Abstract
The genome is constantly subjected to chemical alterations that have the potential to cause genetic mutation, chromosomal rearrangements and, in the case of multi-cellular organisms, cancer. Particular vulnerability exists during DNA replication, when the two DNA strands of a chromosome separate to form templates for the synthesis of sister chromatids. Attempted replication across a damaged or nicked DNA template can result in the formation of a double-strand break (DSB), arguably the most dangerous of DNA lesions. DSBs can also arise directly at any cell cycle stage following exposure to ionizing radiation or radiomimetic agents. To combat these recurrent threats of genomic instability, numerous distinct enzyme systems have evolved that sense DNA damage and coordinate its repair. Part of this coordination involves the activation of signal transduction cascades that target repair proteins, trigger DNA damage-dependent cell cycle checkpoints and profoundly affect chromatin neighboring a DSB. Here, we discuss current models of how lesion processing itself helps to coordinate these signals in dividing cells. Recent evidence in yeast of a role for cyclin-dependent kinases in DNA end resection suggests a possible solution to the long-standing puzzle of how DSBR pathway ‘choice’ is regulated through the cell cycle.
Keywords: DSB, recombination, checkpoint, CDK
Introduction
The context in which a double-strand break (DSB) arises is critical for determining how it is processed. At one extreme, DSBs initiated by the recombinase activating gene products, RAG1 and RAG2, within a mammalian immunoglobulin or T-cell receptor locus, are destined to be repaired by nonhomologous end-joining (NHEJ) (Bassing et al., 2002). NHEJ does not engage a neighboring intact DNA molecule, requires little or no homology between the ligated ends and employs an enzyme system almost entirely distinct from that used for homologous recombination (HR) (Krogh and Symington, 2004). RAG1 and RAG2, which are absolutely required for V(D)J rearrangement in mice and humans, play a part in specifying the extreme preference for NHEJ apparent in this process – ‘shepherding’ the DSB toward the NHEJ pathway (Lee et al., 2004).
In the more general case of a DSB arising in an unprogrammed manner, other rules must apply in the selection of DSBR pathways. Early sensors of the DNA break include the Ku heterodimer (Critchlow and Jackson, 1998) and the Mre11-Rad50-Nbs1 (MRN) complex (de Jager et al., 2001; Lisby et al., 2004; Shroff et al., 2004; Stracker et al., 2004). The binding of Ku leads to the engagement of NHEJ, while MRN association with the DSB is permissive of either NHEJ or HR. For HR to occur, significant 5′–3′ exonucleolytic attack of the double-stranded (ds)DNA end is needed, with the generation of a single-stranded (ss)DNA 3′ overhang. Mre11 is one exonuclease that may contribute to this end processing. Although the polarity of this exonuclease is 3′–5′, opposite to that expected for the generation of a 5′ recessed end, it is possible that the full activity in vivo has not yet been modeled in vitro (Krogh and Symington, 2004). Precedent for polarity switching exists in the prokaryotic DSB processing exonuclease complex, RecBCD (Anderson and Kowalczykowski, 1997). An alternative possible candidate is Exo1, which is a 5′–3′ dsDNA exonuclease (Llorente and Symington, 2004). Although Rad52 has some properties suggestive of a role as a ‘gatekeeper’ for channeling DSBR into HR pathways (Haber, 1999; Van Dyck et al., 2001), Rad52 binds preferentially to ssDNA ends, which might arise only subsequent to the ‘decision’ to resect the DNA end for recombination (Ristic et al., 2003). The factors that determine the choice of DSBR pathways are therefore not fully understood.
