Abstract
Zaïre ebolavirus (ZEBOV) infection rapidly outruns the host's immunity and leads to death within a week. Fatal cases have been associated with an aberrant innate, proinflammatory immune response followed by a suppressed adaptive response leading to the rapid depletion of peripheral NK cells and lymphocytes. A critical role for NK cells has been suggested but not elucidated. In this genetic study, we investigated the association of KIR genotype with disease outcome by comparing genotypes of a Gabonese control population, IgG+ contacts, survivors, and fatalities of ZEBOV infection. We showed that the activating KIR2DS1 and KIR2DS3 genes associate with fatal outcome in Ebola virus infection. In addition, this study brings supplemental evidence in favor of the specificity of the IgG+ contact population. The outcome of fulminating Ebola virus infection could depend in part on the host's inherited KIR gene repertoire. This supports a key role for KIRs in disease susceptibility to infections.
Keywords: Ebola, KIR, Natural killer cells, KIR2DS1, KIR2DS3
Ebola virus, from the family Filoviridae, causes severe hemorrhagic fever in humans and primates (Hoenen et al. 2006). The Zaïre ebolavirus (ZEBOV) species associates with an 80% case-fatality rate. ZEBOV infection rapidly outruns the host's immunity and leads to death within a week. Severe cases are associated with aberrant innate, proinflammatory immunity preceding a suppressed adaptive response which leads to the rapid depletion of peripheral natural killer (NK) cells and other lymphocytes (Baize et al. 1999, 2002; Leroy et al. 2000). A critical role for innate host factors has been suggested, but the function of NK cells during ZEBOV infection remains unclear.
We present a genetic study focusing on the influence of a host factor, the killer immunoglobulin-like receptor (KIR) repertoire, in this context of ZEBOV infection. KIRs are members of the Ig-superfamily of type I membrane proteins, expressed on the surface of NK cells and T-cell subsets (Lanier 1998). Encoded by a family of highly polymorphic genes, KIRs bind to HLA class I alleles, and the complex integration of signals drive NK function (Carrington and Martin 2006; Dohring and Colonna 1996).Thus, individual KIR haplotypes differ in number and identity of genes, each haplotype including seven to 12 genes (Wilson et al. 2000; Witt et al. 1999). Two haplotypes are commonly defined: (1) the A haplotype comprises seven KIR genes, including a unique activating KIR gene KIR2DS4, and (2) B haplotypes are variable and characterized by the presence of additional activating KIR genes (Uhrberg et al. 1997; Valiante et al. 1997). Unrelated individuals are therefore unlikely to express identical KIRs (Shilling et al. 2002).
Following the model of major histocompatibility complex studies, disease association studies have revealed associations of KIR genes with disease. To date, these studies have mainly targeted viral infections such as human immunodeficiency virus infection, but also cancer, autoimmune, and inflammatory disorders (Khakoo et al. 2004; Martin et al. 2002; Cook et al. 2006; Yen et al. 2001). Activating genotypes generally appear to be beneficial during viral infections, whereas they constitute more a risk for susceptibility to autoimmunity and certain cancers (Kulkarni et al. 2008). Our concern here was to evaluate the association of KIR genotype with the outcome of human ZEBOV infection, a fulminating disease in which the innate immunity seems critical.
To investigate the influence of the KIR repertoire upon the outcome of human EBOV infection, we compared KIR genotype of laboratory-confirmed ZEBOV survivors to fatalities. Samples from fatalities were previously obtained during the Gabonese epidemics of 2001–2002 (Leroy et al. 2004). Survivors and contacts (IgG+, with no historic of the disease) were sampled during retrospective campaigns (Wauquier et al. 2009; Becquart et al. 2010). Contacts (43% male; mean age, 41; range, 18–70) and controls (41% male; mean age, 36; range, 18–57) were selected to match for age and sex. All blood samples were retrieved with informed and written consent. EBOV-IgG serostatus was determined by ELISA as previously described (Ksiazek et al. 1999). Genomic DNA was extracted using QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) from whole blood or PBMCs isolated using standard density gradient centrifugation with Ficoll-Paque (Eurobio, les Ulis, France). Genotyping was performed by polymerase chain reaction using the KIR typing kit (Miltenyi Biotec, Inc., Auburn, CA, USA) following manufacturer's instructions. Fifteen KIR genes and two pseudogenes were typed. In total, tests were performed on samples from 21 survivors, 15 non-survivors, 68 contacts (IgG+), and 54 controls (IgG−) of same geographical origin. Statistical analysis used chi-squared test or Fisher's exact test to compare results between each groups. A probability level of less than 0.05 was considered statistically significant.
