Abstract
Bacillus subtilis RecO plays a central role in recombinational repair and genetic recombination by (i) stimulating RecA filamentation onto SsbA-coated single-stranded (ss) DNA, (ii) modulating the extent of RecA-mediated DNA strand exchange and (iii) promoting annealing of complementary DNA strands. Here, we report that RecO-mediated strand annealing is facilitated by cognate SsbA, but not by a heterologous one. Analysis of non-productive intermediates reveals that RecO interacts with SsbA-coated ssDNA, resulting in transient ternary complexes. The self-interaction of ternary complexes via RecO led to the formation of large nucleoprotein complexes. In the presence of homology, SsbA, at the nucleoprotein, removes DNA secondary structures, inhibits spontaneous strand annealing and facilitates RecO loading onto SsbA–ssDNA complex. RecO relieves SsbA inhibition of strand annealing and facilitates transient and random interactions between homologous naked ssDNA molecules. Finally, both proteins lose affinity for duplex DNA. Our results provide a mechanistic framework for rationalizing protein release and dsDNA zippering as coordinated events that are crucial for RecA-independent plasmid transformation.
INTRODUCTION
Homologous recombination (HR) is required for the maintenance of genomic stability, to prevent cancer development and to allow organisms to survive DNA damage (1–4). In the nucleotide bound form, the recombinases of the RecA family (bacterial RecA, eukaryotic Rad51 and Dmc1, etc.) are tightly regulated by a specialized class of mediator proteins that ensure that the assembly of a dynamic single-stranded (ss) DNA·recombinase filament occurs only when needed (1,2,4–6). Single-stranded binding proteins (bacterial SsbA/SSB and eukaryotic RPA) protect the ssDNA intermediates generated during HR from environmental insults, remove secondary structures and limit recombinase assembly onto ssDNA (7). A family of mediator proteins (e.g. bacterial RecO[R], eukaryotic Rad52 and BRCA2) have evolved to counteract the limitation exerted by their cognate single-stranded binding protein (SSB/SsbA, RPA) on the recombinase loading onto ssDNA (8–17). These mediator proteins can also catalyze other activities even in the absence of the recombinase. Indeed, Bacillus subtilis RecO plays an essential role not only in RecA-dependent double-strand break (DSB) repair, but also in RecA-independent plasmid transformation (18,19). During natural transformation, RecO anneals complementary plasmid ssDNA molecules in the presence of SsbA, while RecA fails to promote this reaction (19). Note that unless stated otherwise, the indicated genes and products are of B. subtilis origin.
How RecO mediates homology search and annealing of SsbA–ssDNA complex remains elusive. RecO is a 255-residue-long polypeptide that has poor identity, although similar length, with Escherichia coli RecO (RecOEco) protein. Only the N-terminal region of RecO shares a significant level of identity with RecOEco (29% in the first 164 amino acids), but the identity drops to negligible levels when the C-terminal region is considered (20). The crystal structure of Deinococcus radiodurans RecO (RecODra) has revealed that the N-terminus of RecODra adopts an OB fold domain, also present in eukaryotic RPA or BRCA2 protein (21,22). The SsbA protein (counterpart of SSBEco), which is essential for cell proliferation, inhibits the spontaneous annealing of complementary DNA strands (19). SsbA (a 172-residue-long polypeptide) shares a significant degree of identity at their 105 N-terminal residues with other heterologous single-stranded binding proteins (e.g. bacteriophage SPP1 Ssb [SsbSPP1] and SSBEco, 44% and 38% identity, respectively), but little homology toward the C-terminal ends. However, a patch of high identity at their acidic C-terminal end is also detected (78% with SsbSPP1 residues (SsbA residues 153–172) and 63% with SSBEco (161–172)] (7,15).
In this study, we report that SsbA facilitates RecO-mediated strand annealing through the accumulation of non-productive ternary complexes, by protein–protein and protein–ssDNA interactions. Once these complexes are stabilized, they can interact via RecO molecules, leading to the formation of bridged structures, with RecO decreasing the half-life of the SsbA–ssDNA complexes. The annealing process is based on transient and random interactions between naked ssDNA segments facilitated by RecO and inhibited by SsbA. We propose that the ability of RecO to facilitate the dislodging of SsbA from ssDNA and the concomitant strand annealing is a crucial event for RecA-independent plasmid transformation.