When a cell enters S phase with unrepaired DNA damage, attempted replication across the damaged template may result in stalling of the DNA polymerase (Cox et al., 2000; Kowalczykowski, 2000). Depending on the nature of the original DNA lesion and its location on the leading or lagging strand, this stalling event might generate a tract of ssDNA where lagging strand Okazaki fragment synthesis is incomplete or, if the lesion is on the leading strand, simple fork arrest. In Escherichia coli, the arrested fork is subject to fork reversal, generating a cruciate structure that can become a substrate for Holliday junction resolvases, the action of which could generate a DSB (Seigneur et al., 1998). Replication across a nicked DNA template will automatically generate a DSB at the stalled fork. Therefore, the stalled fork may, under some circumstances, be converted to a DSB. It is thought that the local environment of the stalled replication fork favors HR (using the sister chromatid as a template) over NHEJ. The association of the MRN heterotrimer with sites of replication in the mammalian nucleus suggests that this complex may be ready to load onto a replication-related DSB (Mirzoeva and Petrini, 2003). Perhaps, the same is true for other recombination proteins. In Xenopus, the key checkpoint kinase, ATR, associates with chromatin in a replication-dependent manner, even in the absence of replication arrest (Hekmat-Nejad et al., 2000). In addition, the recurrent generation of tracts of ssDNA on the lagging strand could acquire a new role in the context of replication arrest, perhaps channeling repair processes in favor of HR.
ssDNA as ligand
The generation of a tract of ssDNA has long been thought to be a key trigger to certain DNA damage checkpoints (Paulovich et al., 1997), especially those controlled by ATR (MEC1 in Saccharomyces cerevisiae; rad3 in Schizosaccharomyces pombe). A tract of persistent ssDNA will rapidly become coated with RPA, an abundant high-affinity ssDNA-binding hetero-trimeric protein complex that helps to ‘melt’ secondary structure in ssDNA (Figure 1) – a role similar to that of SSB in E. coli. The detailed mechanisms of signal activation by ssDNA/RPA are the subject of intensive current research. The minimum length of homology that will support efficient HR between two episomal plasmids is approximately 200 bp (Rubnitz and Subramani, 1984; Liskay et al., 1987). Presumably, the length of ssDNA/RPA generated during processing of a typical recombinogenic lesion is of the same order of magnitude. The ssDNA/RPA fiber presents a potentially titratable solid-state ligand for the activation of the key checkpoint kinase, ATR, and its downstream signal transducer, CHK1 (Chen and Sanchez, 2004).
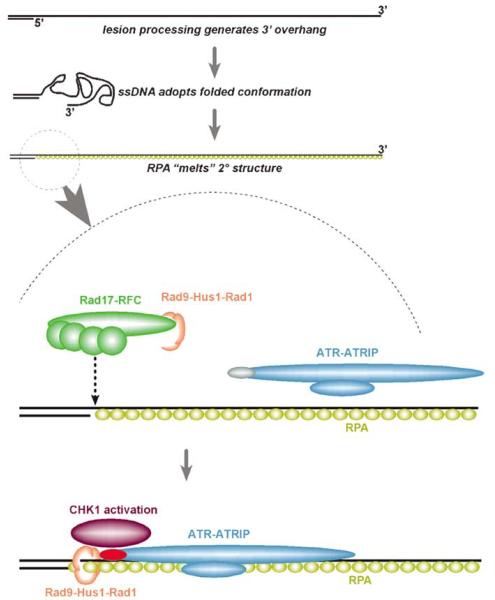
Figure 1.