We studied KIR genotypes in four populations: (1) healthy volunteers from rural regions of Gabon, tested anti-ZEBOV IgG−, were used as controls; (2) healthy and anti-ZEBOV IgG+ individuals, from regions where no cases of ZEBOV infection have ever been reported, were defined as contacts; and (3) patients that had fully recovered from all symptoms (survivors) and (4) fatalities had all been laboratory-confirmed during the epidemics. Percentages of individuals carrying each particular KIR gene were calculated (Table 1), and among all participants, 33 different KIR genotypes were distinguished (Fig. 1).
Table 1.
Percentages of carriers of inhibitory, activating, and pseudogene KIR genes in a control Gabonese population and in contacts (IgG+), survivors, and fatalities of Ebola virus infection
| Control | Control | Contacts | Survivors | Fatalities | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Senegal | (n = 54) | (n = 68) | (n = 21) | (n = 15) | |||||||||
| (n = 90) | n | % | p | n | % | p | n | % | p | n | % | p | |
| 2DL1 | 100 | 54 | 100.0 | 68 | 100.0 | 21 | 100.0 | 13 | 86.7 | * | |||
| 2DL2 | 55 | 35 | 64.8 | 40 | 58.8 | 12 | 57.1 | 11 | 73.3 | ||||
| 2DL3 | 90 | 43 | 79.6 | 56 | 82.4 | 17 | 81.0 | 9 | 60.0 | ||||
| 2DL4 | 100 | 54 | 100.0 | 68 | 100.0 | 21 | 100.0 | 15 | 100.0 | ||||
| 2DL5all | 52 | 39 | 72.2 | 48 | 70.6 | 13 | 61.9 | 13 | 86.7 | ||||
| 2DL5A | 31 | 57.4 | 34 | 50.0 | 11 | 52.4 | 9 | 60.0 | |||||
| 2DL5B | 38 | 70.4 | 48 | 70.6 | 13 | 61.9 | 10 | 66.7 | |||||
| 3DL1 | 99 | 54 | 100.0 | 66 | 97.1 | 21 | 100.0 | 15 | 100.0 | ||||
| 3DL2 | 100 | 54 | 100.0 | 68 | 100.0 | 21 | 100.0 | 15 | 100.0 | ||||
| 3DL3 | 100 | 54 | 100.0 | 68 | 100.0 | 21 | 100.0 | 15 | 100.0 | ||||
| 2DS1 | 13 | 10 | 18.5 | 14 | 20.6 | 1 | 4.8 | 7 | 46.7 | *, ** | |||
| 2DS2 | 42 | 31 | 57.4 | 35 | 51.5 | 9 | 42.9 | 10 | 66.7 | ||||
| 2DS3 | 24 | 19 | 35.2 | 21 | 30.9 | 2 | 9.5 | * | 8 | 53.3 | ** | ||
| 2DS4del | 36 | 66.7 | 47 | 69.1 | 15 | 71.4 | 11 | 73.3 | |||||
| 2DS4ins | 44 | 81.5 | 41 | 60.3 | * | 19 | 90.5 | 11 | 73.3 | ||||
| 2DS5 | 30 | 18 | 33.3 | 27 | 39.7 | 10 | 47.6 | 7 | 46.7 | ||||
| 3DS1 | 4 | 4 | 7.4 | 8 | 11.8 | 2 | 9.5 | 1 | 6.7 | ||||
| 2DP1 | 100 | 54 | 100.0 | 68 | 100.0 | 21 | 100.0 | 15 | 100.0 | ||||
| 3DP1 | 100 | 54 | 100.0 | 68 | 100.0 | 21 | 100.0 | 15 | 100.0 | ||||
*p < 0.05 between studied group and controls; **p < 0.05 between survivors and fatalities
Fig. 1.
