Abstract
ABA is a major phytohormone that regulates a broad range of plant traits and is especially important for adaptation to environmental conditions. Our understanding of the molecular basis of ABA responses in plants improved dramatically in 2009 and 2010, banner years for ABA research. There are three major components; PYR/PYL/ RCAR (an ABA receptor), type 2C protein phosphatase (PP2C; a negative regulator) and SNF1-related protein kinase 2 (SnRK2; a positive regulator), and they offer a double negative regulatory system, [PYR/PYL/RCAR—| PP2C—| SnRK2]. In the absence of ABA, PP2C inactivates SnRK2 by direct dephosphorylation. In response to environmental or developmental cues, ABA promotes the interaction of PYR/PYL/RCAR and PP2C, resulting in PP2C inhibition and SnRK2 activation. This signaling complex can work in both the nucleus and cytosol, as it has been shown that SnRK2 phosphorylates basic-domain leucine zipper (bZIP) transcription factors or membrane proteins. Several structural analyses of PYR/PYL/RCAR have provided the mechanistic basis for this ‘core signaling’ model, by elucidating the mechanism of ABA binding of receptors, or the ‘gate–latch–lock’ mechanism of interaction with PP2C in inhibiting activity. On the other hand, intercellular ABA transport had remained a major issue, as had intracellular ABA signaling. Recently, two plasma membrane-type ABC transporters were identified and shed light on the influx/efflux system of ABA, resolving how ABA is transported from cell to cell in plants. Our knowledge of ABA responses in plants has been greatly expanded from intracellular signaling to intercellular transport of ABA.
Keywords: ABA, Protein phosphorylation, Receptor, Signal transduction, Transport
Introduction
ABA is a phytohormone that plays critical roles in adaptive responses to stresses such as drought and high salinity. It accumulates in plant cells under water stress, promotes stomatal closure in guard cells and regulates the expression of many genes, the products of which may protect vegetative tissues from dehydration or high osmotic pressure. In addition, ABA plays a central role in many developmental stages, such as seed maturation and dormancy. Thus, ABA can be considered to be a water stress-related phytohormone that contributes to the dehydration and/or desiccation tolerance of cells. The importance of ABA to global agriculture is widely accepted. Accordingly, numerous studies have attempted to understand the cellular and molecular responses to ABA in plants, including aspects related to sensing, signaling, metabolism and transport.
Before 2009, our knowledge of the cellular and molecular basis of ABA responses was nebulous, although numerous factors related to ABA responses had been reported (Hirayama and Shinozaki 2007, Hirayama and Shinozaki 2010). Inconveniently, the previous model of ABA signaling was very complicated because each ABA receptor and/or binding protein was randomly placed in different cellular locations in parallel, and their relationships to the ABA signaling pathway were complicated (McCourt and Creelman 2008). Although some ABA-binding proteins (receptors) were previously reported, such as G-protein-coupled receptors (GCR2, GTG1/2) and Mg-chelatase (ABAR), it remained to be determined how they regulate ABA responses in plants (Shen et al. 2006, Liu et al. 2007, Pandey et al. 2009). However, the situation totally changed in 2009, and the ABA signaling model was dramatically updated. The major breakthrough was made by two findings: (i) the discovery of PYR/PYL/RCAR as a new type of soluble ABA receptor (Ma et al. 2009, Park et al. 2009); and (ii) the identification of a protein phosphatase–kinase complex [type 2C protein phosphatase (PP2C)–SNF1-related protein kinase 2 (SnRK2)] as downstream components of PYR/PYL/RCARs (Umezawa et al. 2009, Vlad et al. 2009). After this remarkable achievement, several studies offered a double negative regulatory system of ABA signaling consisting of four stages: ABA receptors (PYR/PYL/RCAR), protein phosphatases (PP2C), protein kinases (SnRK2) and their downstream targets (Fujii et al. 2009, Umezawa et al. 2009). Finally, structural studies on PYR/PYL/RCAR fully supported this model. (Melcher et al. 2009, Miyazono et al. 2009, Nishimura et al. 2009, Santiago et al. 2009a, Yin et al. 2009).
On the other hand, the intercellular mechanism of ABA responses should be quite important, as should intracellular signaling. Cell to cell ABA transport, which remained unclear, was recently clarified by the discovery of plasma membrane-bound ABA transporters (Kang et al. 2010, Kuromori et al. 2010). This discovery made it important to identify the regulatory networks of intercellular signaling.
In this review, we outline a brief history and recent progress in investigations on the molecular basis of the regulatory network of sensing, signaling and transport of ABA, as well as an overview of the general process of ABA signaling and its physiological functions. There are recent reviews of the newly identified soluble ABA receptor and its downstream signaling networks (Cutler et al. 2010, Hubbard et al. 2010, Klingler et al. 2010, Raghavendra et al. 2010).
Emerging ‘Core Components’ of ABA signaling
Type 2C protein phosphatase (PP2C): a global negative regulator of ABA signaling
In the mid 1990s, a genetic screen for ABA-insensitive Arabidopsis mutants identified two genes, ABA-INSENSTIVE1 (ABI1) and ABI2, encoding PP2Cs (Leung et al. 1994, Meyer et al. 1994, Leung et al. 1997, Rodriguez et al. 1998). The mutants, abi1-1 or abi2-1, showed overall ABA insensitivity in various tissues and developmental stages, suggesting that PP2C acts as a global regulator of ABA signaling. Among the many PP2C family members in plants, ABI1 and ABI2 belong to a subgroup, group A, consisting of nine members in Arabidopsis (Fig. 1) (Schweighofer et al. 2004). Since their discovery, ABA signaling regulators such as HOMOLOGY TO ABI1 (HAB1) and HAB2 have been isolated based on sequence similarity to ABI1 (Saez et al. 2004). ABA-HYPERSENSITIVE GERMINATION1 (AHG1) and AHG3/AtPP2CA were cloned from genetic screens of Arabidopsis (Kuhn et al. 2006, T. Yoshida et al. 2006, Nishimura et al. 2007) and a yeast complementation test (Kuromori and Yamamoto 1994). Even though abi1-1 and abi2-1 were originally isolated based on their ABA insensitivity, all of the loss-of-function type mutants of group A PP2Cs exhibited significant ABA hypersensitivity, indicating that they are major negative regulators of ABA signaling (Hirayama and Shinozaki 2007). Negative regulatory roles for PP2C in ABA signaling have been demonstrated in some other plant species, suggesting that PP2C functions are well conserved (Fig. 1) (Gonzalez-Garcia et al. 2003, Komatsu et al. 2009, Tougane et al. 2010). This concept was further supported by double or triple PP2C knockout mutants with an elevated hypersensitivity to ABA (Nishimura et al. 2007, Rubio et al. 2009).
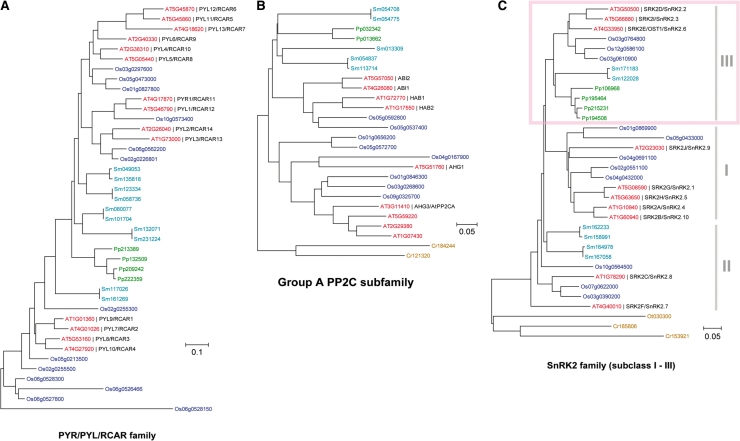
Fig. 1.
Phylogenetic trees of the core components in ABA signaling. Phylogenetic trees of PYR/PYL/RCAR (A), group A PP2C (B) and the SnRK2 family (C) from several plant species. Amino acid sequences for each family were retrieved from Arabidopsis (red), rice (blue), Selaginella moellendorffii (azure blue), Physcomitrella patens (green), Ostreococcus tauri and Chlamydomonas reinhardtii (yellow ocher) using the SALAD database (http://salad.dna.affrc.go.jp/CGViewer/). The tree was drawn using the Neighbor–Joining method in the MEGA 4.0 program.
Group A PP2Cs are functionally redundant at the molecular level, but they have distinctive roles in different tissues and organs, as indicated by tissue-specific expression patterns. For example, ABI1 is expressed in various tissues, including seeds and guard cells, but AHG1 and AHG3/AtPP2CA are expressed predominantly in seeds, reflecting their mutant phenotypes (T. Yoshida et al. 2006, Nishimura et al. 2007, Umezawa et al. 2009). In these cases, the subcellular localization patterns of ABI1 and AHG1/3 are clearly different, as ABI1 is localized to the cytosol and nucleus, but AHG1 and AHG3 are specifically localized in the nucleus (Umezawa et al. 2009). This is consistent with previous observations that ABI1 broadly regulates various ABA responses in tissues from seeds to guard cells (Leung et al. 1994, Meyer et al. 1994), but AHG1 and AHG3 mainly function in seeds, mostly to regulate gene expression in the nucleus (T. Yoshida et al. 2006, Nishimura et al. 2007). This is quite reasonable because transcriptional changes are dynamic in seed development, dormancy and germination (Nakabayashi et al. 2005).
As described above, the physiological functions of PP2Cs were clearly determined genetically in the mid 2000s. However, the molecular basis of PP2C-dependent negative regulation of ABA signaling was poorly understood before 2009. There was another major, long-standing question to be answered for group A PP2Cs. Based on the negative regulatory roles of group A PP2Cs, phenotypic changes in abi1-1 and abi2-1, with strong ABA insensitivity, were enigmatic. The mutation of abi1-1 and abi2-1 occurs in the catalytic domain of PP2C, with a well-conserved glycine being converted to aspartate. The same mutation in other group A PP2Cs also induces strong ABA insensitivity in plants (Robert et al. 2006, T. Yoshida et al. 2006). Although abi1-1 and abi2-1 mutants were widely used for ABA signaling studies, it was a mystery as to how this type of mutation induces dominant ABA insensitivity.
SNF1-related protein kinase 2 (SnRK2): a global positive regulator of ABA signaling
The identification and characterization of PP2C indicated the importance of protein phosphorylation events in ABA signaling. In line with this concept, several protein kinases were isolated and characterized as ABA signaling factors (Hirayama and Shinozaki 2007, Hirayama and Shinozaki 2010). The SnRK2 family was identified as ABA-activated protein kinases from fava bean ABA-ACTIVATED PROTEIN KINASE (AAPK) (Li et al. 2000) and Arabidopsis SRK2E/OPEN STOMATA1 (OST1)/SnRK2.6 (Mustilli et al. 2002, Yoshida et al. 2002). PKABA1 was isolated from wheat and is an ABA-inducible gene encoding an SnRK2 member (Anderberg and Walker-Simmons 1992). There are 10 SnRK2 members in Arabidopsis, designated SRK2A–SRKJ (Yoshida et al. 2002) or SnRK2.1–SnRK2.10 (Hrabak et al. 2003), and they are categorized into three subclasses: I, II and III (Kobayashi et al. 2004). In rice, there are 10 SnRK2 members designated SAPK1–SAPK10 (Kobayashi et al. 2004). Each subclass of SnRK2 is highly conserved among higher plants (Fig. 1).