ssDNA and RPA act as a ligand for activation of the ATR-CHK1 checkpoint pathway. The figure shows a schematic of the steps required for ATR checkpoint activation. First, resection of the DSB generates ssDNA with a 3′ overhang. RPA rapidly coats the ssDNA tail and removes secondary structure. This nucleoprotein filament is recognized, independently, by ATR/ATRIP and by Rad17-RFC. The latter loads the Rad9-Hus1-Rad1 complex onto ssDNA/RPA. Whether the 9-1-1 complex slides along dsDNA or along ssDNA/RPA is not yet proven. ATR is activated by binding to ssDNA and phosphorylates the C-terminus of Rad9. Together, ATR/ATRIP and the 9-1-1 complex synergize to generate maximal activation of CHK1. This may be facilitated by Claspin, Cut5 (not shown) and possibly other mediators not yet identified. ATR and/or CHK1 deliver the DNA damage response signal to downstream targets (not shown)
The activation of ATR, which exists as a heterodimer with ATRIP, is stimulated by its recruitment to the ssDNA/RPA fiber (Zou and Elledge, 2003). Recruitment of ATR/ATRIP is RPA dependent, but the precise structural elements of ATR/ATRIP that are required for recruitment are not known. ATR possesses a C-terminal PI3-kinase-like kinase domain that, when activated, phosphorylates CHK1 as well as many other targets of the DNA damage response (Osborn et al., 2002; Abraham, 2004; Shechter et al., 2004). Interestingly, in Xenopus egg extracts, ATR kinase activity is stimulated only weakly by binding to ssDNA, but strongly by gapped dsDNA (Kumagai et al., 2004). An important cofactor in these effects is claspin, a replication protein that becomes associated with chromatin after origin firing and remains associated during replication (Lee et al., 2003; Kumagai et al., 2004; Lin et al., 2004). The association of claspin with chromatin is not dependent on RPA, but claspin is required for the optimal activation of ATR kinase activity and of CHK1. Another cofactor, Cut5, has recently been found to activate CHK1 by facilitating the RPA-mediated recruitment of ATR to chromatin (Parrilla-Castellar and Karnitz, 2003).
A second important element of the early signaling of ssDNA lesions is the Rad9-Hus1-Rad1 (‘9-1-1′) complex (Parrilla-Castellar et al., 2004). This heterotrimer resembles the trimeric DNA polymerase processivity factor, PCNA, and is loaded onto ssDNA in an ATR-independent and RPA-dependent manner by a larger complex, Rad17-RFC (Figure 1) (Zou et al., 2002, 2003). Rad17-RFC shares components with the p140-RFC clamp loading complex that loads PCNA onto DNA (Griffith et al., 2002). Unlike p140-RFC, which recognizes 3′ recessed RNA primer ends, Rad17-RFC has some affinity for 5′ recessed ends and binds most strongly to a gapped DNA template (Bermudez et al., 2003). Like PCNA, the 9-1-1 complex is topologically bound in a ring around the DNA fiber. Although hydrodynamic experiments have not yet revealed clear evidence that the 9-1-1 complex can slide on DNA (as PCNA can), electron microscopy seems to support the idea that the 9-1-1 complex has at least limited processivity on dsDNA after Rad17-RFC-mediated loading onto a nicked circular plasmid (Bermudez et al., 2003). Purified human Rad9 has been shown to have 3′–5′ exonuclease activity in vitro, although the significance of this for DNA lesion processing is not well understood (Bessho and Sancar, 2000). Rad9 also possesses a unique C-terminal tail (not present in Hus1, Rad1 or PCNA), which is phosphorylated on multiple residues by kinases including ATR and ATM in the DNA damage response. The Rad9 C-terminal tail is required for optimal activation of CHK1 (Roos-Mattjus et al., 2003). Recently, paralogs of Rad9 and Hus1 (Rad9B and Hus1B, respectively) have been identified (Parrilla-Castellar et al., 2004). Their role in the DNA damage response is unknown.