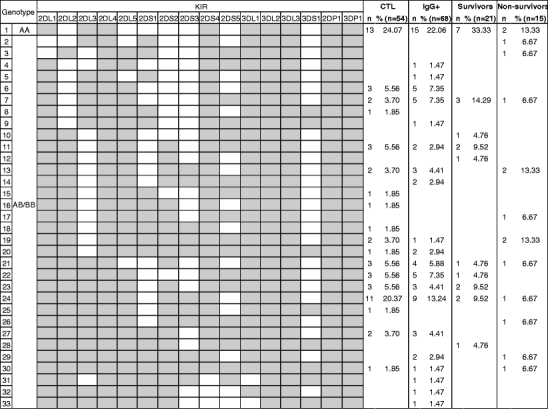
KIR genotype profiles observed in the Gabonese population, contacts (IgG+) group, survivors, and fatalities of Ebola virus infection
Percentages of KIR gene carriers in the Gabonese control population were in accordance with those of another African population from Senegal (Table 1), although Gabon is situated in the Central African forests and mainly inhabited by the Bantu, an ethnic population that highly differs from the sub-Saharan population of Senegal (Denis et al. 2005). The genotype most represented was the unique AA homozygous genotype (Fig. 1). Contacts were otherwise similar to controls only differing by a significantly lower percentage of activating KIR2DS4 carriers (Table 1). Only 9.5% of survivors (vs 35.2% in controls) had the activating KIR2DS3 gene. The frequency of AA profiles, containing only one activating gene (2DS4), was highest in survivors (33.3%) but lowest in fatalities (13.3%) (Fig. 1). Fatal cases differed from controls by three genes. A higher proportion of non-survivors possessed the activating KIR2DS1, whereas two non-survivors lacked the inhibitory KIR2DL1. Interestingly, KIR2DS1 and KIR2DS3 genes were also significantly more frequent in fatalities (46.8% and 53.3%, respectively) than in survivors (4.6% and 9.5%, respectively; Table 1), thus indicating a potential effect of these activating KIRs in fatal outcome. In the same way, KIR2DS1 has been previously associated with susceptibility to psoriasis but also protection against spontaneous abortion or development of Hodgkin's lymphoma (Luszczek et al. 2004; Hiby et al. 2008; Besson et al. 2007). By contrast, KIR2DS3 was significantly decreased among patients with microscopic polyangiitis (Miyashita et al. 2006).
Finally, we found that the mean number of activating KIR genes per genotype was lowest in survivors (2.1) and highest in fatalities (3.2; data not shown). These values were average for controls and contacts at 2.47 and 2.52, respectively. Controls, contacts, survivors, and fatalities presented similar mean number of inhibitory KIR genes per genotype with 7.15, 7.19, 6.95, and 6.93, respectively. These observations suggest that certain KIR combinations could provide a permissive immune microenvironment fostering fatal outcome.
Our study brings additional evidence in favor of the specificity of the IgG+ contact population. If IgG positivity was to be the result of cross-reaction with antibodies against other common infections, this population would present the same KIR repertoire as the controls. On the contrary, contacts were characterized by KIR distribution that differed from controls, survivors, and fatalities, defining a fourth category, implying that this population results from either (1) simple exposure to the virus (abortive infection), meaning that the KIR repertoire here includes repertoires of both survivors and fatalities, or (2) ZEBOV infection, then the KIR repertoire in this population contributes to an asymptomatic, “hyper-resistant” phenotype.
An activating KIR profile was associated with fatal outcome. This deleterious effect seems linked to KIR2DS1 and KIR2DS3 genes that were by far more present amongst non-survivors. KIR genes exhibit considerable sequence diversity (Garcia et al. 2003). Fifteen alleles of KIR2DS1 and 13 alleles of KIR2DS3 have been described (http://www.ebi.ac.uk/ipd/kir/index.html). This may influence expression, ligand binding, cytolysis, and cytokine secretion, as described for other KIRs (Yawata et al. 2006). Furthermore, KIRs are clonally expressed on NK cells in a stochastic manner so that each NK cell clone expresses only a portion of its KIR genome. Therefore, a substantial fraction of a patient's NK cells may not express KIR2DS1 and KIR2DS3 even though they carry the corresponding genes and thus still ignore ZEBOV-infected targets, explaining the survival of certain patients which carry these activating KIRs (Valiante et al. 1997).
It is generally thought that in the context of ZEBOV infection, it is the over-activation of the immune response that is responsible for the rapid depletion in NK cells and lymphocytes. Our results support this hypothesis, suggesting that the activity of KIR2DS1 and KIR2DS3, in conjunction with the appropriate ligands, may favor the activation status of the host's NK cells and participate in the physiopathological process by excessively destroying infected cells.