Although SnRK2 was originally identified as an ABA- activated protein kinase, other studies demonstrated that hyperosmotic stress also enabled the activation of SnRK2s (Mikolajczyk et al. 2000, Monks et al. 2001, Umezawa et al. 2004). One osmotic stress-activated SnRK2, SRK2C/SnRK2.8, positively regulates drought tolerance by the hyperinduction of stress-responsive gene expression in overexpressing plants (Umezawa et al. 2004). Comprehensive analysis of Arabidopsis and rice SnRK2s revealed that each subclass of SnRK2 has a different activation pattern in relation to ABA and osmotic stress (Boudsocq et al. 2004, Kobayashi et al. 2004). Subclass I SnRK2s are rapidly activated by osmotic stress within 1 min, but they are not activated by ABA. In contrast, subclass II and III members are activated by both ABA and osmotic stress. Subclass III SnRK2s are strongly activated by ABA, in contrast to the weak activation of subclass II members.
SnRK2 contains a well-conserved kinase catalytic domain and a relatively diverse C-terminal domain (Yoshida et al. 2002, Hrabak et al. 2003). The C-terminal domain is further divided into two parts, domains I and II (R. Yoshida et al. 2006). Although domain I is relatively similar in all SnRK2s, domain II is clearly different among SnRK2 subgroups, especially in the ‘acidic patch’ region. For example, the acidic patch is rich in aspartate in subclasses II and III but rich in glutamate in subclass I members. Several studies have suggested that the C-terminal region could be a regulatory domain of SnRK2. For example, only aspartate-rich SnRK2s (subclasses II and III) can be activated by ABA (Boudsocq et al. 2004, Kobayashi et al. 2004). Further analysis demonstrated that deletion of domain II impaired ABA-dependent activation, suggesting that this domain involving the acidic patch is required for ABA responsiveness (R. Yoshida et al. 2006). Alternatively, Kobayashi et al. (2004) swapped C-terminal fragments of rice SAPK1 and SAPK2, and found that SnRK2 activation patterns were precisely defined by this region.
Among the Arabidopsis SnRK2 family, subclass III contains three kinases, SRK2D/SnRK2.2, SRK2E/OST1/SnRK2.6 and SRK2I/SnRK2.3, and they are strongly activated by ABA, as described above. These kinases are rapidly activated by ABA within 30 min, suggesting their possible roles in initial ABA signaling (R. Yoshida et al. 2006). The physiological functions of SnRK2 were first determined in guard cells. SRK2E/OST1/SnRK2.6 positively regulates stomatal closure in response to ABA (Mustilli et al. 2002, Yoshida et al. 2002), as well as fava bean AAPK (Li et al. 2000). The other two kinases, SRK2D/SnRK2.2 and SRK2I/SnRK2.3, mainly function in tissues that are separate from guard cells, such as seeds and vegetative tissues (Fujii et al. 2007). Therefore, it was suggested that subclass III SnRK2s are global and positive regulators of ABA signaling.
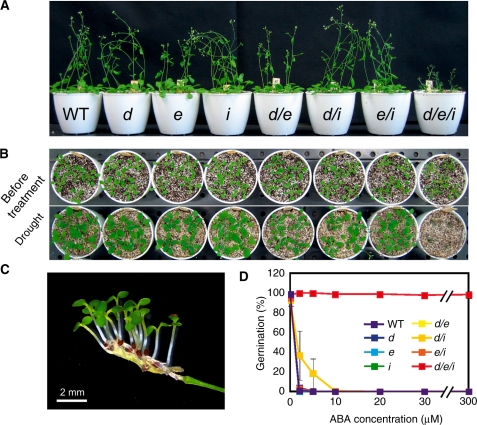
Recently, the importance of subclass III SnRK2s was highlighted by the establishment of a triple knockout mutant in Arabidopsis (Fujii and Zhu 2009, Fujita et al. 2009, Nakashima et al. 2009, Umezawa et al. 2009). The triple mutant, srk2d srk2e srk2i (srk2dei), identical to snrk2.2 snrk2.3 snrk2.6, lacks most ABA responses, including seed dormancy, germination, post-germination growth, ABA-responsive gene expression and stomatal movements, to name a few (Fig. 2). They can germinate even in concentrations of hundreds of micromolar ABA, and, symbolically, they show a strong viviparous phenotype (Fujita et al. 2009, Nakashima et al. 2009). Furthermore, protein kinase activities induced by ABA are largely impaired in srk2dei (Fujii and Zhu 2009, Umezawa et al. 2009). Therefore, it was concluded that subclass III SnRK2s act as a central hub in ABA signaling (also see ‘AREB/ABF bZIP-type transcription factors’). Although subclass III members are fundamentally redundant throughout various tissues and developmental stages, each member may still have a spatially weighted role in some tissues or cell types, as described above.
Fig. 2.
Subclass III SnRK2 protein kinases are essential for the control of drought tolerance and germination. (A) The srk2dei (snrk2.2 snrk2.3 snrk2.6) triple mutants exhibited greatly reduced tolerance to drought stress. (B) Viviparous seeds in attached siliques of an srk2dei mutant grown in high humidity. (C) Extreme ABA insensitivity of srk2dei mutant plants. These figures are modified from Nakashima et al. (2009) and Fujita et al. (2009).
PYR/PYL/RCAR proteins: soluble ABA receptors
Several ABA-binding proteins have been isolated and characterized, such as G-protein-coupled receptors (Liu et al. 2007, Pandey et al. 2009) and an Mg-chelatase H subunit (Shen et al. 2006). Although these proteins at the time were suggested to play important roles in ABA responses, their physiological and molecular connections to well-known signaling factors such as PP2C and SnRK2 still remained elusive. Subsequently, novel ABA-binding proteins, PYR/PYL/RCAR, were identified as soluble ABA receptors by two independent research groups based on different approaches: chemical genetics and biochemistry (Ma et al. 2009, Park et al. 2009). Their discovery was a real breakthrough in ABA signaling studies. Because several reviews mention the details of PYR/PYL/RCARs (Cutler et al. 2010, Hubbard et al. 2010, Klingler et al. 2010, Raghavendra et al. 2010), we briefly summarize this family in this section.
Park et al. (2009) screened chemical libraries and found ‘pyrabactin’ as a selective agonist of ABA. After a genetic screen against pyrabactin, the PYRABACTIN RESISTANCE1 (PYR1) gene was identified. Another group isolated an ABI1-interacting protein by yeast two-hybrid screening, named REGULATORY COMPONENT OF ABA RECEPTOR1 (RCAR1) (Ma et al. 2009). PYR1 and RCAR1 both encode START-domain/Bet V allergen superfamily proteins, typically containing a hydrophobic ligand pocket. They belong to the same gene family consisting of 14 members in the Arabidopsis genome, named PYR1 and PYR1-like (PYL) 1–13 or RCAR1–RCAR14 (Fig. 1C). Two other groups also identified the same family of proteins as PP2C-interacting partners (Santiago et al. 2009b, Nishimura et al. 2010).
One major finding was that PYR/PYL/RCAR proteins can directly bind to ABA (Ma et al. 2009, Park et al. 2009). Each PYR/PYL/RCAR binds (+)-S-ABA or (−)-R-ABA with a different Kd value, and some members can recognize ABA chirality (Ma et al. 2009, Park et al. 2009, Santiago et al. 2009b, Szostkiewicz et al. 2010). Another important discovery was that PYR/PYL/RCAR proteins interact with group A PP2C in an ABA-dependent manner (Ma et al. 2009, Park et al. 2009). An ABA-bound form of PYR/PYL/RCAR exposes an interface that interacts with group A PP2C, as described in ‘Structural Basis of ABA Perception and Signaling’. This interaction inhibits the protein phosphatase activity of PP2C, suggesting that PYR/PYL/RCARs behave like a negative regulatory subunit of PP2C. This appears to be the first discovery to shed light on PP2C regulatory proteins. Because PP2C is a major negative regulator of ABA signaling, it seems that the ABA-bound PYR/PYL/RCAR can shift ABA signaling status to ‘active’. These results clearly indicate that this type of PYR/PYL/RCAR acts as a soluble ABA receptor in plants, and that this family is at the head of the negative regulatory pathways in ABA signaling (Ma et al. 2009, Park et al. 2009).
The functions of PYR/PYL/RCARs were further confirmed by the strong ABA-insensitive phenotype of the pyr1 pyl1 pyl2 pyl4 quadruple mutant (Park et al. 2009). Conversely, overexpression of RCAR1/PYL9, PYL5/RCAR8 or PYL8/RCAR3 produces enhanced ABA responses or elevated drought tolerance in Arabidopsis (Ma et al. 2009, Santiago et al. 2009b, Saavedra et al. 2010).
Establishment of the core signaling pathway
The PP2C–SnRK2 complex: a central regulatory module
As described above, SnRK2 and PP2C have been very well, but separately, studied, although both proteins are known as global regulators in ABA signaling. In fact, PP2C and SnRK2 have some symmetrical features, such as that of a negative regulator and a positive regulator, or catalyzing protein dephosphorylation and phosphorylation, respectively. Recently, several lines of evidence have linked SnRK2 and PP2C. First, most SnRK2 activities induced by ABA are impaired in abi1-1 mutants (Mustilli et al. 2002, R. Yoshida et al. 2006, Umezawa et al. 2009). This suggests that PP2C might be involved in the upstream regulation of the ABA-responsive activation of SnRK2. The first concrete evidence for this was derived from a yeast two-hybrid analysis in which a physical interaction between ABI1 and SRK2E/OST1/SnRK2.6 was demonstrated (R. Yoshida et al. 2006). Deletion analysis revealed that ABI1 binds to a C-terminal ‘domain II’ of SRK2E/OST1/SnRK2.6. Further analysis revealed that all group A PP2Cs interact with subclass III SnRK2s in various combinations (Umezawa et al. 2009). Thus, the major positive and negative regulators in ABA signaling are physically connected at the gene family level.
Importantly, the molecular processes underlying PP2C and SnRK2 were recently decoded by two independent research groups. Umezawa et al. (2009) clearly demonstrated that group A PP2C directly inactivates and dephosphorylates subclass III SnRK2 in vitro, and determined the phosphorylation sites in SnRK2 with a liquid chromatography–tandem mass spectometry (LC-MS/MS) system. In response to ABA, SnRK2 is activated in association with the internal phosphorylation of multiple serine/threonine residues (involving Ser175) in its kinase activation loop. The same sites are dephosphorylated by group A PP2C, resulting in the inactivation of SnRK2. On the other hand, Vlad et al. (2009) screened substrates of a PP2C HAB1 or HAB1G246D using a combinatorial peptide library, and identified Ser175 in SRK2E/OST1 as a target site of PP2C. Therefore, there is convincing evidence that SnRK2 is a direct target of PP2C, providing clear insight into the PP2C-dependent negative regulation of ABA signaling, as well as the upstream regulation of ABA-activated SnRK2.
Interestingly, abi1-1 mutant proteins may also interact with subclass III SnRK2s, and dephosphorylate and inactivate SnRK2 in a fashion comparable with wild-type proteins (Umezawa et al. 2009, Vlad et al. 2009). This implies that abi1-1 proteins negatively regulate SnRK2s similarly to wild-type proteins, which leads to the question of what is affected in the abi1-1 mutant. To get an answer to this question, see ‘Decoding the long-standing mystery of the abi1-1 mutation’.