ssDNA as substrate
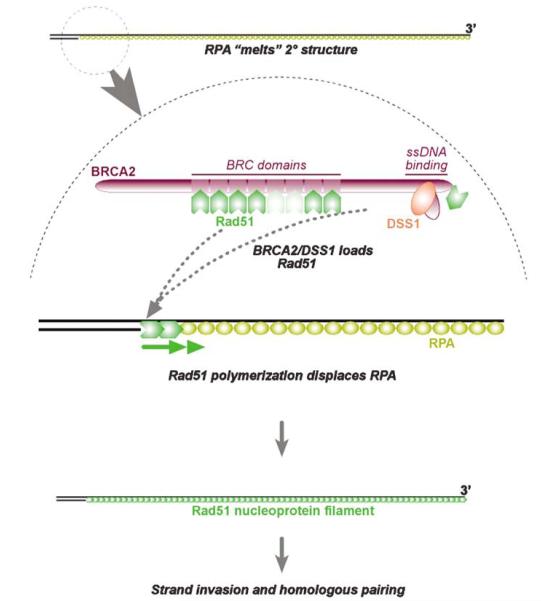
With admirable economy, the same ssDNA/RPA structure that serves as a platform for signal initiation becomes a substrate of recombination enzymes (Figure 2). If the ssDNA/RPA tract has a 3′-end, such as would arise during processing of a DSB for recombination, it is modified by BRCA2/Rad51 and associated recombination proteins, with the displacement of RPA by Rad51, which then polymerizes along the ssDNA (Wyman et al., 2004). The resulting Rad51 nucleoprotein filament, assisted by other recombination proteins, is competent for strand invasion of a homologous duplex, leading to homologous recombinational repair of the DSB. The displacement of RPA by Rad51 may, in this dynamic situation, be a ‘watershed’ event that separates ssDNA as the ligand in a signal transduction cascade from ssDNA as the substrate of recombinational repair. Whether there are specific coupling factors that play a part in terminating ssDNA/RPA-dependent signaling and switching to Rad51 loading is not known. Perhaps, BRCA1 is such a factor, since it receives DNA damage signals in the context of ATR (or ATM) activation, colocalizes with sites of ssDNA and DSB processing, and forms a biochemical association with BRCA2/Rad51 (Scully and Livingston, 2000).
Figure 2.
ssDNA and RPA act as a substrate that is processed for HR. BRCA2/DSS1 helps to load Rad51 onto ssDNA and Rad51 polymerizes along the ssDNA to form a Rad51 nucleoprotein filament. As it does so, RPA is displaced from ssDNA. The factors that regulate BRCA2 recruitment and the putative ‘switch’ of ssDNA/RPA from signaling to Rad51 loading functions are unknown. The Rad51 nucleoprotein filament invades a homologous dsDNA molecule, assisted by other recombination proteins (not shown)
Although ssDNA/RPA may have fundamentally similar properties wherever it appears, the context in which it forms may affect how it is processed. For example, it is unclear what is the fate of a tract of ssDNA generated at stalled forks that is not degraded to a DSB and therefore lacks a 3′-end. Such a situation could well arise following lagging strand DNA polymerase stalling during S phase and has been documented experimentally following treatment of cells with hydroxyurea (HU) (Sogo et al., 2002). HU treatment depletes the nucleotide pool and generates ‘frozen’ forks, with the formation of extensive tracts of ssDNA on the lagging strand. However, even prolonged treatment in HU generates very few DSBs in wild-type cells (Sogo et al., 2002; Lomonosov et al., 2003). The ssDNA gaps generated by HU treatment activate ATR and appear to cause recruitment of BRCA1, BRCA2 and Rad51 to the stalled fork. It therefore seems possible that BRCA2/Rad51 can displace RPA from unbroken ssDNA at an HU-treated fork and load Rad51. However, it is not clear that HR (sister chromatid recombination) is engaged unless the ssDNA tract is converted to a DSB – a rare, presumably stochastic, event. The ssDNA tract generated by HU treatment therefore appears to be a ‘frozen’ recombination intermediate that is perhaps not yet committed to completion of the recombination process.
Role for CDKs in regulating resection of a DSB
A DSB arising during G1 could, in theory, be processed to produce a 3′ overhang coated with RPA, triggering similar ATR-driven checkpoint and recombinational responses to those described above. However, striking new evidence suggests that a DSB encountered in G1 in yeast is relatively immune to resection of the end into ssDNA. Ira et al. (2004) analysed resection of a DSB induced at the MAT locus by the HO endonuclease and examined the consequent triggering of DNA damage checkpoint and recombination functions. G1-arrested cells failed to initiate efficient end resection of an HO endonuclease-induced DSB, failed to load RPA and Rad51 onto the DSB and were defective for Mec1 activation and recombination. Since this effect correlated with low levels of activity of the major cyclin-dependent kinase, CDK1/Cdc28, Ira et al. went on to test whether CDK1 activity itself determines whether the HO endonuclease-induced DSB can undergo end resection. Remarkably, if CDK1 activity is suppressed in G2 cells, either by overexpression of the CDK1 inhibitor, Sic1, or by use of a mutant form of CDK1 that can be blocked specifically by use of the ATP analog inhibitor, 1-NMMPP1, the cell is unable to resect an HO endonuclease-induced DSB. Similarly, checkpoint activation (measured as the activation of the ATR ortholog, Mec1) and recombination at the MAT locus are suppressed. Under these conditions, Mre11 persists for an abnormal duration at the broken MAT locus – consistent with the idea that the processing of the DSB has stalled. This suggests that CDK1 controls Mre11-associated exonuclease function at the DSB, but not the recruitment of Mre11 to DNA ends. Suppression of CDK1 activity can therefore convert a recombination competent G2 cell into one resembling the recombination defective G1 state.