This study presents evidence that KIR2DS1 and KIR2DS3 are associated with fatal outcome in Ebola virus infection, supporting a key role for KIRs in disease susceptibility to infections. It is therefore suggested that NK cells, rapidly efficient after infection, may be critical for either pathogenesis or recovery from this fatal disease.
Acknowledgments
We are very grateful to C. Gasquet and D. Nkoghe for their help in obtaining the samples. We also thank P. Yaba and A. Délicat for technical assistance during this work. This work was funded by a grant from l'Agence National pour la Recherche (ANR) appel MIME 2006, nºANR-06-MIME-022-01. CIRMF is supported by the government of Gabon, Total-Fina-Elf Gabon, and the Ministère des affaires Etrangères, France.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Baize S, Leroy EM, Georges-Courbot MC, Capron M, Lansoud-Soukate J, Debre P, Fisher-Hoch SP, McCormick JB, Georges AJ. Defective humoral responses and extensive intravascular apoptosis are associated with fatal outcome in Ebola virus-infected patients. Nat Med. 1999;5(4):423–426. doi: 10.1038/7422. [DOI] [PubMed] [Google Scholar]
- Baize S, Leroy EM, Georges AJ, Georges-Courbot MC, Capron M, Bedjabaga I, Lansoud-Soukate J, Mavoungou E. Inflammatory responses in Ebola virus-infected patients. Clin Exp Immunol. 2002;128(1):163–168. doi: 10.1046/j.1365-2249.2002.01800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becquart P, Wauquier N, Mahlakoiv T, Nkoghe D, Padilla C, Souris M, Ollomo B, Gonzalez JP, De Lamballerie X, Kazanji M, Leroy EM. High prevalence of both humoral and cellular immunity to Zaire ebolavirus among rural populations in Gabon. PLoS One. 2010;5(2):e9126. doi: 10.1371/journal.pone.0009126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson C, Roetynck S, Williams F, Orsi L, Amiel C, Lependeven C, Antoni G, Hermine O, Brice P, Ferme C, Carde P, Canioni D, Briere J, Raphael M, Nicolas JC, Clavel J, Middleton D, Vivier E, Abel L. Association of killer cell immunoglobulin-like receptor genes with Hodgkin's lymphoma in a familial study. PLoS One. 2007;2(5):e406. doi: 10.1371/journal.pone.0000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington M, Martin MP. The impact of variation at the KIR gene cluster on human disease. Curr Top Microbiol Immunol. 2006;298:225–257. doi: 10.1007/3-540-27743-9_12. [DOI] [PubMed] [Google Scholar]
- Cook M, Briggs D, Craddock C, Mahendra P, Milligan D, Fegan C, Darbyshire P, Lawson S, Boxall E, Moss P. Donor KIR genotype has a major influence on the rate of cytomegalovirus reactivation following T-cell replete stem cell transplantation. Blood. 2006;107(3):1230–1232. doi: 10.1182/blood-2005-03-1039. [DOI] [PubMed] [Google Scholar]
- Denis L, Sivula J, Gourraud PA, Kerdudou N, Chout R, Ricard C, Moisan JP, Gagne K, Partanen J, Bignon JD. Genetic diversity of KIR natural killer cell markers in populations from France, Guadeloupe, Finland, Senegal and Reunion. Tissue Antigens. 2005;66(4):267–276. doi: 10.1111/j.1399-0039.2005.00473.x. [DOI] [PubMed] [Google Scholar]
- Dohring C, Colonna M. Human natural killer cell inhibitory receptors bind to HLA class I molecules. Eur J Immunol. 1996;26(2):365–369. doi: 10.1002/eji.1830260215. [DOI] [PubMed] [Google Scholar]
- Garcia CA, Robinson J, Guethlein LA, Parham P, Madrigal JA, Marsh SG. Human KIR sequences 2003. Immunogenetics. 2003;55(4):227–239. doi: 10.1007/s00251-003-0572-y. [DOI] [PubMed] [Google Scholar]
- Hiby SE, Regan L, Lo W, Farrell L, Carrington M, Moffett A. Association of maternal killer-cell immunoglobulin-like receptors and parental HLA-c genotypes with recurrent miscarriage. Hum Reprod. 2008;23(4):972–976. doi: 10.1093/humrep/den011. [DOI] [PubMed] [Google Scholar]