The ABA signalosome: PYR/PYL/RCAR– PP2C–SnRK2
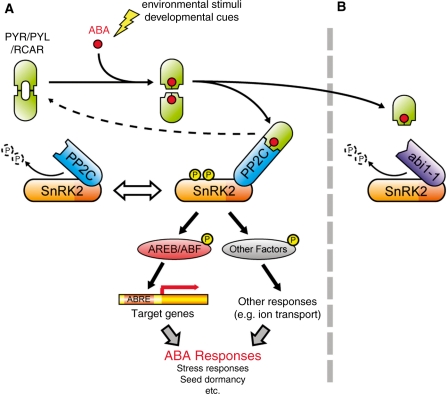
In 2009, our knowledge of ABA signaling dramatically increased with the discovery that PYR/PYL/RCAR negatively regulates PP2C, and PP2C negatively regulates SnRK2. These findings showed that the signaling complex of the receptors, PP2C and SnRK2 offers a double negative regulation system in ABA signaling. This hypothesis was supported by the observation that SnRK2 activities are significantly reduced in the PYR/PYL-quadruple mutant (Park et al. 2009). Finally, these components were successfully reconstituted in vitro, in which PYR/PYL/RCAR proteins inhibited the PP2C-dependent negative regulation of SnRK2 in an ABA-modulated fashion (Umezawa et al. 2009). This means that an ABA signal transduction pathway was regenerated in vitro, allowing the establishment of a simple model of ABA signaling in plants (Fig. 3). Normally, PP2C inactivates SnRK2 by dephosphorylation and ABA signals are silent. Once ABA is induced by environmental conditions or developmental cues, the ABA-bound PYR/PYL/RCAR receptors interact with PP2C and inhibit its phosphatase activity. SnRK2 is then released from negative regulation by PP2C, turning on ABA signals by phosphorylation of downstream factors such as AREB/ABF basic-domain leucine zipper (bZIP) proteins, S anion channels and others (see ‘The PYR/PYL/RCAR–PP2C–SnRK2 Complex-Mediated Signaling Network in Plants’). This model is considered a core component system in ABA signaling, because all components regulate global ABA responses in plants. Therefore, at least one ABA signal transduction pathway consists of just four steps, from perception to gene expression or another output (Fig. 3) (Umezawa et al. 2009). The whole pathway, from receptors to AREB/ABFs, was completely confirmed by an elegant transient expression system in Arabidopsis mesophyll protoplasts (Fujii et al. 2009).
Fig. 3.
Proposed model of the major ABA signaling pathway. PYR/PYL/RCAR, PP2C and SnRK2 form a signaling complex referred to as the ‘ABA signalosome’. (A) Under normal conditions, PP2C negatively regulates SnRK2 by direct interactions and dephosphorylation of multiple residues of SnRK2. Once abiotic stresses or developmental cues up-regulate endogenous ABA, PYR/PYL/RCAR binds ABA and interacts with PP2C to inhibit protein phosphatase activity. In turn, SnRK2 is released from PP2C-dependent regulation and activated to phosphorylate downstream factors, such as the AREB/ABF bZIP-type transcription factor or membrane proteins involving ion channels. (B) In contrast, the abi1-1-type mutated protein lacks PYR/PYL/RCAR binding, resulting in the constitutive inactivation of SnRK2, even in the presence of ABA, and strong insensitivity to ABA in the abi1-1 mutant.
Even though the basic mechanism of the PYR/PYL/RCAR–PP2C–SnRK2 pathway has been clarified, several questions still remain. A major question is whether this complex is stable in plant cells. Fujii et al. (2009) proposed that this complex formation might be plastic, based on a yeast three-hybrid analysis in which the interactions of PYL5/8, ABI1/2 and HAB1 with SRK2E/OST1/SnRK2.6 were disrupted in response to ABA. In contrast, co-immunoprecipitation analyses from Arabidopsis protoplasts or Agrobacterium-infiltrated Nicotiana benthamiana leaves detected no significant changes in PP2C–SnRK2 interactions with or without ABA. This suggests that the PP2C–SnRK2 complex can be formed consistently in vivo (Umezawa et al. 2009, Nishimura et al. 2010), although a type of association/deassociation cycle might be involved in complex formation. Therefore, this issue is currently a matter of controversy. Another question is what is the mechanism of SnRK2 activation induced by ABA. Although autophosphorylation is a possible regulator, as shown with recombinant SnRK2 proteins derived from Escherichia coli (Belin et al. 2006), further evidence is required to confirm autophosphorylation in vivo. In fact, in vivo activation of SnRK2 is not inhibited by staurosporine, although SnRK2 is sensitive to this inhibitor in vitro, suggesting some staurosporine-resistant kinase(s) might be involved in upstream regulation (Boudsocq et al. 2007). Further analysis is necessary to understand this mechanism.
Decoding the long-standing mystery of the abi1-1 mutation
The identification of the ABA signaling complex provided a clear explanation for why abi1-1 is dominantly insensitive to ABA. As described above, abi1-1-mutated proteins can interact normally with subclass III SnRK2s, as well as wild-type proteins (Umezawa et al. 2009, Vlad et al. 2009). In contrast, PYR/PYL/RCAR cannot interact with abi1-1 proteins, suggesting that abi1-1 might not be controlled by ABA receptors (Ma et al. 2009, Park et al. 2009). Therefore, there was a long-standing, controversial question about the protein phosphatase activity of abi1-1. Recent studies have clearly demonstrated that abi1-1 has a low affinity for artificial substrates, such as casein (Umezawa et al. 2009); however, abi1-1 efficiently dephosphorylates and inactivates SnRK2 (Umezawa et al. 2009, Vlad et al. 2009). Consistent with these results, in vitro reconstitution assays further demonstrated that abi1-1 PP2Cs constitutively inactivate SnRK2s, even in the presence of PYR1 and ABA (Umezawa et al. 2009). Thus, the abi1-1 mutation enables the maintenance of a negative state in ABA signaling by the constitutive inactivation of SnRK2, resulting in strong ABA insensitivity in plants (Fig. 3). This model clearly explains the dominant phenotype of abi1-1 plants, and resolves the long-standing mystery of the abi1-1 mutation.
Combinatorial variants of the ABA signalosome
The signaling complex of PYR/PYL/RCAR, group A PP2C and subclass III SnRK2 is emerging as a new signaling system in ABA responses. This is a double negative regulatory system, and the mechanism is very simple yet sophisticated. This system probably varies widely in plant cells, as there are 14 PYR/PYL/RCARs, nine PP2Cs and three essential SnRK2s in Arabidopsis alone (Fig. 1) (Ma et al. 2009, Park et al. 2009, Umezawa et al. 2009), with up to 378 potential combinations of the ABA signalosome. In addition, nine AREB/ABFs may be involved in ABA-responsive gene expression (Uno et al. 2000, Fujita et al. 2005), resulting in >3,000 possible combinations for transcriptional regulation.
Although the many ABA signalosome variants enable a wide range of ABA responses in plants, in vivo combinations of the signalosome might be limited due to multiple determinants, including spatio-temporal limitations such as tissue or organ specificities, stress-responsive gene expression patterns, subcellular localization and preferences in protein–protein interactions. Each gene of a core component has a distinct expression pattern. For example, two SnRK2s, SRK2D/SnRK2.2 and SRK2I/SnRK2.3, are expressed predominantly in seeds or vegetative tissues (Fujii et al. 2007), whereas SRK2E/OST1/SnRK2.6 is strongly expressed in guard cells and in vascular tissues to an extent (Mustilli et al. 2002, Yoshida et al. 2002), consistent with their physiological functions. PYR/PYL/RCARs and PP2Cs also show tissue-specific expression patterns (Nishimura et al. 2007, Park et al. 2009). Notably, they are transcriptionally regulated by ABA, but SnRK2 genes show stable expression, leading to the idea that PYR/PYL/RCAR and PP2C may have evolved to the strict control of SnRK2 activity in planta. As with many PP2C genes induced by ABA, SnRK2 can be quickly inactivated after removal of ABA, enabling fast recovery from stressful conditions.
Each PP2C or SnRK2 has selective protein–protein interactions, as shown by the various combinations of PP2C–SnRK2 complexes that can be formed (Umezawa et al. 2009). In general, protein–protein interactions are limited by subcellular localization, which may also be important to the composition of the ABA signaling complex. PYR/PYL/RCARs and SnRK2s are localized in the cytosol or nucleus, but there are two types of PP2C localization: ABI1 and ABI2 are found in the cytosol and nucleus, but AHG1 and AHG3 are specific to the nucleus. Therefore, this suggests that the composition of the receptor complex should be different between the cytosol and nucleus.
Other factors could affect the variation in core ABA signaling, e.g. some PYR/PYL/RCARs are stereoselective for ABA isomers, as shown with RCAR1/PYL9, which binds (+)-S-ABA with a low dissociation constant of 0.7 μM, in contrast to (−)-R-ABA or trans-ABA (Ma et al. 2009). In addition, a yeast two-hybrid analysis also suggested that each PYR/PYL/RCAR has a different stereoselectivity (Park et al. 2009). Moreover, two closely related receptors, RCAR1 and RCAR3, have different selectivities and sensitivities to ABA (Szostkiewicz et al. 2010). These differences may affect the formation of the signaling complex, and modulate the fine-tuning of ABA signaling.
In conclusion, the combinatorial variation in the ABA signalosome in plant cells is restricted by spatio-temporal or biochemical determinants, affecting their access to downstream factors. Further analyses will clarify how those determinants control the fine-tuning of the formation of ABA signalsomes in plants.
Structural basis of ABA perception and signaling
Understanding of the mechanism of ABA action in plant intracellular signaling was greatly improved by the discovery of PYR/PYL/RCAR as a type of soluble ABA receptor, and its downstream protein phosphorylation system (Ma et al. 2009, Park et al. 2009, Umezawa et al. 2009, Vlad et al. 2009). Although the structures of several PYR/PYL/RCAR-homologous proteins, such as Bet v 1 l from birch pollen and CSBP from Vigna radiata, were previously known, it remained unclear how the receptor perceives the ABA molecule and how ABA binding to the receptor leads to the inhibition of PP2C. Soon after its discovery, five independent groups reported their structures in three different states: the apo-state (ABA-free state), the ABA-bound state and the ternary complex formed by further binding of PP2C (Melcher et al. 2009, Miyazono et al. 2009, Nishimura et al. 2009, Santiago et al. 2009a, Yin et al. 2009). These structures clearly elucidated the mechanism of action of ABA. In this section, we describe in detail the mechanisms of ABA perception and ABA-induced PP2C inhibition by PYR/PYL/RCARs.
