorylation does not inactivate its E3 ligase activity per se, since MDM2 self-ubiquitination and MDMX ubiquitination functions are retained. The selective inhibition of p53 poly-ubiquitination is accomplished through disrupting MDM2 oligomerization that may provide a scaffold for processive elongation of poly ubiquitin chains. These findings suggest a novel model of p53 activation and a general mechanism of E3 ligase regulation by phosphorylation.
Keywords: p53, MDM2, MDMX, ATM, phosphorylation, oligomerization, ubiquitination
Regulation of p53 by MDM2 and MDMX
The most notable feature of the p53 tumor suppressor is its stabilization and nuclear accumulation after exposure to many stress signals. This leads to induction of over one hundred downstream transcriptional targets that inhibit cell cycle progress, induce apoptosis, and regulate energy metabolism.1 A critical regulator of p53 and perhaps the major factor responsible for the dynamic characteristics of p53 is the MDM2 protein. MDM2 is best known as an ubiquitin E3 ligase for p53 that promotes p53 degradation in normal cells. Although additional E3 ligases (Pirh2, Cop1) have also been shown to degrade p53 in cell culture,2,3 mouse models of MDM2 gene knock out provided unequivocal evidence that MDM2 function is indispensable for controlling p53 activity at all stages of life.4,5 The role of MDM2 as an important regulator of p53 stability is also validated by MDM2 knock down, and by small molecule inhibitors that disrupt p53-MDM2 binding.6
The MDM2 homolog MDMX is also emerging as another important regulator of p53.7 The physiological role of MDMX was revealed by the embryonic lethality of MDMX null mice, which can be rescued by knockout of p53.8–10 Tissue-specific knockout of MDMX generally result in mild phenotypes compared to MDM2,5,11,12 suggesting a supplemental or developmental stage-specific function in p53 regulation. MDMX has weak intrinsic E3 ligase activity and does not directly promote p53 degradation.13 Although biochemical experiments suggest that MDMX can stimulate the ability of MDM2 to ubiquitinate p53 through hetero dimerization,14,15 the impact of MDMX expression on p53 stability is moderate at best.16 Instead, MDMX regulates p53 mainly by formation of inactive p53-MDMX complexes.16–18 Therefore, it appears that p53 stability is mainly regulated by MDM2.
MDM2 promotes p53 degradation by forming a stable complex through N terminal domains. After binding to p53, the MDM2 C terminal RING domain recruits ubiquitin-conjugating enzyme E2 that performs covalent modification of p53 lysine residues. In addition to E3 ligase activity, MDM2 also interacts with the proteosome subunit C8 and may deliver substrates directly to the proteasome.19 As expected, both the p53-binding and RING domains of MDM2 are critical for p53 degradation. However, the central acidic region of MDM2 (residue 200–300) is also critical for ubiquitination of p53 through unknown mechanisms.20,21 The acidic domain has features of a disordered region that contains binding sites for most of the MDM2-binding proteins identified to date, including transcription regulators,22,23 de-ubiquitinating enzyme HAUSP,24 ribosomal proteins,25 and the tumor suppressor ARF.26
Stress Activation of p53
Numerous studies have established that different cellular stress and damage signals converge on MDM2 and MDMX to cause p53 activation. Oncogene-induced ARF expression induces p53 accumulation by binding MDM2 acidic domain and inhibiting p53 ubiquitination.27 Inhibitors of rRNA transcription (such as ActD, 5-FU and growth factor deprivation) induce ribosomal stress, which stimulates MDM2 interaction with several ribosomal proteins (such as L5, L11 and L23) that also block p53 ubiquitination.28–31 These proteins all interact with the MDM2 acidic domain, highlighting the importance of the acidic domain in sensing such growth-related stress signals.
Perhaps the most extensively studied p53 activation pathway is DNA damage signaling. A critical player in p53 DNA damage response is the ATM kinase.32 ATM is activated within minutes after DNA double-strand break and phosphorylates numerous substrates involved in cell cycle regulation and DNA repair.33 ATM activation of Chk2 kinase further amplifies the signal and expands the number of target proteins. Loss of ATM prevents the rapid accumulation of p53 after IR and abrogates the p53-mediated cell cycle arrest response.32 The importance of the ATM-mediated response is not limited to direct DNA damage by irradiation. A range of physiologically-relevant stresses can also trigger ATM activation either directly or by inducing DNA damage, including chemotherapy, oncogene activation, oxidative stress, heat shock, and acidic pH.
The Role of p53 Phosphorylation in DNA Damage Response
MDM2-p53 binding has been extensively studied as a target of regulation by DNA damage. Several studies showed that DNA double-strand breaks induce phosphorylation of p53 S15 by ATM or DNA-PK.34,35 ATM also activates Chk2, which in turn phosphorylates p53 on S20 which is part of the MDM2 binding site.36,37 These findings suggested a model that p53 phosphorylation on the N terminus disrupts MDM2 binding and results in p53 stabilization.