Mre11 can be phosphorylated by CDK1 on certain cdk consensus sites. However, mutation of these sites does not abolish checkpoint activation, suggesting that end resection is also intact (Ira et al., 2004). This, in turn, suggests that the direct target of CDK1 in the control of end resection is something other than Mre11. Indeed, several other recombination proteins possess consensus cdk phosphorylation sites, raising the possibility that CDKs regulate multiple targets in the checkpoint and recombination response.
This work suggests an elegant and simple solution to the problem posed earlier regarding the ‘choice’ made between NHEJ and HR and its alteration during the cell cycle. During G1, the absence of CDK1 activity will disallow DSB end resection, ATR/Mec1 checkpoint activation and HR. Perhaps NHEJ predominates by default. Once CDK1 is activated as cells transition toward S phase, the environment becomes permissive of end resection and HR becomes the favored DSBR pathway. This CDK1-dependent mechanism might be reinforced by the relatively low levels of ATR and recombination proteins in G1 and by the absence of chromatin-bound Claspin. It is not clear how general this effect is beyond the specific case of the MAT locus, nor whether the CDK-mediated control of end resection is conserved across species. It is apparent from this work that ssDNA generated at stalled replication forks may not be subject to control by CDK1, since generation of ssDNA in this case does not arise by DNA end resection. However, the particular interest of the work of Ira et al. resides in the possibility it raises that signals concerned primarily with cell growth and division might also ‘shape’ the cell’s response to DSBs by regulating the efficiency of HR.
Acknowledgements
We thank members of the Scully lab for helpful discussions. This work was supported by awards R01 CA95175, ACS grant RSG-04-198-01-MGO and a Pew Scholars Award (to RS).
References
- Abraham RT. DNA Repair (Amst.) 2004;3:883–887. doi: 10.1016/j.dnarep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Anderson DG, Kowalczykowski SC. Genes Dev. 1997;11:571–581. doi: 10.1101/gad.11.5.571. [DOI] [PubMed] [Google Scholar]
- Bassing CH, Swat W, Alt FW. Cell. 2002;109(Suppl):S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- Bermudez VP, Lindsey-Boltz LA, Cesare AJ, Maniwa Y, Griffith JD, Hurwitz J, Sancar A. Proc. Natl. Acad. Sci. USA. 2003;100:1633–1638. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho T, Sancar A. J. Biol. Chem. 2000;275:7451–7454. doi: 10.1074/jbc.275.11.7451. [DOI] [PubMed] [Google Scholar]
- Chen Y, Sanchez Y. DNA Repair (Amst.) 2004;3:1025–1032. doi: 10.1016/j.dnarep.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- Critchlow SE, Jackson SP. Trends Biochem. Sci. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- de Jager M, van Noort J, van Gent DC, Dekker C, Kanaar R, Wyman C. Mol. Cell. 2001;8:1129–1135. doi: 10.1016/s1097-2765(01)00381-1. [DOI] [PubMed] [Google Scholar]
- Griffith JD, Lindsey-Boltz LA, Sancar A. J. Biol. Chem. 2002;277:15233–15236. doi: 10.1074/jbc.C200129200. [DOI] [PubMed] [Google Scholar]
- Haber JE. Nature. 1999;398:665–667. doi: 10.1038/19423. [DOI] [PubMed] [Google Scholar]