ABA perception by PYR/PYL/RCAR and structural changes
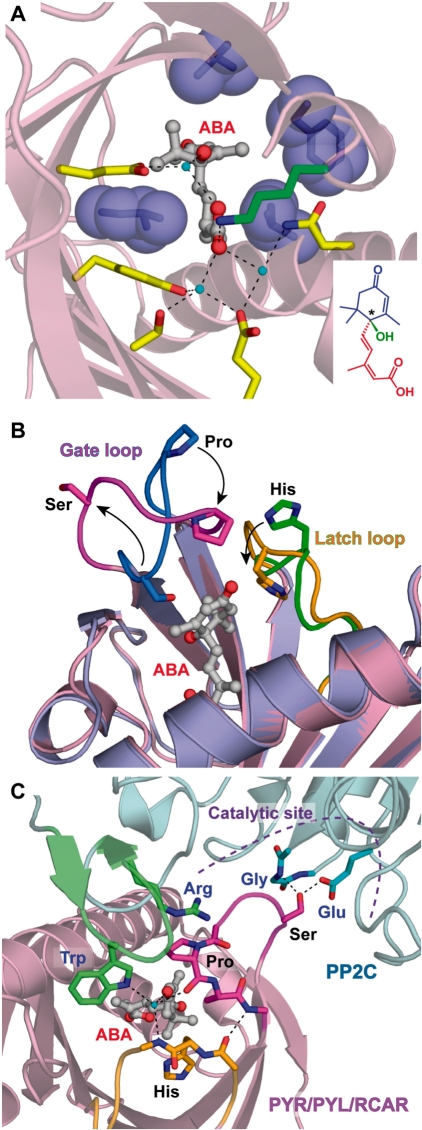
PYR/PYL/RCAR possesses a large, internal, water-filled cavity, in which an ABA molecule can be nearly completely trapped with its carboxyl group oriented toward the center of the receptor molecule. This structure defines the binding mode of ABA and explains its stereoselectivity (Fig. 4A). The carboxyl group of ABA forms an ion pair with the side chain of a lysine residue (PYL1 K86, PYL2 K64 and PYR1 K59) and a water-mediated hydrogen bond network with side chains of five polar residues. This network also links the carboxyl group with a hydroxyl group attached to the chiral carbon, which defines the location of the pentadienoic acid moiety and the hydroxyl group in the cavity. All other residues in the cavity are hydrophobic and form van der Waals contacts with cyclohexene and pentadienoic acid moieties. In particular, the conformation of the cyclohexene group is defined by several hydrophobic residues. PYR/PYL/RCAR shows a higher affinity for the (+)stereoisomer of ABA than for the biologically less active (−)stereoisomer (Melcher et al. 2009, Miyazono et al. 2009). The flipped dimethyl group in (−)-R-ABA would cause steric hindrance between the dimethyl group and the narrow pocket that accommodates the monomethyl group (Melcher et al. 2009). Thus, the arrangement of the hydrophobic residues surrounding the cyclohexene moiety defines the stereoselectivity of ABA isomers. These residues, and the position of the water molecules, are well conserved in PYR/PYL/RCAR.
Fig. 4.
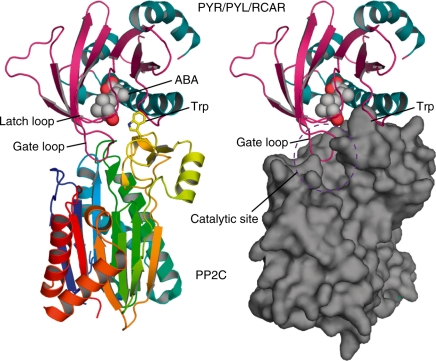
Structural analysis of PYR/PYL/RCAR ABA receptors. (A) Stereoselective ABA-binding mode of the PYR/PYL/RCAR proteins. A lysine residue (green) directly interacts with ABA, and polar residues (yellow) form a water-mediated hydrogen bond network with ABA. Water molecules and hydrogen bonds are shown by cyan spheres and dashed lines, respectively. Hydrophobic residues (blue) are localized around dimethyl and monomethyl groups of the cyclohexene moiety. For the structural formula of ABA in the inset, the cyclohexene moiety, pentadienoic acid moiety and hydroxyl group from the chiral carbon (shown by an asterisk) are colored blue, red and green, respectively. This figure was created using Protein Data Bank (PDB) coordinates of ABA-bound PYL1 (3JRS). (B) Open-to-closed gating mechanism of the PYR/PYL/RCAR proteins. Gate and latch loops dramatically shift to the closed conformation (magenta and orange, respectively) from the open conformation (blue and green, respectively) upon ABA binding. This figure was created using the PDB coordinates of apo-PYL1 (3KAY) and ABA-bound PYL1 (3JRS). (C) ABA-dependent mechanism for PP2C inhibition. Conserved tryptophan and arginine residues on the additional antiparallel β-sheet of PP2C (green) contact the gate and latch loops of the PYR/PYL/RCAR proteins. The gate loop is locked by these interactions and seals the catalytic site of PP2C to inhibit phosphatase activity competitively. This figure was created using PDB coordinates from apo-PYL1 (3KAY), ABA-bound PYL1 (3JRS) and the complex of ABA-bound PYL1 and ABI1 (3JRQ).
ABA perception by PYR/PYL/RCAR induces structural changes essential for ABA signal transduction. Superposition of the apo- and ABA-bound structures in the receptors shows the conformational differences in the two conserved loops (Fig. 4B; Melcher et al. 2009, Nishimura et al. 2009, Santiago et al. 2009a, Yin et al. 2009). One of the two loops connects β-strand 3 with β-strand 4 (referred to as the ‘gate loop’). Upon ABA binding, a proline residue on the loop (PYL1 P115, PYL2 P92 and PYR1 P88) moves toward the cyclohexene moiety of ABA to close the gate on the cavity, whereas the serine residue on the loop (PYL1 S112, PYL2 S89 and PYR1 S85) is flipped out of the ABA-occupied cavity. The other loop connects β-strand 5 with β-strand 6 (referred to as the ‘latch loop’). The imidazole ring of the histidine residue on the loop (PYL1 H142, PYL2 H119 and PYR1 H115) turns into the cavity to form van der Waals contacts with the cyclohexene moiety of ABA, which induces the conformational change in the loop. The latch loop locks the closed gate loop by a hydrogen bond and van der Waals contacts. These loops in the closed conformation then provide the surface for the interaction with group A PP2Cs, including ABI1 and HAB1 (Melcher et al. 2009, Miyazono et al. 2009, Yin et al. 2009). Thus, PYR/PYL/RCAR is allosterically regulated by ABA and switches on ABA signal transduction using an open-to-closed gating mechanism. In fact, multiple conformations of these loops are observed in the ABA-bound structures of the receptors, although the surface of the loops is often utilized for intermolecular contact in the crystals. Most residues on these loops very weakly contribute to ABA binding (Miyazono et al. 2009). Hence, the closed conformations of the gate and latch loops are expected to be insufficiently stabilized even if ABA is trapped in the cavity of its receptors (Melcher et al. 2009, Miyazono et al. 2009).
Structural change by ABA perception is also shown in the homodimer assembly of PYL2 (Yin et al. 2009). Apo- and ABA-bound PYL2 form a homodimer by the gate loop. The dimerization of the receptors can be observed in their crystal structures and confirmed in solution by small-angle X-ray scattering (SAXS) (Nishimura et al. 2009) and size-exclusion chromatography (Yin et al. 2009). The relative orientation of the two protomers is modified by conformational changes in the gate loop upon ABA binding (Yin et al. 2009). Although the functional significance of dimer formation remains unclear, the contact between protomers seems to stabilize the gate loop with conformational plasticity in the open or closed state. Yin et al. (2009) discussed the fixation of the gate loop in the dimer interface as one of the possible reasons why PP2C does not bind to the loop of the apo-receptor proteins via an induced fit mechanism. The homodimer assembly may be required to strictly regulate the ABA-dependent switching of signal transduction by PYR/PYL/RCAR.
ABA-induced inhibition mechanism of PP2C
The crucial role of PYR/PYL/RCAR in ABA signaling is to inhibit the phosphatase activity of PP2C in an ABA-dependent manner (Ma et al. 2008, Park et al. 2008). The inhibition mechanism of the ABA-bound receptor is well defined by the ternary complex structures of PYL1–ABA–ABI1 (Miyazono et al. 2009, Yin et al. 2009) and PYL2–ABA–HAB1 (Melcher et al. 2009). The catalytic site of PP2C is sealed by the closed gate loop of PYR/PYL/RCAR (Figs. 4C, 5). The serine residue (PYL1 S112 and PYL2 S89) exposed upon ABA binding forms a hydrogen bond between its side chain and one of the conserved catalytic residues of PP2C (ABI1 E142 and HAB1 E203). Thus, the ABA-bound receptor is capable of competitively inhibiting the phosphatase activity of PP2C by using the gate loop like a plug. The serine residue also contacts the glycine residue on the active site loop of PP2C (ABI1 G180 and HAB1 G246) through the hydrogen bond. The mutation to an aspartic acid residue at this position is known as the abi1-1 mutation (Koornneef et al. 1984, Leung et al. 1994, Meyer et al. 1994). This structural observation explains why the mutation hinders contact between the gate loop of the ABA-bound receptor and the active site of PP2C (Melcher et al. 2009, Miyazono et al. 2009, Yin et al. 2009).
Fig. 5.
Overall structure of the complex of ABA-bound PYL1 and PP2C. PP2C in the left and right diagrams is represented by a ribbon and surface model, respectively. This figure was created using PDB coordinates for the complex of ABA-bound PYL1 and ABI1 (3JRQ).
The additional antiparallel β-sheet of PP2C provides the major binding interface with the ABA-bound PYR/PYL/RCAR (Melcher et al. 2009, Miyazono et al. 2009, Yin et al. 2009). There are two crucial residues for binding the closed gate and latch loops of the receptor (Figs. 4C, 5; Miyazono et al. 2009). A conserved tryptophan residue (ABI1 W300 and HAB1 W385) inserts its indole ring between the two closed loops of the ABA-bound receptor. The indole imine group forms a water-mediated hydrogen bond with the two closed loops and the carbonyl group of ABA (Melcher et al. 2009, Miyazono et al. 2009, Yin et al. 2009). In addition, a conserved arginine residue (ABI1 R304 and HAB1 R389) seems to hold down the gate loop of the receptor by stacking between its guanidinium group and a conserved proline residue on the loop (PYL1 P115 and PYL2 P92). The gate loop is further locked in the closed conformation by these interactions, which properly positions the gate loop in the active site of PP2C (Melcher et al. 2009, Miyazono et al. 2009). This ‘gate–latch–lock’ mechanism by PP2C also enhances the ABA binding affinity of PYR/PYL/RCAR (Ma et al. 2009). These structural features mainly ensure the binding specificity of ABA-bound receptors toward group A PP2C.
Mechanistic basis of hormone-induced signal transduction
Structural studies of phytohormone receptors, auxin receptor TIR1 (Tan et al. 2007) and gibberellin receptor GID1 (Murase et al. 2008, Shimada et al. 2008), have led to the proposition of two different modes for the hormone-induced enhancement of protein–protein interactions resulting in the degradation of the target repressor protein. Auxin binds directly to TIR1 and mediates continuous contact between TIR1 and the transcription repressor Aux/IAA (Tan et al. 2007). Auxin binding does not induce significant conformational changes in the receptor, except for rearranging the side chains of some residues. Thus, auxin promotes TIR1–Aux/IAA interactions by acting as a ‘molecular glue’, and TIR1 is a co-receptor of auxin with Aux/IAA. In contrast, gibberellin induces conformational changes in GID1 leading to the covering of its gibberellin- binding pocket (Murase et al. 2008, Shimada et al. 2008). The closed lid of GID1 provides the binding interface for the transcription repressor DELLA. Thus, gibberellin functions as an ‘allosteric effector’ to promote the interaction between GID1 and DELLA.