However, biochemical analysis indicated that p53 S15 and S20 phosphorylation do not prevent MDM2 binding in vitro.38 Mutation of multiple phosphorylation sites including S15 and S20 do not abrogate p53 stabilization after DNA damage in cell culture.39–41 Mouse models showed that blocking p53 phosphorylation on S18 and S23 (equivalents of S15 and S20 in human p53) partially reduced p53 accumulation after DNA damage, and causes partial defects in apoptosis and tumor suppression.42 S23 single site mutation also caused partial defect in p53 stabilization by gamma irradiation and increased the incidence of B-cell lymphoma.43 Single site mutation of S18 had no significant effect on p53 stabilization or tumor suppression, despite poor activation of certain p53 target genes after DNA damage.44 These studies showed that p53 phosphorylation contributes to its stabilization, but also implicate the presence of additional signaling mechanisms.
The Role of MDMX Phosphorylation After DNA Damage
Befitting their roles as major regulators of p53, MDM2 and MDMX have emerged as important signaling targets by the ATM pathway. To date, the function of MDMX phosphorylation in p53 response has been extensively characterized. The results showed that ATM triggers MDMX degradation after DNA damage, eliminating its inhibitory effect on p53. We and others found that MDMX can be ubiquitinated and degraded by MDM2.45–47 However, this reaction is tightly controlled in vivo and significantly accelerated by stress signals such as DNA damage,46 ARF expression47 or ribosomal stress.48
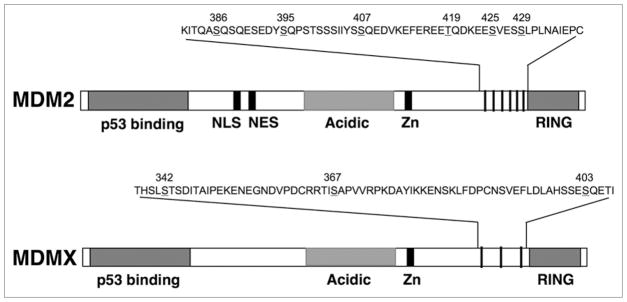
After DNA damage, MDMX is phosphorylated at several serine residues near the RING domain in an ATM-dependent manner (Fig. 1). Phosphorylation of MDMX led to increased binding, ubiquitination, and degradation by MDM2.49 ATM modifies S403,50 and Chk2 modifies S342 and S367.49 Chk2 phosphorylation of S367 creates a binding site for 14-3-3. 14-3-3 binding promotes MDMX translocation to the nucleus by activating a cryptic NLS in the RING domain,51 or sequesters phosphorylated MDMX in the nucleus.52 14-3-3 may also stimulate MDMX degradation by displacing deubiquitinating enzyme HAUSP,52 which plays a role in MDMX degradation after DNA damage.53,54 The physiological function of MDMX phosphorylation sites has been validated by knock-in mouse model.55 Blocking MDMX phosphorylation in vivo dampens p53 activation after DNA damage and accelerates lymphoma development upon overexpression of c-Myc oncogene.
Figure 1.
Phosphorylation sites on MDM2 and MDMX. Schematic diagram shows the relative positions of phosphorylation sites near the C terminus of MDM2 and MDMX.
MDM2 Phosphorylation After DNA Damage
MDM2 phosphorylation has also been implicated in mediating p53 activation. DNA damage induces MDM2 phosphorylation on serine 395 by ATM,56 on serine 407 by ATR,57 and on tyrosine 394 by c-Abl.58 Phospho-mimic mutations of these sites inhibit MDM2’s ability to regulate p53 degradation or nuclear export.56,59 The WIP1 phosphatase has been shown to inhibit p53 damage response, possibly in part by dephosphorylating MDM2 serine 395.60 Several phosphorylation sites (targets of GSK3β) in the MDM2 acidic domain are downregulated by DNA damage,61 and alanine substitution of S256 reduces MDM2-mediated ubiquitination of p53.62
Recent studies suggested two mechanisms of how MDM2 phosphorylation regulates p53 degradation. One study showed that MDM2 interaction with the scaffold protein Daxx and deubiquiting enzyme HAUSP is stimulated by DNA damage, which promote p53 de-ubiquitination.63 A second study reported that DNA damage induces MDM2 degradation in an phosphorylation-dependent fashion,64 suggesting that elimination of MDM2 leads to p53 stabilization. However, an earlier report showed that X-ray induces loss of MDM2 signal in western blot through ATM-dependent masking of the SMP14 epitope.65 It is note-worthy that SMP14 was one of the MDM2 antibody used in the Stommel study. Several subsequent studies that observed MDM2 downregulation also mentioned the use of SMP14. Therefore, additional work is needed to determine whether MDM2 undergoes accelerated self-degradation after DNA damage.
