Abstract
The IMGT/HLA Database (www.ebi.ac.uk/imgt/hla/) specialises in sequences of polymorphic genes of the HLA system, the human major histocompatibility complex (MHC). The HLA complex is located within the 6p21.3 region on the short arm of human chromosome 6 and contains more than 220 genes of diverse function. Many of the genes encode proteins of the immune system and these include the 21 highly polymorphic HLA genes, which influence the outcome of clinical transplantation and confer susceptibility to a wide range of non-infectious diseases. The database contains sequences for all HLA alleles officially recognised by the WHO Nomenclature Committee for Factors of the HLA System and provides users with online tools and facilities for their retrieval and analysis. These include allele reports, alignment tools and detailed descriptions of the source cells. The online IMGT/HLA submission tool allows both new and confirmatory sequences to be submitted directly to the WHO Nomenclature Committee. The latest version (release 1.7.0 July 2000) contains 1220 HLA alleles derived from over 2700 component sequences from the EMBL/GenBank/DDBJ databases. The HLA database provides a model which will be extended to provide specialist databases for polymorphic MHC genes of other species.
INTRODUCTION
The IMGT, international ImMunoGeneTics Database, is a series of integrated databases specialising in the immunoglobulins, T cell receptors (TcR) and the major histocompatibility complexes (MHC) of all vertebrate species. The IMGT project includes IMGT/LIGM-DB (1) which contains Ig and TcR sequences, and IMGT/HLA (2) which contains sequences of the human MHC.
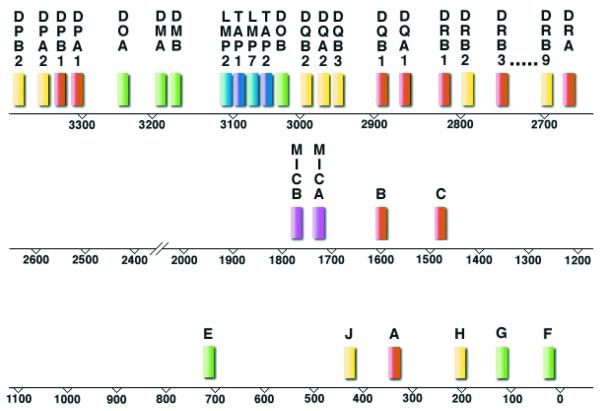
The IMGT/HLA Database is a specialist database for allelic sequences of highly polymorphic genes in the HLA system, the human MHC. This complex of ∼4 Mb is located within the 6p21.3 region of the short arm of human chromosome 6 and contains in excess of 220 genes. Many of these genes encode proteins of the immune system, in particular ones controlling the responses of T lymphocytes and natural killer cells. Genes included in the HLA nomenclature, and which comprise the HLA system, are those involved in antigen presentation to T cells, or are non-functional genes related to them. The core of the HLA system consists of the 21 highly polymorphic HLA genes (Fig. 1) which influence the outcome of cell and organ transplants and for which certain alleles are associated with progression of infectious disease and susceptibility to a wide range of chronic, non-infectious diseases (3,4). Sequences for more than 1200 different alleles of these genes have been determined. The naming of new allelic sequences and their quality control is the responsibility of the WHO Nomenclature Committee for Factors of the HLA System (5).
Figure 1.
Simplified map of the HLA complex. The highly polymorphic (classical) HLA genes are represented in red, HLA pseudogenes in yellow and monomorphic or oligomorphic (non-classical) HLA genes are shown in green. The MICA and MICB genes which are related in structure to the HLA-A, -B and -C genes and, like HLA genes, have been shown to be highly polymorphic, are shown in pink and have been included in the database. The remaining TAP and LMP genes, turquoise and blue, respectively, encode transporter proteins and proteasome subunits, respectively. Both TAP and LMP gene products contribute to antigen presentation by HLA-A, -B and -C molecules.