ABA induces conformational changes in PYR/PYL/RCAR, creating a continuous binding interface with PP2C, and does not directly bind to PP2C (Melcher et al. 2009, Miyazono et al. 2009, Yin et al. 2009). These structural findings show that PYR/PYL/RCAR is the direct ABA receptor, similar to the gibberellin receptor, GID1, rather than the auxin co-receptor TIR1, and ABA regulates the receptor protein as an ‘allosteric effector’. However, there are different modes in the interaction between the hormone-bound receptor and its continuous binding partner. Gibberellin-bound GID1 induces a coil-to-helix conformational transition in its DELLA binding partner, which stimulates the recognition of DELLA by the ubiquitin ligase E3 complex (Murase et al. 2008). In contrast, PP2C refines the closed conformation of the ABA-bound receptor by assisting with ABA loading into the receptor, which allows the gate loop of the receptor to bind to the active site of PP2C (Melcher et al. 2009, Miyazono et al. 2009, Yin et al. 2009). Although the common action of plant hormones toward their soluble receptors is to induce the interaction between the receptor and its binding partner, the mechanisms of action are quite distinct for each receptor.
Recent structural investigations have led to major progress in understanding hormone signaling. The mechanistic basis of the soluble receptor provides a rational framework for future design of alternative ligands and for engineering plants to control plant cellular functions. In particular, elucidating the structural basis of ABA signaling will help us to understand plant stress biology and to develop abiotic stress-resistant crops.
The PYR/PYL/RCAR–PP2C–SnRK2 complex-mediated signaling network in plants
To understand the details of ABA signaling, it is important to determine the downstream targets of the PYR/PYL/RCAR–PP2C–SnRK2 complex. A series of studies have identified several proteins that interact with PP2C, including AtHB6, KAT2, GPX and SWI3B [see reviews by Hirayama and Shinozaki (2007, 2010)]. Recently, Nishimura et al. (2010) screened a number of proteins in a pull-down assay of ABI1 that involved PYR/PYL/RCAR, SnRK2 and H+-ATPase. However, not all of the proteins were tested to determine if they are directly targeted by group A PP2C dephosphorylation. Thus, further analysis is required to determine the direct targets. In contrast, several SnRK2 substrates have already been reported, such as bZIP transcription factors and other membrane proteins. These proteins have been shown to be directly phosphorylated by SnRK2 in vivo and in vitro. In the following section, recently reported SnRK2 substrates are reviewed and their function or regulation in stress responses and seed development are discussed.
AREB/ABF bZIP-type transcription factors
ABA accumulates in plant cells under stress conditions, such as drought and high salinity, and induces the expression of many ABA-responsive genes (Seki et al. 2002). By analyzing the promoters of ABA-inducible genes, it was found that ABA- responsive gene expression requires multiple cis-elements, designated as ABA-responsive elements (ABREs; PyACGTGG/TC), or the combination of an ABRE with a coupling element (Zhang et al. 2005, Gomez-Porras et al. 2007). The ABRE- binding (AREB) proteins or ABRE-binding factors (ABFs) were isolated by using ABRE sequences as bait in yeast one-hybrid screenings (Choi et al. 2000, Uno et al. 2000). The AREB/ABFs encode bZIP transcription factors and belong to the group A subfamily, which is composed of nine homologs in the Arabidopsis genome that harbor one C-terminal and three N-terminal conserved domains (Jakoby et al. 2002). Another bZIP protein, ABA-INSENSITIVE 5 (ABI5), was identified through genetic screening of ABA-insensitive germination (Finkelstein and Lynch 2000, Lopez-Molina and Chua 2000). Among the group A bZIP subfamily, AREB1/ABF2, AREB2/ABF4 and ABF3 are induced by dehydration, high salinity and ABA treatment in vegetative tissues (Fujita et al. 2005), and plants overexpressing these factors show enhanced drought stress tolerance (Kang et al. 2002, Kim et al. 2004, Fujita et al. 2005). These independent analyses imply that the three AREB/ABF transcription factors are functionally redundant. To elucidate the role of these AREB/ABF transcription factors in stress responses in vegetative tissues, Yoshida et al. (2010) generated an areb1 areb2 abf3 triple mutant. Large-scale transcriptome analysis, which showed that stress-responsive gene expression is remarkably impaired in the triple mutant, revealed novel AREB/ABF downstream genes in response to water stress, including many late embryogenesis abundant (LEA) class and group A PP2C genes and transcription factors. The areb1 areb2 abf3 triple mutant is more resistant to ABA than other single and double mutants with respect to primary root growth, and it displays reduced drought tolerance. These results indicate that AREB1, AREB2 and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent gene expression in ABA signaling under stress conditions. In contrast, AREB/ABF-type bZIP proteins, such as AREB3 and EEL, are expressed in the nuclei of developing seeds and play important roles in seed development (Bensmihen et al. 2002, Bensmihen et al. 2005). These factors regulate the expression of LEA class genes, including AtEm1 and AtEm6, during seed maturation (Bensmihen et al. 2002).
AREB/ABF bZIP proteins require post-translational modification for their activation (Uno et al. 2000). Several studies have demonstrated that AREB/ABFs are highly phosphorylated in response to ABA, and this phosphorylation is sufficient for their activation (Uno et al. 2000, Kagaya et al. 2002, Furihata et al. 2006). Several SnRK2s, such as SRK2D/SnRK2.2, SRK2E/OST1/SnRK2.6 and SRK2I/SnRK2.3 (SRK2D/E/I), can phosphorylate AREB/ABF polypeptides in vitro (Furihata et al. 2006, Fujii et al. 2007). Bimolecular fluorescence complementation (BiFC) analysis has shown that SRK2D/E/I and AREB1 co-localize and interact in plant cell nuclei (Fujita et al. 2009). In that study, 75% of the AREB/ABF target genes that showed reduced expression levels in the srk2dei triple mutant compared with wild-type plants after ABA treatment were also ABA responsive. These data indicate that most AREB/ABF target genes overlap with SRK2D/E/I targets in response to ABA. Fujii and Zhu (2009) and Umezawa et al. (2009) also reported that a large part of the ABA-activated protein kinase activities are eliminated in the srk2dei mutant, and the expression of the examined ABA-induced genes is completely blocked in the triple mutant. These results indicate that SRK2D/E/I regulates AREB/ABFs in ABA signaling in response to water stress. Furthermore, srk2dei mutant seeds had greatly reduced phosphorylation activity in in-gel kinase experiments using bZIP transcription factors, including ABI5 (Nakashima et al. 2009). Microarray experiments have revealed that 48% of the down-regulated genes in abi5 seeds are suppressed in srk2dei mutant seeds (Nakashima et al. 2009). This indicates that in ABA signaling, SRK2D/E/I controls gene expression which partly depends on the phosphorylation of ABI5 during seed maturation. Likewise, a wheat SnRK2 ortholog, PKABA1, phosphorylates the wheat AREB1 ortholog, TaABF, and the rice SnRK2 orthologs, SAPK8, SAPK9 and SAPK10, phosphorylate the AREB1 ortholog TRAB1, in vitro (Johnson et al. 2002, Kagaya et al. 2002, Kobayashi et al. 2005). Thus, the system of regulation of bZIP transcription factors by SnRK2 should be conserved among plant species.
Membrane proteins
ABA responses include not only transcriptional regulation but also rapid physiological changes in stomatal closure. In this process, ion currents via the plasma membrane or tonoplast play a major part in regulation (Kim et al. 2010). Recently, several membrane proteins were identified as SnRK2 substrates. One is a slow anion channel, SLAC1, which has a central role in guard cells (Negi et al. 2008, Vahisalu et al. 2008). SLAC1 is phosphorylated and activated by SRK2E/OST1/SnRK2.6 under the control of PP2C (Geiger et al. 2009, Lee et al. 2009). Another target of SnRK2 is KAT1, an inward-rectifying potassium channel, which is also important for stomatal movements (Pilot et al. 2001), as well as anion channels. Biochemical analysis has shown that SRK2E/OST1 can phosphorylate Thr306 of KAT1, and that the modification of Thr306 greatly affects KAT1 activity (Sato et al. 2009). ABA-activated SnRK2 may inhibit KAT1 by phosphorylation to promote stomatal closure. SnRK2 can also phosphorylate an NADPH oxidase, AtrbohF, suggesting a possible role in reactive oxygen species (ROS) signaling (Sirichandra et al. 2009). Another type of NADPH oxidase, AtrbohD, was shown to be synergistically activated by Ca2+ and phosphorylation (Ogasawara et al. 2008), suggesting that phosphorylation could be involved in the common regulatory system of ROS production (Trouverie et al. 2008).
These results indicate that plasma membrane is one of the major sites for SnRK2 function in guard cells. However, the regulation of membrane proteins is not simple. For example, SLAC1 is also phosphorylated by another type of kinase, calcium-dependent protein kinase (CDPK23) (Geiger et al. 2010). Other CDPKs, CPK3 and CPK6, also function in the regulation of the anion channel in guard cells (Mori et al. 2006). Likewise, CDPKs could be involved in KAT1 regulation (Sato et al. 2010). This is reasonable because calcium ions are well-known second messengers in intercellular signal transduction in guard cells (Kim et al. 2010). These observations suggest that the Ca2+-dependent pathway could be another signal transduction path in the regulation of ion homeostasis in guard cells, although the relationship between SnRK2s and CDPKs remains unclear. Furthermore, SLAC1 is thought to be downstream of mitogen-activated protein kinases (MAPKs; Jammes et al. 2009); thus multiple pathways could be involved in the ABA-dependent regulation of stomatal movements.
It is noteworthy that most of the studies described above largely depended on in vitro assays to determine protein kinase substrates. The problem with this is that in vitro assays do not always reflect in vivo events, because the in vitro condition is unrestricted by spatio-temporal controls. Further analyses are required to confirm the aforementioned results, and it is necessary to collect in vivo evidence of protein phosphorylation/dephosphorylation to clarify the targets of SnRK2 and PP2C in plant cells.
Evolution of the core ABA signaling pathway
In general, achievement of drought tolerance is thought to be one of the critical steps in the evolution of land plants. Plant evolution can be traced from green algae, mosses and ferns to angiosperms, from which genomic sequences are available for comparative analysis. Umezawa et al. (2009) and Mizoguchi et al. (2010) attempted to find SnRK2 family genes by surveying complete sequence data sets from Chlamydomonas reinhardtii, Physcomitrella patens, Selaginella moellendorffii and several angiosperm genomes, such as Arabidopsis, rice and poplar. Interestingly, all subclasses of SnRK2s were well conserved among higher plants, although P. patens only had subclass III SnRK2s (Fig. 1). Subclass II SnRK2s were found in Selaginella, which lacks subclass I members. Chlamydomonas reinhardtii had a different type of SnRK2 compared with higher plants. These results clearly enable an overview of SnRK2 in plant evolution: subclass III is an ancient form of plant SnRK2, with subclass II appearing around the emergence of ferns. Subclass I SnRK2s are the most recent form, arising before angiosperms. Consistent with this hypothesis, physiological and molecular functions of subclass II SnRK2s partly overlap with those of subclass III (Mizoguchi et al. 2010).