Regardless of whether MDM2 undergoes a transient downregulation after DNA damage, a significant change in MDM2 activity must also occur to allow p53 stabilization. DNA damage rapidly induces MDM2 expression due to p53 activation. For at least 12 hours after DNA damage, stabilized p53 co-exists with high level MDM2 in the cell, indicating that MDM2 has lost the ability to promote p53 degradation. In the past few years, we investigated the effect of DNA damage on MDM2 biochemical activity. The results have led to new insights on the molecular basis of this important signaling pathway, which is discussed below.
MDM2 Phosphorylation is Necessary for p53 Stabilization
In addition to the previously identified phosphorylation sites near the MDM2 RING domain (S395, S407), our mass spectrometric analysis of MDM2 from irradiated cells revealed several novel sites (S386, T419, S425 and S429). Two of these sites were confirmed to be ATM targets and were strongly induced by DNA damage.66 The location of these sites on MDM2 show an interesting parallel to the ATM and Chk2 sites on MDMX (Fig. 1), suggesting that this region has conserved regulatory functions. Mutational analysis suggested that the MDM2 phosphorylation sites have significant redundancy in regulating p53 degradation. Phospho-mimic mutation of a single site can strongly inhibit p53 degradation. Interestingly, alanine substitution of all 6 sites resulting in MDM2-6A mutant that is hyper active in p53 degradation. Furthermore, MDM2-6A ectopic expression blocked p53 accumulation after DNA damage despite normal p53 acetylation and S15 phosphorylation.66 These results demonstrated that MDM2 phosphorylation is critical for p53 stabilization by ATM signaling.
Because of the functional redundancy of the ATM sites on MDM2, alanine substitution of up to 3 sites showed no phenotype. From hindsight it is clear that exhaustive mutagenesis was a necessary approach, given that a single ATM site is sufficient for DNA damage response. Therefore, comprehensive mapping of all major phosphorylation sites on MDM2 was essential for evaluating their functional significance in p53 stabilization. Obviously, these findings will need to pass the ultimate test in mouse models in the future.
ATM Regulates MDM2 Oligomerization and E3 Function
How does MDM2 phosphorylation prevent degradation of p53? We found that phosphorylation inhibits the ability of MDM2 to promote p53 poly ubiquitination. Furthermore, ATM does not cause a general inhibition of MDM2 E3 ligase activity, since p53 mono ubiquitination, MDM2 self-ubiquitination and MDMX ubiquitination are not affected. Because proteasome degradation of substrates requires conjugation of poly ubiquitin chains,67 this finding provides an attractive explanation for p53 stabilization. The results also explain a previous observation that ionizing irradiation does not eliminate low MW forms of ubiquitinated p53 (mono ubiquitination).68
What is the molecular basis of p53 poly ubiquitination by MDM2? Important clues can be obtained from the literature. A recent study suggests that E3 dimerization is important for promoting substrate poly ubiquitination.69 Interestingly MDM2 RING domain produced in E. coli assembles into oligomers.70 Furthermore, recombinant MDM2 C terminal fragments have a tendency to precipitate out of solution.71 These observations suggest that MDM2 RING domain oligomerization may be key to its p53 poly ubiquitination activity, which is regulated by ATM phosphorylation.
This hypothesis was born out by our experiments. MDM2 C terminal fragment behaved as high MW oligomeric complex in gel filtration chromatography. Phosphorylation by ATM or phospho-mimic substitution inhibited oligomer formation during gel filtration, and also blocked RING domain interaction in an in vitro mixing assay.66 These results suggest that ATM-mediated phosphorylation inhibits MDM2 RING domain homo-dimerization or oligomerization, preventing the formation of a scaffold for synthesis of poly ubiquitin chains on p53.
The structural basis of MDM2 oligomerization remains unclear. The co-crystal structure of MDM2-MDMX RING heterodimer revealed the presence of an interface that may also mediate MDM2 homo dimerization,71 but does not indicate what structure could be responsible for higher-order interaction. Structure of BRCA1-BARD1RINGdomainheterodimershowed that sequences flanking the RING domains also engage in dimer formation.72 Results by Poyurovsky et al. indicated that the MDM2 RING domain together with additional N and C terminal sequences (400–491) are sufficient for oligomerization.70 The MDM2 sequence containing ATM phosphorylation sites may mediate additional interactions to stabilize the RING domain oligomer. Such interaction may be the target for regulation by phosphorylation.