MATERIALS AND METHODS
Enzymes, reagents and protein purification
All chemicals were p.a. grade and purchased from Merck Darmstadt, Germany. DNA restriction and modification enzymes and nucleotides were from Boehringer Mannheim, Germany. Ultrapure acrylamide was from Serva, Heidelberg, Germany.
RecO, SsbA and SsbSPP1 proteins were purified as described (15,23). RecO and SsbSPP1 concentrations were expressed as moles of protein dimers and SsbA as tetramers.
EcoRI-cleaved 3197-bp pGEM 3Zf(+) vector was partially resected with T7 exonuclease (Invitrogen, Spain) to generate 5′-resected duplex DNA (or short 3′-ssDNA tails), as previously described (24). The EcoRI–AflIII-cleaved 440-bp pGEM 3Zf(+) DNA segment was gel purified as described (19). DNA concentrations were established using the molar extinction coefficients of 8780 and 6500 M−1 cm−1 at 260 nm for ssDNA and dsDNA, respectively. Unless otherwise stated, DNA concentrations are given as moles of nucleotides (nt).
DNA substrates and binding reactions
Linear 440-bp [γ-32P]-ssDNA (7 µM) or AflIII-cleaved 2958-bp pBluescriptII KS(−) DNA (0.5 µM) was heat denatured as described (19). The 440-nt ssDNA fragment was incubated with variable SsbA, SsbSPP1 or RecO concentrations for 40 min at 30°C in buffer A [50 mM Tris–HCl (pH 7.5), 50 mM NaCl, 1 mM DTT, 2 mM EDTA, 50 µg/ml BSA, 5% glycerol] or buffer B [50 mM Tris–HCl (pH 7.5), 50 mM NaCl, 1 mM DTT, 10 mM magnesium acetate, 50 µg/ml BSA, 5% glycerol]. The ssDNA was pre-incubated with SsbA or SsbSPP1 (90 nM) for 10 min at 30°C in buffer A or B. Variable amounts of RecO were added and incubated for 40 min. Alternatively, a fix RecO concentration was incubated for variable times. The reaction mixture was deproteinized, run in a 6% polyacrylamide gel electrophoresis (PAGE), visualized and quantified as described (19).
A heat-denatured 2958-nt DNA (0.5 µM) or 5′-resected duplex DNA (0.65 µM in bp, with ∼7.8% of 3′-tailed ssDNA) was pre-incubated with SsbA (10 nM) for 10 min at 30°C in buffer C [5 mM HEPES (pH7.5), 20 mM NaCl, 5 mM MgCl2]. Then, the nucleoprotein complexes were incubated for fix (60 min) or variable times. Samples were incubated for 10 min at 30°C and diluted with buffer D [5 mM HEPES (pH7.5), 50 µM spermidine] before depositing on freshly cut mica as described (25).
The rate of dissociation of SsbA (4 nM) from 60-nt long [γ-32P]-poly[dT] ssDNA (90 nM) in buffer E [50 mM Tris–HCl (pH 7.5), 30 mM NaCl, 1 mM DTT, 10 mM EDTA, 50 µg/ml BSA, 5% glycerol] in the presence of a 20-fold excess of cold poly[dT] ssDNA and RecO (500 nM) was measured by using alkali-treated filters (millipore, HAWP 45 µm) as described (26).
Atomic force microscopy analyses
Atomic force microscopy (AFM) observations were performed on a Nanoscope IIIa (Digital Instruments) in air using the tapping mode. The cantilever (OMCL-AC160TS-W2, Olympus) was 160 µm in length with a spring constant of 33–62 N/m. The scanning frequency was 2–3 Hz, and images were captured with the height mode in a 512 × 512 pixel format. The obtained images were plane-fitted and flattened by the computer program accompanying the imaging module. The ‘tip effect’ was removed using the apparent size of DNA as a reference. Volume analysis was carried out using the Image SXM software (27). Image processing of the topographs and height measurements were performed as described (25).