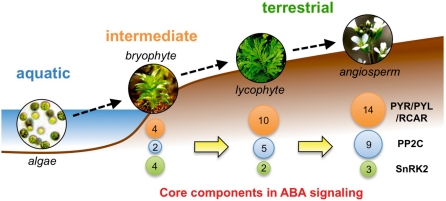
Some other components of ABA signaling are conserved in bryophytes. For example, group A PP2Cs function as signaling factors for ABA responses in mosses and liverworts (Rensing et al. 2008, Komatsu et al. 2009, Tougane et al. 2010). Fig. 1 shows phylogenetic trees for PYR/PYL/RCAR, group A PP2Cs and SnRK2s in various species from green algae to higher plants, allowing for discussion of the evolution of the core ABA signaling pathway. Physcomitrella possesses a small subset of the core components, consisting of four receptors, two group A PP2Cs and four subclass III SnRK2s, but there are no components in Chlamydomonas (Fig. 1). Selaginella also equips ABA-responsive mechanisms (Liu et al. 2008), consistent with a subset of core components. This strongly suggests that a basic ABA signaling pathway was already established in bryophytes. The bryophytes are intermediate between aquatic plants and land plants, and the establishment of the core ABA signaling pathway had a great impact on the movement of plants to land, especially in achieving drought tolerance (Fig. 6).
Fig. 6.
Evolution of core components of ABA signaling. As shown in Fig. 1, PYR/PYL/RCAR, group A PP2C and subclass III SnRK2 are conserved from bryophytes. The development of an ABA signaling system seems to be highly correlated with the evolution from aquatic to terrestrial plants. As representatives, component numbers of bryophyte, lycophyte and angiosperm were obtained from Physcomitrella patens, Selaginella moellendorffii and Arabidopsis thaliana, respectively.
ABA transporters for intercellular signaling
Based on the physiological studies of ABA function to date, the translocation and communication of this phytohormone between cells, organs and tissues play important roles in whole plant physiological responses (Schachtman and Goodger 2008, Wilkinson and Davies 2010). For example, ABA is a key regulator of leaf stomatal conductance: under drought conditions, ABA concentrations increase in the apoplast, leading to stomatal closure (Schachtman and Goodger 2008, Wilkinson and Davies 2010). ABA is predominantly biosynthesized and metabolized in vascular tissues, but acts in the stomatal responses of distant guard cells (Cheng et al. 2002, Koiwai et al. 2004, Okamoto et al. 2009). Indeed, some reports suggest that there are systemic and dynamic changes in gene expression related to ABA or stress responses (Christmann et al. 2007, Endo et al. 2008). Most genes and factors identified so far in ABA signaling are mainly involved in ABA intracellular regulation (Hirayama and Shinozaki 2007, Hirayama and Shinozaki 2010); however, ABA intercellular regulation is not well studied in any plant species. Thus, the molecular basis of ABA transport needs to be investigated to understand whole plant ABA intercellular communication.
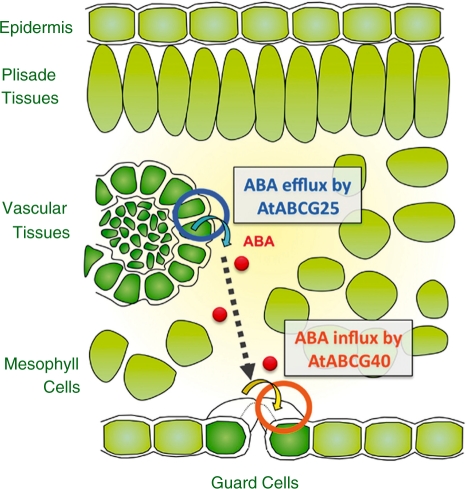
Recently, it was reported that one of the ATP-binding cassette (ABC) transporter genes, AtABCG25, encodes a protein responsible for ABA transport and response in Arabidopsis (Kuromori et al. 2010). The atabcg25 mutants were originally isolated by genetically screening for ABA sensitivity during the greening of cotyledons. AtABCG25 was expressed mainly in vascular tissues, where ABA is predominantly biosynthesized. The fluorescent protein-fused AtABCG25 was localized at the plasma membrane in plant cells.
The ABC transporter is conserved in many model species from E. coli to humans and was reported to transport various metabolites or signaling molecules, involving phytohormones, in an ATP-dependent manner (Higgins 1992, Rea 2007, Nagashima et al. 2008). In membrane vesicles derived from AtABCG25-expressing insect cells, AtABCG25 exhibited ATP-dependent ABA transport activity. Furthermore, the AtABCG25-overexpressing plants had higher leaf temperatures, implying an influence on stomatal regulation. These results suggest that AtABCG25 is an exporter of ABA through the plasma membrane and is involved in the intercellular ABA signaling pathway.
Another ABC transporter in Arabidopsis, AtABCG40, was independently reported to function as an ABA importer in plant cells (Kang et al. 2010). atabcg40 mutants were selected by testing seed germination and stomatal movements in 13 of 15 Arabidopsis ABC transporter gene knockout mutants (atabcg29–atabcg41). AtABCG40 was expressed in the leaves of young plantlets and in primary and lateral roots; in leaves, the expression was the highest in guard cells. Plasma membrane localization was shown by ABCG40::sGFP expression driven by the native promoter in Arabidopsis guard cells. In addition, uptake of ABA by yeast and BY2 cells expressing AtABCG40 increased, whereas ABA uptake by the protoplasts of atabcg40 plants decreased, compared with control cells. In loss-of- function atabcg40 mutants, the stomata closed more slowly in response to ABA, resulting in reduced drought tolerance. In response to exogenous ABA, the up-regulation of ABA- inducible genes was strongly delayed in atabcg40 plants, indicating that ABCG40 is necessary for timely responses to ABA. These results suggest that AtABCG40 is an importer of ABA through plasma membranes, and integrates ABA- dependent signaling and transport processes.
In both cases, stereospecificity for transport was shown by experiments using ABA stereoisomers. Interestingly, the Km saturation kinetics of ATP-dependent ABA transport do not differ greatly (260 nM and 1 μM for AtABCG25 and AtABCG40, respectively), although different assay systems were used to calculate activity (Kang et al. 2010, Kuromori et al. 2010). These findings strongly suggest that AtABCG25 and AtABCG40 are responsible for active control of ABA transport between plant cells. From two reports (Kang et al. 2010, Kuromori et al. 2010), a simple model can be proposed: ABA is exported from ABA-biosynthesizing cells to the apoplastic area, and then imported from the apoplast into guard cells (Fig. 7).
Fig. 7.
Schematic view of hypothetical ABA intercellular transmission. This diagram is an Arabidopsis leaf section showing two distinct cell types: vascular tissues, including vascular parenchyma cells, and guard cells on the leaf epidermis. AtABCG25 might function in ABA efflux from ABA-biosynthesizing vascular cells, and ABA would diffuse into apoplastic areas. AtABCG40 might function in ABA influx into guard cells to facilitate stomatal closure.
This model is also consistent with recent reports that some ABA receptors that trigger ABA signaling in cells are soluble and localized to the cytosol (Ma et al. 2009, Park et al. 2009), and suggests the potential importance of an ABA transporter that could deliver ABA in a regulated fashion to initiate rapid and controlled responses to various stress conditions (Kang et al. 2010). The investigation of the ABA transport mechanism has just begun and it provides a novel impetus for examining ABA intercellular regulation.
Conclusion and perspectives
There was major progress in the study of ABA during 2009–2010. The major ABA signaling pathway was established and the ABA transport system was discovered. These discoveries provided new insight into how plants respond to ABA at the intra- and intercellular levels. The overview of ABA sensing, signaling and transport described in this review is summarized in Fig. 8. ABA-related studies are entering a new stage, and many questions are beginning to arise. A major question is the complexity of ABA signaling networks in plants. Although a core signaling model has been established, it is not clear whether this model can explain all ABA responses. Numerous ABA signaling factors have been reported, involving other ABA receptors, protein kinases/phosphatases, transcription factors, RNA-binding proteins, chromatin remodeling factors, protein degradation enzymes and so on (Hirayama and Shinozaki 2007, Hirayama and Shinozaki 2010). It is necessary to determine whether these factors are dependent on or independent of the core pathway. In particular, some ABA-binding proteins, different from PYR/PYL/RCAR, are of major interest because it is not fully understood how they regulate downstream factors. For example, one of the ABA-binding proteins, ABAR/ChlH/GUN5, is a chloroplast protein regulated by circadian rhythms (Legnaioli et al. 2009), and it regulates multiple ABA-related transcription factors via a series of WRKY proteins (Shang et al. 2010). Several studies reported that WRKY transcription factors have significant roles in the regulation of ABA-responsive genes, involving bZIPs (Jiang and Yu 2009, Ren et al. 2010, Shang et al. 2010), suggesting some connections to the PYR/PYL/RCAR–PP2C–SnRK2 pathway. Likewise, the core pathway serves as a guide for positioning and ordering in understanding the significance of each factor. Studies suggest that the plasma membrane could be an ABA perception site, and we already know of two types of plasma membrane ABA receptors, GCR2 and GTG1/2. Further analysis will clarify how these receptors regulate ABA signaling in plants. It is also well known that ABA signaling cross-talks with other phytohormones such as gibberellin, jasmonic acid, ethylene and salicylic acid. It should be clarified whether the core ABA signaling pathway is involved in such cross-talk or not.
Fig. 8.
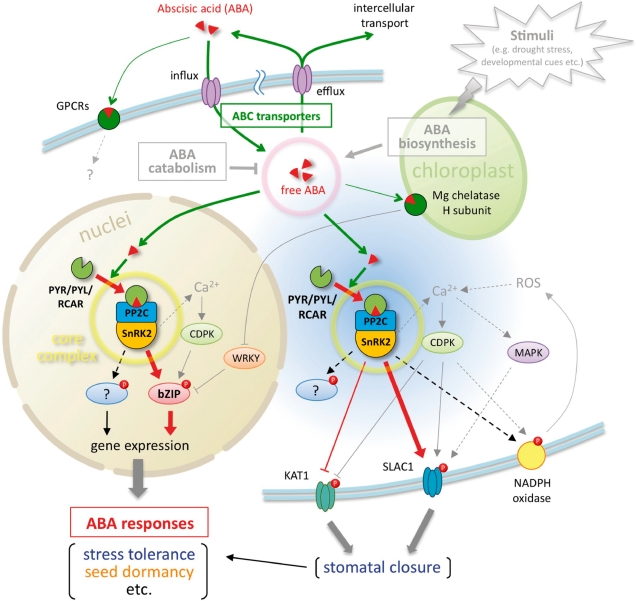
Overview of ABA sensing, signaling and transport. The ABA-related components described in this review are summarized. PYR/PYL/RCAR, PP2C and SnRK2 form a core signaling complex (yellow circle), which functions in at least two sites. One is the nucleus, in which the core complex directly regulates ABA-responsive gene expression by phosphorylation of AREB/ABF-type transcription factors. The other is the cytoplasm, and the core complex can access the plasma membrane and phosphorylate anion channels (SLAC1) or potassium channels (KAT1) to induce stomatal closure in response to ABA. Other substrates of SnRK2s have yet to be identified. The principal mechanism of ABA sensing or signaling in the core signaling complex is illustrated in Fig. 2. In contrast, the endogenous ABA level is a major determinant of ABA sensing that is maintained by ABA biosynthesis, catabolism or transport. The ABA transport system consists of two types of ABC transporter for influx or efflux. Although ABA biosynthesis and catabolism were not the focus of this review, these types of regulation are well described in other reviews (e.g. Nambara and Marion-Poll 2005, Hirayama and Shinozaki 2010). ABA movements are indicated by green lines and arrows, and major signaling pathways are indicated by red lines and arrows. Dotted lines indicate indirect or unconfirmed connections.