How efficient is MDM2 phosphorylation by ATM in vivo? Taking advantage of phosphorylation-induced mobility shift of an MDM2 proteolytic fragment, we determined that IR induces nearly 100% phosphorylation of MDM2 even in overexpression conditions.66 This explains the strong inhibition of RING domain oligomerization during gel filtration analysis. The observation underscores the efficiency of ATM modification, possibly attributed to the abundance of activated ATM and affinity of the MDM2 target sites. The redundancy of ATM sites may also ensure a complete shut down of the MDM2 feedback loop after DNA damage, despite MDM2 induction by p53.
MDM2 Oligomerization and p53 Poly Ubiquitination
How does MDM2 RING domain oligomerization lead to p53 poly ubiquitination? Previous studies showed that E3 ligase stimulates ubiquitination mainly by binding both substrate and E2 conjugating enzymes, bringing the reactive E2-ubiquitin thioester bond to close proximity with a substrate lysine. Nucleophilic attack by the lysine epsilon amino group results in the transfer of ubiquitin from E2 to the substrate lysine.73–75 This “induced-proximity” model explains the substrate specificity of E3 ligases, but does not show how subsequent reactions synthesize poly ubiquitin chains necessary for substrate degradation.
To degrade p53, MDM2 must promote synthesis of poly ubiquitination chains (>4 subunits) on p53. However, p53 has 20 lysine residues and ~6–10 of these can be mono ubiquitinated by MDM2, as shown using lysine-free ubiquitin and mass spectrometry analysis. Therefore, in each round of reaction, ubiquitin can be transferred to free lysine residues on p53, or to a previously conjugated ubiquitin. Random modification of all accessible p53 lysines or random extension of all short ubiquitin chains would be inefficient for p53 degradation. Therefore, MDM2 may have a mechanism to selectively elongating a single ubiquitin chain after p53 mono ubiquitination, i.e., behaving as a processive enzyme.
Classic processive enzymes that synthesize long polymers (DNA or RNA polymerases, ribosomes) achieve un-interrupted elongation of a single product by staying engaged to the template and substrate while allowing two-dimensional sliding. This is accomplished either topologically (by the PCNA toroid during DNA replication) or through extensive but weak surface contact (between RNA polymerase and DNA during transcription).76
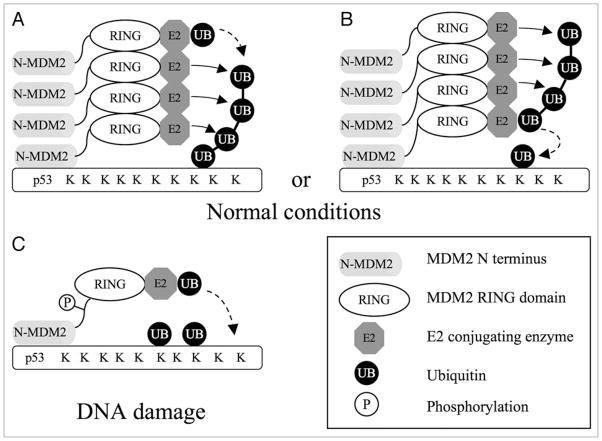
By analogy, MDM2 may gain processivity through formation of dimers and oligomers (Fig. 2). MDM2-p53 binding initiates p53 mono ubiquitination. MDM2 RING domain oligomer may favor subsequent synthesis of poly ubiquitin chain by recruiting multiple E2 molecules. It has been shown that UbcH5 family (the major E2 for MDM2) has a secondary non-covalent ubiquitin binding site.77,78 Low-affinity ubiquitin binding by multiple E2s may attract the RING-E2 complex to the ubiquitin chain. As such, ubiquitin transfer is more likely to occur to ubiquitin instead of p53. Long ubiquitin chains will have higher affinity for RING-E3 oligomer through multiple binding, thus achieving a self-reinforcing chain elongation reaction. Previous study showed that low concentrations of MDM2 (=1 × [p53]) can only mono-ubiquitinate p53 in vitro, whereas excess MDM2 (=54 × [p53]) induces p53 poly ubiquitination.79 This may be due to mass-driven oligomerization of MDM2 at high concentration, conferring a poly ubiquitination activity.
Figure 2.
A model of MDM2 regulation by phosphorylation. (a) in the absence of DNA damage, MDM2 RING domain forms oligomers that recruit multiple E2s, increasing the processivity of ubiquitin chain elongation. Ubiquitin is preferentially transferred to another proximal ubiquitin, thus building poly-ubiquitin chain on p53 through sequential transfer reactions. (B) alternatively, oligomerization of MDM2 stabilizes E2 oligomers, allowing poly-ubiquitin chain to be pre-assembled on E2 before transfering en bloc to p53. (C) after DNA damage, phosphorylation of MDM2 causes conformational changes that prevent RING domain oligomerization. Monomeric RING domain retains the ability to promote mono ubiquitination of different lysines on p53.