The IMGT/HLA database is a repository for the sequences officially recognised by the Nomenclature Committee (5) and was developed from the HLA database originally held at the Imperial Cancer Research Fund and, since 1996, at the Anthony Nolan Research Institute (ANRI). In contrast to the earlier HLA databases the IMGT/HLA Database is accessible via the World Wide Web and the sequences are accompanied by a number of tools and facilities for their analysis.
IMGT/HLA organisation and content
The database contains entries for all HLA alleles officially named by the WHO HLA Nomenclature Committee. These entries are derived from expertly annotated copies of the original EMBL/GenBank/DDBJ entries. The IMGT/HLA Database is based on a relational database model system and is maintained using the ORACLE® database management system. All IMGT/HLA entries have a unique accession number specific to IMGT/HLA. The EMBL/GenBank/DDBJ accession numbers are not used as primary identifiers in IMGT/HLA because many single alleles are derived from multiple sequence entries. The IMGT/HLA also assigns standardised keywords for all entries. The July 2000 release (Version 1.7.0) contained 1220 HLA alleles derived from over 2700 component EMBL/GenBank/DDBJ and GSDB sequences.
Database distribution
The first public release of the IMGT/HLA Database was made on the 16th December 1998 and was included on the European Bioinformatics Institute (EBI) web server as part of the IMGT project. The IMGT/HLA Database is available in a number of release formats. Previous versions were limited to static text pages. The IMGT/HLA data are now available via the EBI web site, EBI’s SRS search engine, EMBL CD-ROM and public FTP.
The IMGT/HLA web site was designed to provide a centralised source of both HLA sequences and tools for their analysis. The main tools allow the users to perform sequence alignments, single allele queries, BLAST searches and phylogenetic analysis. The web site combines custom built tools designed for the IMGT/HLA Database with existing tools already available from the EBI.
The IMGT/HLA web site is the main access point for most users of the database. The web site comprises a number of tools and facilities to enable the user to retrieve data and to perform analysis of the HLA sequences. It has been designed to provide the user with a single centralised resource that provides not only the sequence data, but also the tools with which to analyse the data and the necessary background information to support the work. It can be split into three main areas. First, information and help pages that provide background on the database provide in-depth help on the tools and data available and documentation of the IMGT/HLA flat-file format. The second area includes the tools designed specifically for the IMGT/HLA Database. These allow the user to perform sequence analysis and retrieval. The final pages are links to sequence-analysis tools at the EBI, including SRS, BLAST, FASTA, ClustalW and DNAplot.
The IMGT/HLA Database provides a number of tools for the online analysis of HLA allele sequences. These tools are unique to the IMGT/HLA Database web site. The allele-query tool allows users to retrieve the sequence of any officially named allele. These data include information on the source individual/cell, ethnic origin, references, official nucleotide and protein sequence. The output form provides this information complete with hypertext links to other online resources, including MEDLINE and cross-references to other sequence databases. The sequences are provided in a format similar to that produced previously (6,7).
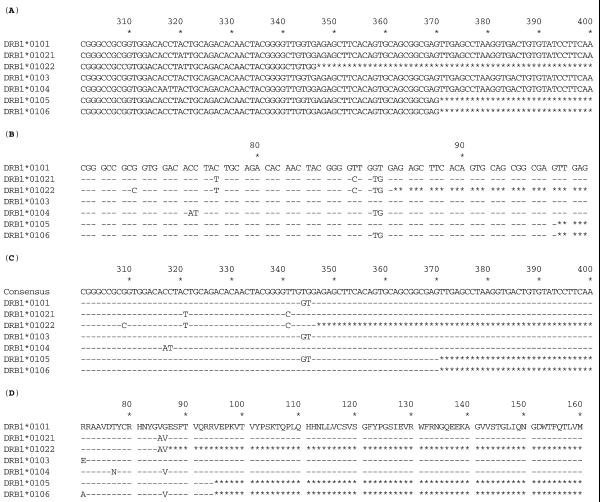
The sequence-alignment tool provided online follows the same format as the text alignments still available from the ANRI web pages (http://www.anthonynolan.org.uk/HIG/). The main difference is that the user has more control over the selection of what is to be aligned (Fig. 2). The alignment tool does not perform a sequence alignment each time it is used, but it extracts pre-aligned sequences, allowing for faster access. The tool allows the user to specify which sequences are aligned from an entire locus, for example DRB1, 3, 4 and 5, down to a small set of sequences such as the DRB1*0101, 0103, 0104 subset of DRB1*01 alleles. Additionally the sequences can be displayed as complete nucleotide sequence, partial sequences of single exons or the amino acid sequence of the encoded protein. The second change is in the display options, as the user can now specify how the output should appear; options include highlighting mismatches, splitting the sequence into codons and defining the reference sequence used. This provides a level of flexibility previously unavailable to the user.