As described in this review, the PYR/PYL/RCAR–PP2C–SnRK2 pathway is highly redundant, suggesting that combinatorial variants of the complex have distinctive roles in ABA signaling among different tissues, organs and cell types. Further functional analyses of each combination are required to elucidate the fine details of ABA signal transduction. On the other hand, there is another question regarding the downstream events of PYR/PYL/RCAR–PP2C–SnRK2. Although several direct substrates or interacting proteins of PP2C or SnRK2 have been described, other substrates have remained elusive. Screening of SnRK2 substrates is a major challenge to fully understanding ABA signaling in plants. Precise knowledge of SnRK2 substrates will tell us a lot about the regulation and points of signal divergence or convergence, as well as points of cross-talk with other signaling pathways.
In addition to intracellular ABA signaling, intercellular ABA transport is emerging as a new area in ABA research. Presently, two ABA transporters are known; however, they belong to a large family of ABC transporters in Arabidopsis (Kang et al. 2010, Kuromori et al. 2010). It is possible that some other ABC transporters are also involved in cell to cell ABA transport. The next question is how ABA transport activity is regulated. Further analyses are required to clarify whether ABC transporters are regulated by environmental signals and/or ABA itself. In addition to intercellular ABA transport, long-distance signals may be involved in ABA responses at the whole plant level, i.e. from roots to shoots. Another challenging issue in ABA signaling is to understand where plants sense environmental stimuli, such as water deficits, and how they convert this to ABA signals.
In conclusion, our knowledge of ABA responses is rapidly expanding as more tools for manipulating ABA responses become available. We are entering the next phase in considering the agricultural or biotechnological uses of ABA signaling factors. For example, structural analyses could shed light on the detailed interface of ABA in the ligand pocket of PYR/PYL/RCAR receptors, possibly leading to protein engineering or chemical design to control ABA signaling artificially. ABA- induced post-transcriptional or post-translational modifications of proteins can be used for genetic engineering of plants (Umezawa et al. 2006). Many trials will be necessary to utilize the core ABA signaling pathway or ABA transport system for future crop improvement.
Funding
This work was supported in part by a Grant-in-Aid from the Targeted Proteins Research Program (TPRP), Grant-in-Aid for Scientific Research (C) and Grant-in-Aid for Young Scientists (B) of the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan, and the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (BRAIN).
Acknowledgments
The authors thank Dr. Eiji Nambara of University of Toronto and Dr. Takashi Hirayama of Okayama University for their valuable suggestions and comments.
Glossary
Abbreviations
- ABC transporter
ATP-binding cassette transporter
- ABF
ABRE-binding factor
- ABI1/2/5
ABA insensitive 1/2/5
- ABRE
ABA-responsive element
- AHG1/3
ABA-hypersensitive germination 1/3
- AREB
ABRE-binding protein
- AUX/IAA
auxin/indole-3-acetic acid
- bZIP
basic-domain leucine zipper
- GID1
gibberellin insensitive dwarf 1
- HAB1/2
homology to ABA 1/2
- LEA
late embryogenesis abundant
- PP2C
protein phosphatase 2C
- PYL
PYR1-like
- PYR1
pyrabactin resistance 1
- RCAR1
regulatory component of ABA receptor 1
- ROS
reactive oxygen species
- SLAC1
slow anion channel-associated 1
- SnRK2
sucrose non-fermenting 1-related protein kinase 2
- START
StAR-related lipid transfer
- TIR1
transport inhibitor response 1.
References
- Anderberg RJ, Walker-Simmons MK. Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases. Proc. Natl Acad. Sci. USA. 1992;89:10183–10187. doi: 10.1073/pnas.89.21.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin C, de Franco P, Bourbousse C, Chaignepain S, Schmitter J, Vavasseur A, et al. Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol. 2006;141:1316–1327. doi: 10.1104/pp.106.079327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensmihen S, Giraudat J, Parcy F. Characterization of three homologous basic leucine zipper transcription factors (bZIP) of the ABI5 family during Arabidopsis thaliana embryo maturation. J. Exp. Bot. 2005;56:597–603. doi: 10.1093/jxb/eri050. [DOI] [PubMed] [Google Scholar]
- Bensmihen S, Rippa S, Lambert G, Jublot D, Pautot V, Granier F, et al. The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell. 2002;14:1391–1403. doi: 10.1105/tpc.000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M, Barbier-Brygoo H, Lauriere C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 2004;279:41758–41766. doi: 10.1074/jbc.M405259200. [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Droillard M, Barbier-Brygoo H, Laurière C. Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol. Biol. 2007;63:491–503. doi: 10.1007/s11103-006-9103-1. [DOI] [PubMed] [Google Scholar]
- Cheng W, Endo A, Zhou L, Penney J, Chen H, Arroyo A., et al. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell. 2002;14:2723–2743. doi: 10.1105/tpc.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY. ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- Christmann A, Weiler EW, Steudle E, Grill E. A hydraulic signal in root-to-shoot signalling of water shortage. Plant J. 2007;52:167–174. doi: 10.1111/j.1365-313X.2007.03234.x. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- Endo A, Koshiba T, Kamiya Y, Nambara E. Vascular system is a node of systemic stress responses. Plant Signal. Behav. 2008;3:1138–1140. doi: 10.4161/psb.3.12.7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell. 2000;12:599–610. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park S, et al. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu J. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Zhu J. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl Acad. Sci. USA. 2009;106:8380–8385. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, et al. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell. 2005;17:3470–3488. doi: 10.1105/tpc.105.035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, et al. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009;50:2123–2132. doi: 10.1093/pcp/pcp147. [DOI] [PubMed] [Google Scholar]
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, et al. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl Acad. Sci. USA. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Marten I, Ache P, Matschi S, et al. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl Acad. Sci. USA. 2010;107:8023–8028. doi: 10.1073/pnas.0912030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase–phosphatase pair. Proc. Natl Acad. Sci. USA. 2009;106:21425–21430. doi: 10.1073/pnas.0912021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Porras J, Riano-Pachon D, Dreyer I, Mayer J., Mueller-Roeber B. Genome-wide analysis of ABA-responsive elements ABRE and CE3 reveals divergent patterns in Arabidopsis and rice. BMC Genom. 2007;8:260. doi: 10.1186/1471-2164-8-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Garcia MP, Rodriguez D, Nicolas C, Rodriguez PL, Nicolas G, Lorenzo O. Negative regulation of abscisic acid signaling by the Fagus sylvatica FsPP2C1 plays a role in seed dormancy regulation and promotion of seed germination. Plant Physiol. 2003;133:135–144. doi: 10.1104/pp.103.025569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins CF. ABC transporters: from microorganisms to man. Annu. Rev. Cell. Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 2007;12:343–351. doi: 10.1016/j.tplants.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 2010;61:1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x. [DOI] [PubMed] [Google Scholar]
- Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003;132:666–680. doi: 10.1104/pp.102.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev. 2010;24:1695–1708. doi: 10.1101/gad.1953910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, et al. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–111. doi: 10.1016/s1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- Jammes F, Song C, Shin D, Munemasa S, Takeda K, Gu D, et al. MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc. Natl Acad. Sci. USA. 2009;106:20520–20525. doi: 10.1073/pnas.0907205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Yu D. Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 2009;9:96. doi: 10.1186/1471-2229-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RR, Wagner RL, Verhey SD, Walker-Simmons MK. The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol. 2002;130:837–846. doi: 10.1104/pp.001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Hobo T, Murata M, Ban A, Hattori T. Abscisic acid-induced transcription is mediated by phosphorylation of an abscisic acid response element binding factor, TRAB1. Plant Cell. 2002;14:3177–3189. doi: 10.1105/tpc.005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Hwang J, Lee M, Kim Y, Assmann SM, Martinoia E, et al. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl Acad. Sci. USA. 2010;107:2355–2360. doi: 10.1073/pnas.0909222107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Choi H, Im M, Kim SY. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell. 2002;14:343–357. doi: 10.1105/tpc.010362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kang J, Cho D, Park JH, Kim SY. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 2004;40:75–87. doi: 10.1111/j.1365-313X.2004.02192.x. [DOI] [PubMed] [Google Scholar]
- Kim T, Böhmer M, Hu H, Nishimura N, Schroeder JI. Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 2010;61:561–591. doi: 10.1146/annurev-arplant-042809-112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingler JP, Batelli G, Zhu J. ABA receptors: the START of a new paradigm in phytohormone signalling. J. Exp. Bot. 2010;61:3199–3210. doi: 10.1093/jxb/erq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T, et al. Abscisic acid-activated SnRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 2005;44:939–949. doi: 10.1111/j.1365-313X.2005.02583.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T. Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell. 2004;16:1163–1177. doi: 10.1105/tpc.019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwai H, Nakaminami K, Seo M, Mitsuhashi W, Toyomasu T, Koshiba T. Tissue-specific localization of an abscisic acid biosynthetic enzyme, AAO3, in Arabidopsis. Plant Physiol. 2004;134:1697–1707. doi: 10.1104/pp.103.036970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu K, Nishikawa Y, Ohtsuka T, Taji T, Quatrano R, Tanaka S, et al. Functional analyses of the ABI1-related protein phosphatase type 2C reveal evolutionarily conserved regulation of abscisic acid signaling between Arabidopsis and the moss Physcomitrella patens. Plant Mol. Biol. 2009;70:341–357. doi: 10.1007/s11103-009-9476-z. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol. Plant. 1984;61:377–383. [Google Scholar]
- Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI. The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 2006;140:127–139. doi: 10.1104/pp.105.070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Miyaji T, Yabuuchi H, Shimizu H, Sugimoto E, Kamiya A, et al. ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc. Natl Acad. Sci. USA. 2010;107:2361–2366. doi: 10.1073/pnas.0912516107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Yamamoto M. Cloning of cDNAs from Arabidopsis thaliana that encode putative protein phosphatase 2C and a human Dr1-like protein by transformation of a fission yeast mutant. Nucleic Acids Res. 1994;22:5296–5301. doi: 10.1093/nar/22.24.5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S. A protein kinase–phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl Acad. Sci. USA. 2009;106:21419–21424. doi: 10.1073/pnas.0910601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnaioli T, Cuevas J, Mas P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 2009;28:3745–3757. doi: 10.1038/emboj.2009.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J. Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science. 1994;264:1448–52. doi: 10.1126/science.7910981. [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell. 1997;9:759–71. doi: 10.1105/tpc.9.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang X, Watson MB, Assmann SM. Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science. 2000;287:300–303. doi: 10.1126/science.287.5451.300. [DOI] [PubMed] [Google Scholar]