RESULTS
RecO anneals complementary strands complexed with SsbA
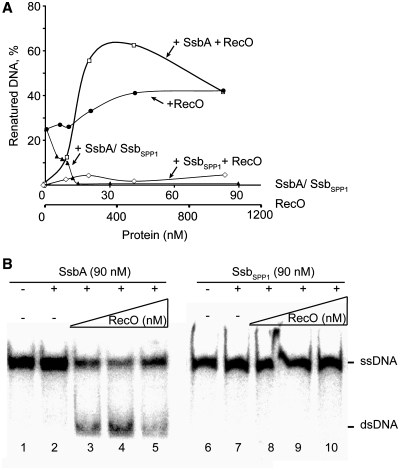
Spontaneous strand annealing of complementary homologous 440-nt-long ssDNAs was measured to be ∼16% and ∼24% in the presence or absence, respectively, of Mg2+. The spontaneous renaturation was suppressed upon addition of SsbA, both in the absence or presence of Mg2+ (Figure 1A and Supplementary Figure S1A). The heterologous SsbSPP1, used as a non-specific control, also inhibited spontaneous annealing both in the absence or presence of Mg2+ (Figure 1A and Supplementary Figure S1A).
Figure 1.
RecO anneals complementary strands complexed with SsbA protein. (A) A 440-nt long [γ-32P]-ssDNA (7 µM, in nt) pre-incubated with a fix amount of SsbA or SsbSPP1 (90 nM) and incubated with increasing concentrations of RecO (125, 250, 500 and 1000 nM) or incubated with variable amount of SsbA or SsbSPP1 (5, 7.5, 10, 15, 30, 60 and 90 nM) for 40 min at 30°C in buffer A. The products were separated and quantified as described in ‘Materials and Methods’ section. Inhibition spontaneous annealing by SsbA or SsbSPP1 was similar, therefore only the data for the former are shown. (B) The ssDNA was pre-incubated with SsbA or SsbSPP1 (90 nM, 1 protein/78-nt) in buffer A. Increasing concentrations of RecO (125, 250, 500 and 1000 nM, 1 RecO/28-, 14 nt or 7 nt) were added and the reaction incubated for 40 min at 30°C.
The addition of RecO facilitated the accumulation of annealed complementary DNA strands, both in the absence or presence of Mg2+ (Figures 1 and S1). To determine whether RecO could overcome the inhibitory effect of SsbA or SsbSPP1, the ssDNA probe was pre-incubated with SsbA or SsbSPP1 (90 nM), followed by the addition of increasing concentrations of RecO (125–1000 nM) (Figure 1B). SsbA facilitated RecO-mediated annealing of complementary strands (Figure 1). The extent of RecO-mediated strand annealing increased with increasing RecO concentrations, but at a ratio of 1 RecO to 7 nt the extent of annealing was significantly reduced (Figure 1). This effect, however, was overcome by increasing SsbA concentrations (1 SsbA/26 nt) (data not shown).
When the ssDNA was coated by SsbSPP1, RecO was unable to catalyze annealing of complementary strands (Figures 1 and S1). It is likely that the primary role of SsbA is to facilitate specific RecO–SsbA interaction that should be involved in the annealing of complementary strands, rather than removing DNA secondary structures and/or inhibiting non-productive RecO binding to ssDNA.
To further characterize the annealing reaction, ssDNA was pre-incubated with SsbA or SsbSPP1 (1 protein/78 nt), followed by the addition of a fixed RecO amount for a variable time in the presence of Mg2+ (Figure S1). RecO was able to anneal ∼70% of the SsbA-coated complementary ssDNAs (Figure S1), but it was unable to catalyze annealing of complementary strands when the ssDNA was coated by the non-cognate SsbSPP1 protein (Figure S1). This is consistent with the observation that only SsbA interacts physically with RecO, albeit both SsbA and SsbSPP1 share an acidic C-terminus (15).