After DNA damage, phosphorylated MDM2 monomers may still bind p53 and recruit activated E2, and thus can promote mono ubiquitination of p53. However, recruiting a single E2 by RING monomer may not provide sufficient affinity for the already conjugated ubiquitin, thus each round of ubiquitin transfer will occur at random with lysines on p53 or ubiquitin (Fig. 2). In addition, the termini of short ubiquitin chains that are synthesized by chance will extend out of reach by MDM2, reducing the probability of further elongation. In contrast, MDM2 oligomerization forms a large scaffold that can present E2 at various distances and angles, thus increasing the chance of chain elongation.
In addition to the model proposed above, the data do not rule out other mechanisms of poly ubiquitin chain synthesis. The secondary ubiquitin-binding site on UbcH5 allows formation of oligomers between activated UbcH5~Ubiquitin.77 MDM2 may further stimulate and stabilize UbcH5~Ub oligomer formation and recruitment to p53. UbcH5~Ub oligomer formation may lead to pre-assembly of poly ubiquitin chain on UbcH5, allowing the poly ubiquitination of p53 through a single round of transfer. An example of such mechanism is illustrated by the gp78-Ube2g2 system.80,81 MDM2 oligomerization may also be associated with an ability to allosterically regulate E2 activity.82 Further experiments are needed to investigate these possibilities.
Summary and General Implications
Analysis of MDM2 phosphorylation suggests a novel mechanism of p53 regulation by ATM. The efficiency of MDM2 phosphorylation in vivo and the functional effects suggest that this may be a major mechanism of p53 stabilization after DNA damage. However, p53 stress response is likely to be achieved by multiple mechanisms acting synergistically. Phosphorylation of MDM2 also regulates interaction with HAUSP,63 which provides an active means of de-ubiquitinating p53 and rescues the molecules from degradation. Furthermore, the p300/CBP coactivators promote p53 poly ubiquitination by acting as E4 ligase in the cytoplasm. p300/CBP encode intrinsic E3 ligase activity that can extend the ubiquitin chain initiated by MDM2.83 Presumably, p300/CBP binding to MDM2 or p53 is needed to provide target specificity for the E4 function, which may also be regulated by DNA damage.
Targeting of proteins for proteasomal degradation requires conjugation of poly ubiquitin chain on a substrate lysine. The mechanisms of ubiquitin chain polymerization and its regulation are still under active investigation.84 Recent studies uncovered the importance of E3 dimerization in promoting ubiquitin chain elongation. In the SCFcdc4 E3 ligase complex, dimer formation by SCF protomers is important for poly ubiquitination of substrates, whereas failure to form E3 dimer results in mono ubiquitination.69 Defective SCF(Fbx4) dimerization in cancer prevents ubiquitination of its substrate cyclin D1 and contributes to cyclin D1 overexpression.85 Oligomerization of the E3 ligase gp78 promotes complex formation of its cognate E2 Ube2g2 and pre-assembly of poly ubiquitin chains on the E2.80 Formation of high-order oligomers is a common feature of RING domains.70,86 Inhibition of RING domain self-assembly may be an effective and general mechanism for regulating E3 ligase activity. Understanding the structural basis for this regulation may provide clues for therapeutic targeting of MDM2 and other E3 ligases.
Acknowledgments
This work was supported by grants from the National Institutes of Health (CA109636, CA118219).
References
- 1.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–83. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 2.Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–91. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 3.Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 4.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–6. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 5.Grier JD, Xiong S, Elizondo-Fraire AC, Parant JM, Lozano G. Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Mol Cell Biol. 2006;26:192–8. doi: 10.1128/MCB.26.1.192-198.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 7.Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, Bazuine M, et al. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 1996;15:5349–57. [PMC free article] [PubMed] [Google Scholar]
- 8.Finch RA, Donoviel DB, Potter D, Shi M, Fan A, Freed DD, et al. mdmx is a negative regulator of p53 activity in vivo. Cancer Res. 2002;62:3221–5. [PubMed] [Google Scholar]
- 9.Migliorini D, Lazzerini Denchi E, Danovi D, Jochemsen A, Capillo M, Gobbi A, et al. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol. 2002;22:5527–38. doi: 10.1128/MCB.22.15.5527-5538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parant JM, Reinke V, Mims B, Lozano G. Organization, expression and localization of the murine mdmx gene and pseudogene. Gene. 2001;270:277–83. doi: 10.1016/s0378-1119(01)00432-2. [DOI] [PubMed] [Google Scholar]