- Liu MS, Chien CT, Lin TP. Constitutive components and induced gene expression are involved in the desiccation tolerance of Selaginella tamariscina. Plant Cell Physiol. 2008;49:653–663. doi: 10.1093/pcp/pcn040. [DOI] [PubMed] [Google Scholar]
- Liu X, Yue Y, Li B, Nie Y, Li W, Wu W, et al. A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science. 2007;315:1712–1716. doi: 10.1126/science.1135882. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Chua NH. A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 2000;41:541–547. doi: 10.1093/pcp/41.5.541. [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- McCourt P, Creelman R. The ABA receptors—we report you decide. Curr. Opin. Plant Biol. 2008;11:474–478. doi: 10.1016/j.pbi.2008.06.014. [DOI] [PubMed] [Google Scholar]
- Melcher K, Ng L, Zhou XE, Soon F, Xu Y, Suino-Powell KM, et al. A gate–latch–lock mechanism for hormone signalling by abscisic acid receptors. Nature. 2009;462:602–608. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E. A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science. 1994;264:1452–1455. doi: 10.1126/science.8197457. [DOI] [PubMed] [Google Scholar]
- Mikolajczyk M, Awotunde OS, Muszynska G, Klessig DF, Dobrowolska G. Osmotic stress induces rapid activation of a salicylic acid-induced protein kinase and a homolog of protein kinase ASK1 in tobacco cells. Plant Cell. 2000;12:165–178. [PMC free article] [PubMed] [Google Scholar]
- Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang H, Asano A, et al. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- Mizoguchi M, Umezawa T, Nakashima K, Kidokoro S, Takasaki H, Fujita Y, et al. Two closely related subclass II SnRK2 protein kinases cooperatively regulate drought-inducible gene expression. Plant Cell Physiol. 2010;51:842–847. doi: 10.1093/pcp/pcq041. [DOI] [PubMed] [Google Scholar]
- Monks DE, Aghoram K, Courtney PD, DeWald DB, Dewey RE. Hyperosmotic stress induces the rapid phosphorylation of a soybean phosphatidylinositol transfer protein homolog through activation of the protein kinases SPK1 and SPK2. Plant Cell. 2001;13:1205–1219. doi: 10.1105/tpc.13.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori IC, Murata Y, Yang Y, Munemasa S, Wang Y, Andreoli S, et al. CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca2+- permeable channels and stomatal closure. PLoS Biol. 2006;4:e327. doi: 10.1371/journal.pbio.0040327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K, Hirano Y, Sun T, Hakoshima T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature. 2008;456:459–463. doi: 10.1038/nature07519. [DOI] [PubMed] [Google Scholar]
- Mustilli A, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima A, Uehara Y, Sakai T. The ABC subfamily B auxin transporter AtABCB19 is involved in the inhibitory effects of N-1-naphthyphthalamic acid on the phototropic and gravitropic responses of Arabidopsis hypocotyls. Plant Cell Physiol. 2008;49:1250–1255. doi: 10.1093/pcp/pcn092. [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E. Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J. 2005;41:697–709. doi: 10.1111/j.1365-313X.2005.02337.x. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009;50:1345–1363. doi: 10.1093/pcp/pcp083. [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll. A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- Negi J, Matsuda O, Nagasawa T, Oba Y, Takahashi H, Kawai-Yamada M, et al. CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature. 2008;452:483–486. doi: 10.1038/nature06720. [DOI] [PubMed] [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C., Cutler SR, et al. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science. 2009;326:1373–1379. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, Park S, Wang A, Carvalho PC, et al. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 2010;61:290–299. doi: 10.1111/j.1365-313X.2009.04054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Yoshida T, Kitahata N, Asami T, Shinozaki K, Hirayama T. ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 2007;50:935–949. doi: 10.1111/j.1365-313X.2007.03107.x. [DOI] [PubMed] [Google Scholar]
- Ogasawara Y, Kaya H, Hiraoka G, Yumoto F, Kimura S, Kadota Y, et al. Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by Ca2+ and phosphorylation. J. Biol. Chem. 2008;283:8885–8892. doi: 10.1074/jbc.M708106200. [DOI] [PubMed] [Google Scholar]
- Okamoto M, Tanaka Y, Abrams SR, Kamiya Y, Seki M, Nambara E. High humidity induces abscisic acid 8′-hydroxylase in stomata and vasculature to regulate local and systemic abscisic acid responses in Arabidopsis. Plant Physiol. 2009;149:825–834. doi: 10.1104/pp.108.130823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Nelson DC, Assmann SM. Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell. 2009;136:136–148. doi: 10.1016/j.cell.2008.12.026. [DOI] [PubMed] [Google Scholar]
- Park S, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilot G, Lacombe B, Gaymard F, Chérel I, Boucherez J, Thibaud J, et al. Guard cell inward K+ channel activity in Arabidopsis involves expression of the twin channel subunits KAT1 and KAT2. J. Biol. Chem. 2001;276:3215–3221. doi: 10.1074/jbc.M007303200. [DOI] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signalling. Trends Plant Sci. 2010;15:395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Rea PA. Plant ATP-binding cassette transporters. Annu. Rev. Plant Biol. 2007;58:347–375. doi: 10.1146/annurev.arplant.57.032905.105406. [DOI] [PubMed] [Google Scholar]
- Ren X, Chen Z, Liu Y, Zhang H, Zhang M, Liu Q, et al. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010;63:417–429. doi: 10.1111/j.1365-313X.2010.04248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Robert N, Merlot S, N’Guyen V, Boisson-Dernier A., Schroeder JI. A hypermorphic mutation in the protein phosphatase 2C HAB1 strongly affects ABA signaling in Arabidopsis. FEBS Lett. 2006;580:4691–4696. doi: 10.1016/j.febslet.2006.07.047. [DOI] [PubMed] [Google Scholar]
- Rodriguez PL, Benning G, Grill E. ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett. 1998;421:185–190. doi: 10.1016/s0014-5793(97)01558-5. [DOI] [PubMed] [Google Scholar]
- Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim T, et al. Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol. 2009;150:1345–1355. doi: 10.1104/pp.109.137174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra X, Modrego A, Rodriguez D, Gonzalez-Garcia M.P., Sanz L, Nicolas G, et al. The nuclear interactor PYL8/RCAR3 of Fagus sylvatica FsPP2C1 is a positive regulator of abscisic acid signaling in seeds and stress. Plant Physiol. 2010;152:133–150. doi: 10.1104/pp.109.146381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez-Garcia MP, Nicolas C, Lorenzo O, et al. Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J. 2004;37:354–369. doi: 10.1046/j.1365-313x.2003.01966.x. [DOI] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park S, Jamin M, et al. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009a;462:665–668. doi: 10.1038/nature08591. [DOI] [PubMed] [Google Scholar]
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, et al. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009b;60:575–578. doi: 10.1111/j.1365-313X.2009.03981.x. [DOI] [PubMed] [Google Scholar]
- Sato A, Gambale F, Dreyer I, Uozumi N. Modulation of the Arabidopsis KAT1 channel by an activator of protein kinase C in Xenopus laevis oocytes. FEBS J. 2010;277:2318–2328. doi: 10.1111/j.1742-4658.2010.07647.x. [DOI] [PubMed] [Google Scholar]
- Sato A, Sato Y, Fukao Y, Fujiwara M, Umezawa T, Shinozaki K, et al. Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem. J. 2009;424:439–448. doi: 10.1042/BJ20091221. [DOI] [PubMed] [Google Scholar]
- Schachtman DP, Goodger JQ. Chemical root to shoot signaling under drought. Trends Plant Sci. 2008;13:281–287. doi: 10.1016/j.tplants.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I. Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 2004;9:236–243. doi: 10.1016/j.tplants.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Seki M, Ishida J, Narusaka M, Fujita M, Nanjo T, Umezawa T, et al. Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct. Integr. Genomics. 2002;2:282–291. doi: 10.1007/s10142-002-0070-6. [DOI] [PubMed] [Google Scholar]
- Shang Y, Yan L, Liu Z, Cao Z, Mei C, Xin Q, et al. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell. 2010;22:1909–1935. doi: 10.1105/tpc.110.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Wang X, Wu F, Du S, Cao Z, Shang Y, et al. The Mg-chelatase H subunit is an abscisic acid receptor. Nature. 2006;443:823–826. doi: 10.1038/nature05176. [DOI] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Nakatsu T, Nakajima M, Naoe Y, Ohmiya H, et al. Structural basis for gibberellin recognition by its receptor GID1. Nature. 2008;456:520–523. doi: 10.1038/nature07546. [DOI] [PubMed] [Google Scholar]
- Sirichandra C, Gu D, Hu H, Davanture M, Lee S, Djaoui M, et al. Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 2009;583:2982–2986. doi: 10.1016/j.febslet.2009.08.033. [DOI] [PubMed] [Google Scholar]
- Szostkiewicz I, Richter K, Kepka M, Demmel S, Ma Y, Korte A, et al. Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J. 2010;61:25–35. doi: 10.1111/j.1365-313X.2009.04025.x. [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LIA, Sharon M, Zheng C., Robinson CV, Estelle M, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- Tougane K, Komatsu K, Bhyan SB, Sakata Y, Ishizaki K, Yamato KT, et al. Evolutionarily conserved regulatory mechanisms of abscisic acid signaling in land plants: characterization of ABSCISIC ACID INSENSITIVE1-like type 2C protein phosphatase in the liverwort Marchantia polymorpha. Plant Physiol. 2010;152:1529–1543. doi: 10.1104/pp.110.153387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouverie J, Vidal G, Zhang Z, Sirichandra C, Madiona K, Amiar Z, et al. Anion channel activation and proton pumping inhibition involved in the plasma membrane depolarization induced by ABA in Arabidopsis thaliana suspension cells are both ROS dependent. Plant Cell Physiol. 2008;49:1495–1507. doi: 10.1093/pcp/pcn126. [DOI] [PubMed] [Google Scholar]
- Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K. Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr. Opin. Biotechnol. 2006;17:113–122. doi: 10.1016/j.copbio.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, et al. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl Acad. Sci. USA. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Yoshida R, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K. SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2004;101:17306–17311. doi: 10.1073/pnas.0407758101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K., Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl Acad. Sci. USA. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahisalu T, Kollist H, Wang Y, Nishimura N, Chan W., Valerio G, et al. SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature. 2008;452:487–491. doi: 10.1038/nature06608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, et al. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell. 2009;21:3170–3184. doi: 10.1105/tpc.109.069179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson S, Davies WJ. Drought, ozone, ABA and ethylene: new insights from cell to plant to community. Plant Cell Environ. 2010;33:510–525. doi: 10.1111/j.1365-3040.2009.02052.x. [DOI] [PubMed] [Google Scholar]
- Yin P, Fan H, Hao Q, Yuan X, Wu D, Pang Y, et al. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat. Struct. Mol. Biol. 2009;16:1230–1236. doi: 10.1038/nsmb.1730. [DOI] [PubMed] [Google Scholar]
- Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F., Aronso J, et al. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 2002;43:1473–1483. doi: 10.1093/pcp/pcf188. [DOI] [PubMed] [Google Scholar]
- Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 2006;281:5310–5318. doi: 10.1074/jbc.M509820200. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T., Asami T, et al. ABA-Hypersensitive Germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 2006;140:115–126. doi: 10.1104/pp.105.070128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Ruan J, Ho TD, You Y, Yu T, Quatrano RS. Cis-regulatory element based targeted gene finding: genome-wide identification of abscisic acid- and abiotic stress-responsive genes in Arabidopsis thaliana. Bioinformatics. 2005;21:3074–3081. doi: 10.1093/bioinformatics/bti490. [DOI] [PubMed] [Google Scholar]