Analysis of SsbA–ssDNA complexes
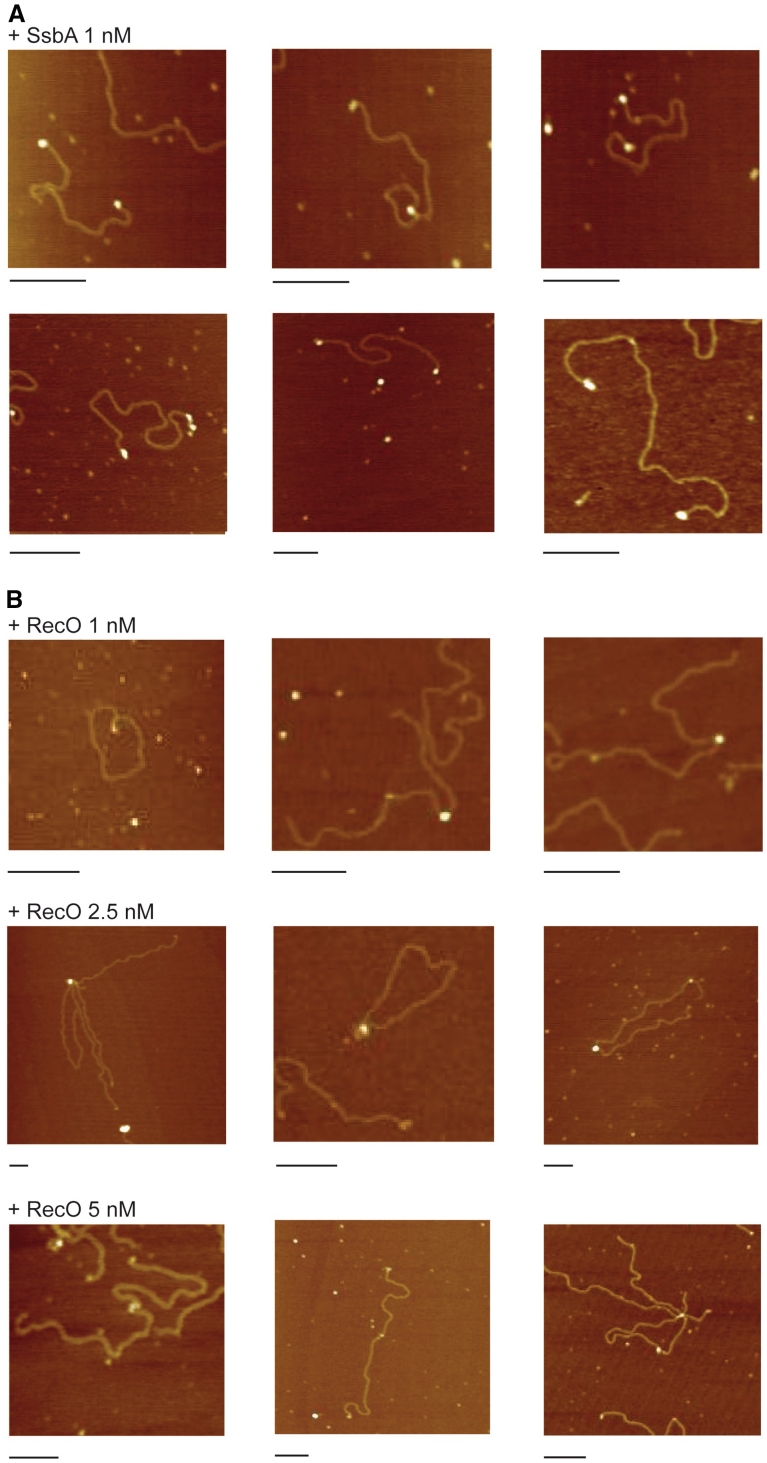
To gain an insight in the mechanism by which RecO mediates homology search and annealing, the recombination intermediates between two non-complementary 3′-tailed duplex DNA, either coated or not by SsbA, were monitored by AFM. The naked ssDNA behaved as a disordered coil that made difficult its measurement. The measure of the duplex region of the 5′-resected DNA was variable, with an average of 876 ± 50 nm and 1027 ± 5 nm for the untreated linear DNA (Figure S2). From these data we can deduce that the DNA molecules had ssDNA ends of ∼80 nm (∼125 nt per termini or ∼7.8% of ssDNA on the duplex molecule) (Figure S2).
SsbA, at a concentration of 1 SsbA tetramer/∼50 nt, formed discrete nucleoprotein complexes with 99% of the ssDNA region (n = 153, Figure 2A). The SsbA particles bound to ssDNA increased the average height of the nucleoprotein complex to 7.8 nm, compared to the naked bush-like ssDNA that was ∼4.3 nm (Figure S3, Table 1). Since the ratio used in these experiments was 1 SsbA/∼50 nt, it is likely that the ssDNA was totally coated considering its binding site size (65 nt) in the presence of Mg2+ (7).
Figure 2.
SsbA–ssDNA and RecO–ssDNA complexes. Selected images of SsbA–ssDNA complexes at 1 SsbA/∼50 nt (1 nM) (A) or RecO–ssDNA complexes 1 RecO/∼50-, ∼25 or ∼10 nt (1, 2.5 and 5 nM) (B) on 5′-resected duplex DNA. Scale bar = 200 nm.