Figure 2.
Alignment formats available from the IMGT/HLA Database.The examples shown are all based on alignment A which shows seven DRB1*01 alleles. Alignment B shows these sequences with the mismatches highlighted and the sequence split into codons. In these alignments a dash (–) indicates identity to the reference sequence and an asterisk (*) denotes an unsequenced base. Alignment C shows how an alternative reference sequence can be used, here for example we have used a DRB1 consensus sequence. Alignment D represents a translation of the nucleotide sequence to produce a protein sequence alignment.
The cell-query tool is used to interrogate the accompanying cell database. The HLA Cell Database contains a description of the primary lymphocytes or cell lines from which each HLA allele in the database was isolated and characterised. These cells are called source cells, and some of this data is only available through this interface and not currently via the IMGT/HLA flat files. This allows the HLA Cell Database to be used as an independent resource to the allele database. The tool can be used to search on HLA type, ethnic origin of the cell or geographical location, and then used to link to the appropriate HLA alleles.
IMGT/HLA flat files
The IMGT/HLA Database is available in flat-file format, which follows the EMBL flat-file format (8). The flat files provide a standard release format, assigning a standardised description and keywords to all entries. The IMGT/HLA Database assigns a unique accession number to every allele. This accession number is generated by the database and is not based on any of the EMBL/GenBank/DDBJ source entries. The accession numbers follow the format HLA00000, where the ‘00000’ represents a numerical code. This accession number is used in EBI tools like BLAST, SRS and the IMGT DNAplot tool. The flat files also contain the first release of the IMGT/HLA features. The sequence features currently used are the same as those used by EMBL/GenBank/DDBJ. As further features are developed, additional documentation will be provided to accompany these releases. The initial feature qualifiers cover source, coding sequence (cds), exon boundaries and the protein translation. The source feature contains only the basic information on each cell.
The IMGT/HLA Database is also available via FTP (see below). The FTP site provides the sequences in a number of predefined file formats including flat-file, FASTA, PIR and MSF formats.
Data submission
The database contains a tool for the online submission of HLA allele sequences. It acts as the main repository for the submission of new and confirmatory sequences to the WHO Nomenclature Committee for Factors of the HLA System. This online tool replaces the previous practice of emailing submissions directly to the committee. The submission tool mails a standardised report on each submission directly to the committee. This incorporates automated analysis and annotation of the sequence to aid in the identification of new alleles. The processed submissions can then be included directly into the IMGT/HLA Database. The submissions tool can be used for both new and confirmatory sequences, and is capable of holding confidential entries until a set time, thus allowing alleles to be named before publication. The submission of new HLA sequences to the IMGT/HLA Database does not replace the submission of these sequences to EMBL/GenBank/DDBJ, as the submission criteria state that the sequences must also have been submitted to these databases.
Future developments
Ongoing development of the database involves an expansion to include intron sequences into both the allele nomenclature and the alignments. Once this is completed for all the loci other motifs such as promoter sequences can be investigated. Further developments to the web site include tools for population genetics analysis and the design of oligonucleotide primers for typing and polymerase chain reaction amplification.