- 11.Xiong S, Van Pelt CS, Elizondo-Fraire AC, Liu G, Lozano G. Synergistic roles of Mdm2 and Mdm4 for p53 inhibition in central nervous system development. Proc Natl Acad Sci USA. 2006;103:3226–31. doi: 10.1073/pnas.0508500103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maetens M, Doumont G, Clercq SD, Francoz S, Froment P, Bellefroid E, et al. Distinct roles of Mdm2 and Mdm4 in red cell production. Blood. 2007;109:2630–3. doi: 10.1182/blood-2006-03-013656. [DOI] [PubMed] [Google Scholar]
- 13.Badciong JC, Haas AL. MdmX is a RING finger ubiquitin ligase capable of synergistically enhancing Mdm2 ubiquitination. J Biol Chem. 2002;277:49668–75. doi: 10.1074/jbc.M208593200. [DOI] [PubMed] [Google Scholar]
- 14.Linares LK, Hengstermann A, Ciechanover A, Muller S, Scheffner M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci USA. 2003;100:12009–14. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu J, Kawai H, Nie L, Kitao H, Wiederschain D, Jochemsen AG, et al. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J Biol Chem. 2002;277:19251–4. doi: 10.1074/jbc.C200150200. [DOI] [PubMed] [Google Scholar]
- 16.Gilkes DM, Chen L, Chen J. MDMX regulation of p53 response to ribosomal stress. EMBO J. 2006;25:5614–25. doi: 10.1038/sj.emboj.7601424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toledo F, Krummel KA, Lee CJ, Liu CW, Rodewald LW, Tang M, et al. A mouse p53 mutant lacking the proline-rich domain rescues Mdm4 deficiency and provides insight into the Mdm2-Mdm4-p53 regulatory network. Cancer Cell. 2006;9:273–85. doi: 10.1016/j.ccr.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 18.Francoz S, Froment P, Bogaerts S, De Clercq S, Maetens M, Doumont G, et al. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci USA. 2006;103:3232–7. doi: 10.1073/pnas.0508476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sdek P, Ying H, Chang DL, Qiu W, Zheng H, Touitou R, et al. MDM2 promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma protein. Mol Cell. 2005;20:699–708. doi: 10.1016/j.molcel.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 20.Kawai H, Wiederschain D, Yuan ZM. Critical contribution of the MDM2 acidic domain to p53 ubiquitination. Mol Cell Biol. 2003;23:4939–47. doi: 10.1128/MCB.23.14.4939-4947.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meulmeester E, Frenk R, Stad R, de Graaf P, Marine JC, Vousden KH, et al. Critical role for a central part of Mdm2 in the ubiquitylation of p53. Mol Cell Biol. 2003;23:4929–38. doi: 10.1128/MCB.23.14.4929-4938.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang C, Ivanov A, Chen L, Fredericks WJ, Seto E, Rauscher FJ, 3rd, et al. MDM2 interaction with nuclear corepressor KAP1 contributes to p53 inactivation. EMBO J. 2005;24:3279–90. doi: 10.1038/sj.emboj.7600791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sui G, Affar el B, Shi Y, Brignone C, Wall NR, Yin P, et al. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–72. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Hu M, Gu L, Li M, Jeffrey PD, Gu W, Shi Y. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol. 2006;4:27. doi: 10.1371/journal.pbio.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003;23:8902–12. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Midgley CA, Desterro JM, Saville MK, Howard S, Sparks A, Hay RT, et al. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene. 2000;19:2312–23. doi: 10.1038/sj.onc.1203593. [DOI] [PubMed] [Google Scholar]
- 27.Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6:663–73. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- 28.Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 2004;23:2402–12. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–87. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 30.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem. 2004;279:44475–82. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 31.Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol. 2004;24:7654–68. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, et al. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–97. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 33.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–68. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 34.Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–7. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 35.Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–34. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 36.Chehab NH, Malikzay A, Appel M, Halazonetis TD. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 2000;14:278–88. [PMC free article] [PubMed] [Google Scholar]
- 37.Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- 38.Bottger V, Bottger A, Garcia-Echeverria C, Ramos YF, van der Eb AJ, Jochemsen AG, et al. Comparative study of the p53-mdm2 and p53-MDMX interfaces. Oncogene. 1999;18:189–99. doi: 10.1038/sj.onc.1202281. [DOI] [PubMed] [Google Scholar]