- Hekmat-Nejad M, You Z, Yee MC, Newport JW, Cimprich KA. Curr. Biol. 2000;10:1565–1573. doi: 10.1016/s0960-9822(00)00855-1. [DOI] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, Haber JE, Foiani M. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC. Trends Biochem. Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- Krogh BO, Symington LS. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Kim SM, Dunphy WG. J. Biol. Chem. 2004;279:49599–49608. doi: 10.1074/jbc.M408353200. [DOI] [PubMed] [Google Scholar]
- Lee GS, Neiditch MB, Salus SS, Roth DB. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- Lee J, Kumagai A, Dunphy WG. Mol. Cell. 2003;11:329–340. doi: 10.1016/s1097-2765(03)00045-5. [DOI] [PubMed] [Google Scholar]
- Lin SY, Li K, Stewart GS, Elledge SJ. Proc. Natl. Acad. Sci. USA. 2004;101:6484–6489. doi: 10.1073/pnas.0401847101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Liskay RM, Letsou A, Stachelek JL. Genetics. 1987;115:161–167. doi: 10.1093/genetics/115.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente B, Symington LS. Mol. Cell. Biol. 2004;24:9682–9694. doi: 10.1128/MCB.24.21.9682-9694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonosov M, Anand S, Sangrithi M, Davies R, Venkitaraman AR. Genes Dev. 2003;17:3017–3022. doi: 10.1101/gad.279003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzoeva OK, Petrini JH. Mol. Cancer Res. 2003;1:207–218. [PubMed] [Google Scholar]
- Osborn AJ, Elledge SJ, Zou L. Trends Cell Biol. 2002;12:509–516. doi: 10.1016/s0962-8924(02)02380-2. [DOI] [PubMed] [Google Scholar]
- Parrilla-Castellar ER, Arlander SJ, Karnitz L. DNA Repair (Amst.) 2004;3:1009–1014. doi: 10.1016/j.dnarep.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Parrilla-Castellar ER, Karnitz LM. J. Biol. Chem. 2003;278:45507–45511. doi: 10.1074/jbc.C300418200. [DOI] [PubMed] [Google Scholar]
- Paulovich AG, Toczyski DP, Hartwell LH. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- Ristic D, Modesti M, Kanaar R, Wyman C. Nucleic Acids Res. 2003;31:5229–5237. doi: 10.1093/nar/gkg729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos-Mattjus P, Hopkins KM, Oestreich AJ, Vroman BT, Johnson KL, Naylor S, Lieberman HB, Karnitz LM. J. Biol. Chem. 2003;278:24428–24437. doi: 10.1074/jbc.M301544200. [DOI] [PubMed] [Google Scholar]
- Rubnitz J, Subramani S. Mol. Cell. Biol. 1984;4:2253–2258. doi: 10.1128/mcb.4.11.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R, Livingston DM. Nature. 2000;408:429–432. doi: 10.1038/35044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneur M, Bidnenko V, Ehrlich SD, Michel B. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- Shechter D, Costanzo V, Gautier J. DNA Repair (Amst.) 2004;3:901–908. doi: 10.1016/j.dnarep.2004.03.020. [DOI] [PubMed] [Google Scholar]
- Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M. Curr. Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- Stracker TH, Theunissen JW, Morales M, Petrini JH. DNA Repair (Amst.) 2004;3:845–854. doi: 10.1016/j.dnarep.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Van Dyck E, Stasiak AZ, Stasiak A, West SC. EMBO Rep. 2001;2:905–909. doi: 10.1093/embo-reports/kve201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman C, Ristic D, Kanaar R. DNA Repair (Amst.) 2004;3:827–833. doi: 10.1016/j.dnarep.2004.03.037. [DOI] [PubMed] [Google Scholar]
- Zou L, Cortez D, Elledge SJ. Genes Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- Zou L, Liu D, Elledge SJ. Proc. Natl. Acad. Sci. USA. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]