The database structure will also form the basis of a standard model for databases of non-human MHC sequences. The tools and flat-file formats will be incorporated to provide a standard for MHC sequence data. When complete this standardised storage and output of data will facilitate cross-species comparison of MHC sequences.
Summary
IMGT/HLA Database provides a centralised resource for everybody interested, either centrally or peripherally, in the HLA system. The database and accompanying tools allow the study of HLA alleles from a single site on the World Wide Web. It should aid in the management and continual expansion of the HLA nomenclature, providing an ongoing resource for the WHO Nomenclature Committee. The database will also act as a model system for the development of similar projects in other species.
Access and Contact
IMGT/HLA homepage: http://www.ebi.ac.uk/imgt/hla/
IMGT/HLA FTP site: ftp://ftp.ebi.ac.uk/pub/databases/imgt/mhc/hla/
IMGT/HLA documentation: http://www.ebi.ac.uk/imgt/hla/docs/release.html
IMGT/HLA submissions: http://www.ebi.ac.uk/imgt/hla/subs/submit.html
Email contact: hladb@ebi.ac.uk
Since release of the IMGT/HLA database in December 1998 the web site has received over 220 000 requests (figures from 16th December 1998 to 31st August 2000).
Acknowledgments
ACKNOWLEDGEMENTS
We would like to acknowledge the support of the following organisations: the American Society for Histocompatibility and Immunogenetics (ASHI), the Anthony Nolan Bone Marrow Trust (ANBMT), Dynal, Genovision, Lifecodes Corporation, the National Marrow Donor Program (NMDP), PE Applied Biosystems, Visible Genetics and the German National Bone Marrow Registry (ZKRD). In particular we thank Dr Janet Hegland at the NMDP for her support and effort in co-ordinating funding for this project. Initial support for the IMGT/HLA Database project was from the Imperial Cancer Research Fund and an EU Biotech grant (BIO4CT960037). We would also like to thank Dr Peter Stoehr and the staff at the European Bioinformatics Institute for their continued support of this project.
References
- 1.Ruiz M., Giudicelli,V., Ginestoux,C., Stoehr,P., Robinson,J., Bodmer,J., Marsh,S.G.E., Bontrop,R., Lemaitre,M., Lefranc,G. et al. (2000) IMGT, the international ImMunoGeneTics database. Nucleic Acids Res., 28, 219–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson J., Malik,A., Parham,P., Bodmer,J.G. and Marsh,S.G.E. (2000) IMGT/HLA database–a sequence database for the human major histocompatibility complex. Tissue Antigens, 55, 280–287. [DOI] [PubMed] [Google Scholar]
- 3.Charron D. (1997) Genetic diversity of HLA: Functional and Medical Implications. EDK, Paris, France. [PubMed]
- 4.Marsh S.G.E., Parham,P. and Barber,L.D. (2000) The HLA FactsBook. Academic Press, London, UK.
- 5.Bodmer J.G., Marsh,S.G.E., Albert,E.D., Bodmer,W.F., Bontrop,R.E., Dupont,B., Erlich,H.A., Hansen,J.A., Mach,B., Mayr,W.R. et al. (1999) Nomenclature for factors of the HLA system, 1998. Tissue Antigens, 53, 407–446. [DOI] [PubMed] [Google Scholar]
- 6.Marsh S.G.E. (1998) HLA class II region sequences, 1998. Tissue Antigens, 51, 467–507. [DOI] [PubMed] [Google Scholar]
- 7.Mason P.M. and Parham,P. (1998) HLA class I region sequences, 1998. Tissue Antigens, 51, 417–466. [DOI] [PubMed] [Google Scholar]
- 8.Baker W., van den Broek,A., Camon,E., Hingamp,P., Sterk,P., Stoesser,G. and Tuli,M.A. (2000) The EMBL nucleotide sequence database. Nucleic Acids Res., 28, 19–23. Updated article in this issue: Nucleic Acids Res. (2001), 29, 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]