- 39.Ashcroft M, Kubbutat MH, Vousden KH. Regulation of p53 function and stability by phosphorylation. Mol Cell Biol. 1999;19:1751–8. doi: 10.1128/mcb.19.3.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blattner C, Tobiasch E, Litfen M, Rahmsdorf HJ, Herrlich P. DNA damage induced p53 stabilization: no indication for an involvement of p53 phosphorylation. Oncogene. 1999;18:1723–32. doi: 10.1038/sj.onc.1202480. [DOI] [PubMed] [Google Scholar]
- 41.Dumaz N, Meek DW. Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J. 1999;18:7002–10. doi: 10.1093/emboj/18.24.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chao C, Herr D, Chun J, Xu Y. Ser18 and 23 phosphorylation is required for p53-dependent apoptosis and tumor suppression. EMBO J. 2006;25:2615–22. doi: 10.1038/sj.emboj.7601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacPherson D, Kim J, Kim T, Rhee BK, Van Oostrom CT, DiTullio RA, et al. Defective apoptosis and B-cell lymphomas in mice with p53 point mutation at Ser 23. EMBO J. 2004;23:3689–99. doi: 10.1038/sj.emboj.7600363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chao C, Hergenhahn M, Kaeser MD, Wu Z, Saito S, Iggo R, et al. Cell type- and promoter-specific roles of Ser18 phosphorylation in regulating p53 responses. J Biol Chem. 2003;278:41028–33. doi: 10.1074/jbc.M306938200. [DOI] [PubMed] [Google Scholar]
- 45.de Graaf P, Little NA, Ramos YF, Meulmeester E, Letteboer SJ, Jochemsen AG. Hdmx protein stability is regulated by the ubiquitin ligase activity of Mdm2. J Biol Chem. 2003;278:38315–24. doi: 10.1074/jbc.M213034200. [DOI] [PubMed] [Google Scholar]
- 46.Kawai H, Wiederschain D, Kitao H, Stuart J, Tsai KK, Yuan ZM. DNA damage-induced MDMX degradation is mediated by MDM2. J Biol Chem. 2003;278:45946–53. doi: 10.1074/jbc.M308295200. [DOI] [PubMed] [Google Scholar]
- 47.Pan Y, Chen J. MDM2 promotes ubiquitination and degradation of MDMX. Mol Cell Biol. 2003;23:5113–21. doi: 10.1128/MCB.23.15.5113-5121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gilkes DM, Chen J. Distinct roles of MDMX in the regulation of p53 response to ribosomal stress. Cell Cycle. 2007;6:151–5. doi: 10.4161/cc.6.2.3719. [DOI] [PubMed] [Google Scholar]
- 49.Chen L, Gilkes DM, Pan Y, Lane WS, Chen J. ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J. 2005;24:3411–22. doi: 10.1038/sj.emboj.7600812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pereg Y, Shkedy D, de Graaf P, Meulmeester E, Edelson-Averbukh M, Salek M, et al. Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proc Natl Acad Sci USA. 2005;102:5056–61. doi: 10.1073/pnas.0408595102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.LeBron C, Chen L, Gilkes DM, Chen J. Regulation of MDMX nuclear import and degradation by Chk2 and 14-3-3. EMBO J. 2006;25:1196–206. doi: 10.1038/sj.emboj.7601032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pereg Y, Lam S, Teunisse A, Biton S, Meulmeester E, Mittelman L, et al. Differential roles of ATM- and Chk2-mediated phosphorylations of Hdmx in response to DNA damage. Mol Cell Biol. 2006;26:6819–31. doi: 10.1128/MCB.00562-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meulmeester E, Maurice MM, Boutell C, Teunisse AF, Ovaa H, Abraham TE, et al. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell. 2005;18:565–76. doi: 10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 54.Meulmeester E, Pereg Y, Shiloh Y, Jochemsen AG. ATM-mediated phosphorylations inhibit Mdmx/Mdm2 stabilization by HAUSP in favor of p53 activation. Cell Cycle. 2005;4:1166–70. doi: 10.4161/cc.4.9.1981. [DOI] [PubMed] [Google Scholar]
- 55.Wang YV, Leblanc M, Wade M, Jochemsen AG, Wahl GM. Increased radioresistance and accelerated B cell lymphomas in mice with Mdmx mutations that prevent modifications by DNA-damage-activated kinases. Cancer Cell. 2009;16:33–43. doi: 10.1016/j.ccr.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maya R, Balass M, Kim ST, Shkedy D, Leal JF, Shifman O, et al. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 2001;15:1067–77. doi: 10.1101/gad.886901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shinozaki T, Nota A, Taya Y, Okamoto K. Functional role of Mdm2 phosphorylation by ATR in attenuation of p53 nuclear export. Oncogene. 2003;22:8870–80. doi: 10.1038/sj.onc.1207176. [DOI] [PubMed] [Google Scholar]