- Hoenen T, Groseth A, Falzarano D, Feldmann H. Ebola virus: unravelling pathogenesis to combat a deadly disease. Trends Mol Med. 2006;12(5):206–215. doi: 10.1016/j.molmed.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, Goedert JJ, Vlahov D, Hilgartner M, Cox S, Little AM, Alexander GJ, Cramp ME, O'Brien SJ, Rosenberg WM, Thomas DL, Carrington M. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305(5685):872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- Ksiazek TG, West CP, Rollin PE, Jahrling PB, Peters CJ. Elisa for the detection of antibodies to Ebola viruses. J Infect Dis. 1999;179(Suppl 1):S192–S198. doi: 10.1086/514313. [DOI] [PubMed] [Google Scholar]
- Kulkarni S, Martin MP, Carrington M. The yin and yang of HLA and KIR in human disease. Semin Immunol. 2008;20(6):343–352. doi: 10.1016/j.smim.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier LL. NK cell receptors. Annu Rev Immunol. 1998;16:359–393. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- Leroy EM, Baize S, Volchkov VE, Fisher-Hoch SP, Georges-Courbot MC, Lansoud-Soukate J, Capron M, Debre P, McCormick JB, Georges AJ. Human asymptomatic Ebola infection and strong inflammatory response. Lancet. 2000;355(9222):2210–2215. doi: 10.1016/S0140-6736(00)02405-3. [DOI] [PubMed] [Google Scholar]
- Leroy EM, Rouquet P, Formenty P, Souquiere S, Kilbourne A, Froment JM, Bermejo M, Smit S, Karesh W, Swanepoel R, Zaki SR, Rollin PE. Multiple Ebola virus transmission events and rapid decline of central African wildlife. Science. 2004;303(5656):387–390. doi: 10.1126/science.1092528. [DOI] [PubMed] [Google Scholar]
- Luszczek W, Manczak M, Cislo M, Nockowski P, Wisniewski A, Jasek M, Kusnierczyk P. Gene for the activating natural killer cell receptor, KIR2DS1, is associated with susceptibility to psoriasis vulgaris. Hum Immunol. 2004;65(7):758–766. doi: 10.1016/j.humimm.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, Wilson M, O’Brien SJ, Carrington M. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to aids. Nat Genet. 2002;31(4):429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- Miyashita R, Tsuchiya N, Yabe T, Kobayashi S, Hashimoto H, Ozaki S, Tokunaga K. Association of killer cell immunoglobulin-like receptor genotypes with microscopic polyangiitis. Arthritis Rheum. 2006;54(3):992–997. doi: 10.1002/art.21653. [DOI] [PubMed] [Google Scholar]
- Shilling HG, Young N, Guethlein LA, Cheng NW, Gardiner CM, Tyan D, Parham P. Genetic control of human NK cell repertoire. J Immunol. 2002;169(1):239–247. doi: 10.4049/jimmunol.169.1.239. [DOI] [PubMed] [Google Scholar]
- Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7(6):753–763. doi: 10.1016/S1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- Valiante NM, Lienert K, Shilling HG, Smits BJ, Parham P. Killer cell receptors: keeping pace with MHC class I evolution. Immunol Rev. 1997;155:155–164. doi: 10.1111/j.1600-065X.1997.tb00948.x. [DOI] [PubMed] [Google Scholar]
- Wauquier N, Becquart P, Gasquet C, Leroy EM. Immunoglobulin g in Ebola outbreak survivors, Gabon. Emerg Infect Dis. 2009;15(7):1136–1137. doi: 10.3201/eid1507.090402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, Beck S, Trowsdale J. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci U S A. 2000;97(9):4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt CS, Dewing C, Sayer DC, Uhrberg M, Parham P, Christiansen FT. Population frequencies and putative haplotypes of the killer cell immunoglobulin-like receptor sequences and evidence for recombination. Transplantation. 1999;68(11):1784–1789. doi: 10.1097/00007890-199912150-00024. [DOI] [PubMed] [Google Scholar]
- Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203(3):633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen JH, Moore BE, Nakajima T, Scholl D, Schaid DJ, Weyand CM, Goronzy JJ. Major histocompatibility complex class I-recognizing receptors are disease risk genes in rheumatoid arthritis. J Exp Med. 2001;193(10):1159–1167. doi: 10.1084/jem.193.10.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]