- 58.Sionov RV, Coen S, Goldberg Z, Berger M, Bercovich B, Ben-Neriah Y, et al. c-Abl regulates p53 levels under normal and stress conditions by preventing its nuclear export and ubiquitination. Mol Cell Biol. 2001;21:5869–78. doi: 10.1128/MCB.21.17.5869-5878.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goldberg Z, Vogt Sionov R, Berger M, Zwang Y, Perets R, Van Etten RA, et al. Tyrosine phosphorylation of Mdm2 by c-Abl: implications for p53 regulation. EMBO J. 2002;21:3715–27. doi: 10.1093/emboj/cdf384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu X, Ma O, Nguyen TA, Jones SN, Oren M, Donehower LA. The Wip1 Phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell. 2007;12:342–54. doi: 10.1016/j.ccr.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 61.Blattner C, Hay T, Meek DW, Lane DP. Hypophosphorylation of Mdm2 augments p53 stability. Mol Cell Biol. 2002;22:6170–82. doi: 10.1128/MCB.22.17.6170-6182.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kulikov R, Boehme KA, Blattner C. Glycogen synthase kinase 3-dependent phosphorylation of Mdm2 regulates p53 abundance. Mol Cell Biol. 2005;25:7170–80. doi: 10.1128/MCB.25.16.7170-7180.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang J, Qu LK, Zhang J, Wang W, Michaelson JS, Degenhardt YY, et al. Critical role for Daxx in regulating Mdm2. Nat Cell Biol. 2006;8:855–62. doi: 10.1038/ncb1442. [DOI] [PubMed] [Google Scholar]
- 64.Stommel JM, Wahl GM. Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. EMBO J. 2004;23:1547–56. doi: 10.1038/sj.emboj.7600145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Toledo SM, Azzam EI, Dahlberg WK, Gooding TB, Little JB. ATM complexes with HDM2 and promotes its rapid phosphorylation in a p53-independent manner in normal and tumor human cells exposed to ionizing radiation. Oncogene. 2000;19:6185–93. doi: 10.1038/sj.onc.1204020. [DOI] [PubMed] [Google Scholar]
- 66.Cheng Q, Chen L, Li Z, Lane WS, Chen J. ATM activates p53 by regulating MDM2 oligomerization and E3 processivity. EMBO J. 2009 doi: 10.1038/emboj.2009.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maki CG, Howley PM. Ubiquitination of p53 and p21 is differentially affected by ionizing and UV radiation. Mol Cell Biol. 1997;17:355–63. doi: 10.1128/mcb.17.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tang X, Orlicky S, Lin Z, Willems A, Neculai D, Ceccarelli D, et al. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 2007;129:1165–76. doi: 10.1016/j.cell.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 70.Poyurovsky MV, Priest C, Kentsis A, Borden KL, Pan ZQ, Pavletich N, et al. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 2007;26:90–101. doi: 10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008;15:841–8. doi: 10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
- 72.Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE. Structure of a BRCA1-BARD1 heterodi-meric RING-RING complex. Nat Struct Biol. 2001;8:833–7. doi: 10.1038/nsb1001-833. [DOI] [PubMed] [Google Scholar]
- 73.Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell. 2003;11:1445–56. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 74.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–9. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 75.Passmore LA, Barford D. Getting into position: the catalytic mechanisms of protein ubiquitylation. Biochem J. 2004;379:513–25. doi: 10.1042/BJ20040198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Breyer WA, Matthews BW. A structural basis for processivity. Protein Sci. 2001;10:1699–711. doi: 10.1110/ps.10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21:873–80. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 78.Saville MK, Sparks A, Xirodimas DP, Wardrop J, Stevenson LF, Bourdon JC, et al. Regulation of p53 by the ubiquitin-conjugating enzymes UbcH5B/C in vivo. J Biol Chem. 2004;279:42169–81. doi: 10.1074/jbc.M403362200. [DOI] [PubMed] [Google Scholar]
- 79.Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–5. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 80.Li W, Tu D, Li L, Wollert T, Ghirlando R, Brunger AT, et al. Mechanistic insights into active site-associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc Natl Acad Sci USA. 2009;106:3722–7. doi: 10.1073/pnas.0808564106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li W, Tu D, Brunger AT, Ye Y. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature. 2007;446:333–7. doi: 10.1038/nature05542. [DOI] [PubMed] [Google Scholar]
- 82.Ozkan E, Yu H, Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc Natl Acad Sci USA. 2005;102:18890–5. doi: 10.1073/pnas.0509418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shi D, Pop MS, Kulikov R, Love IM, Kung AL, Grossman SR. CBP and p300 are cytoplasmic E4 polyubiquitin ligases for p53. Proc Natl Acad Sci USA. 2009;106:16275–80. doi: 10.1073/pnas.0904305106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 85.Barbash O, Zamfirova P, Lin DI, Chen X, Yang K, Nakagawa H, et al. Mutations in Fbx4 inhibit dimerization of the SCF(Fbx4) ligase and contribute to cyclin D1 overexpression in human cancer. Cancer Cell. 2008;14:68–78. doi: 10.1016/j.ccr.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kentsis A, Gordon RE, Borden KL. Control of biochemical reactions through supramolecular RING domain self-assembly. Proc Natl Acad Sci USA. 2002;99:15404–9. doi: 10.1073/pnas.202608799. [DOI] [PMC free article] [PubMed] [Google Scholar]