Table 1.
Volume of SsbA–ssDNA, RecO–ssDNA and ssDNA–SsbA–RecO complexes in a 3′-tailed non-homologous substrate
| Condition | Free DNAa | High (nm)b | Volume (in nm3) and frequenciesc (in %) | ||
|---|---|---|---|---|---|
| SsbA 1 nM | <1 | 7.8 (4.3) | 236 ± 7.4 (99) | – | – |
| RecO 1 nM | ∼4 | 5.7 (3.4) | 102 ± 12 (54) | 317 ± 32 (41) | – |
| RecO 2.5 nM | ∼1 | 4.2 (1.3) | 112 ± 10 (78) | 323 ± 29 (21) | – |
| RecO 5 nM | ∼1 | 4.0 (1.7) | 111 ± 7.1 (75) | 319 ± 32 (24) | – |
| SsbA 1 and RecO 1 nM | <1 | 13.4 (7.2) | 128 ± 12 (33) | 363 ± 30 (66) | – |
| SsbA 1 and RecO 2.5 nM | ∼1 | 16.7 (8.6) | 235 ± 42 (61) | 319 ± 33 (11) | 537 ± 17 (27) |
| SsbA 1 and RecO 5 nM | ∼5 | 10.4 (5.6) | 103 ± 3.1 (74) | 224 ± 48 (25) | – |
aPercentage of protein-free ssDNA at given protein(s) concentrations.
bThe median high, in nm, is indicated, and the IQR are denoted between parenthesis.
cThe volume of the protein–ssDNA complexes are shown and their frequencies represented as percentages.
The observed volume of SsbA in solution was ∼132 nm3 (data not shown), and it is in good agreement with the theoretical volume of tetrameric SsbA (∼120 nm3). The volume of the observed SsbA–ssDNA complexes was ∼236 nm3 (Table 1). It is likely that the ∼125-nt average tailed-end was bound by one or more SsbA molecules. In the latter case, perhaps the broadening of the tip did not permit direct visualization of the ssDNA linker (Figure S3A). Since the expected length of the 3′-ssDNA is variable (see above) we cannot address if SsbA wraps around the ssDNA ends (28). SsbA bound to ssDNA ends failed to bridge two non-complementary DNA molecules by a direct protein–protein interaction.
RecO preferentially binds ssDNA
RecO binding to ssDNA produced large nucleoprotein aggregates that failed to enter the gel, but RecO did not seem to form discrete complexes with dsDNA (Figure 4, data not shown). Addition of a nucleotide cofactor had no effect on the binding (data not shown). To examine the type of complexes RecO formed with linear 5′-resected duplex DNA the reacting products were monitored by AFM. In the presence of 1 RecO/∼50 nt, RecO only formed discrete complexes at the ssDNA ends of the duplex molecule (∼96 %, n = 174) (Figure 2B, Table 1). However, at a ratio of 1 RecO/∼10 nt, RecO formed discrete complexes at ssDNA ends (∼99%), but could also bind duplex DNA regions, albeit at low frequency (7.1%, n = 177) (Figure 2B). We considered the possibility that RecO binding to dsDNA resulted from the enlargement of pre-existing nicks upon T7 exonuclease treatment. Arguing against this possibility was that SsbA binding to these ‘putative enlarged nicks’ was not observed.
Figure 4.
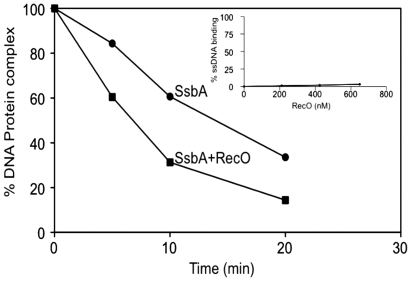
RecO destabilizes SsbA binding to ssDNA. The 60-nt [γ-32P]-poly[dT] DNA was pre-incubated with SsbA (4 nM, 1 SsbA/22 nt) for 15 min at 37°C. Twenty-fold excess of 60-nt cold poly[dT] DNA and RecO (500 nM, 1 RecO/3 nt) were added (time zero) and sampling begun. The reaction was stopped by ice-cold buffer at different times, and filtered through KOH-treated filters. Radioactivity retained on the filter was determined by scintillation counting.
RecO formed discrete globular shaped structures on ssDNA, larger than the expected for monomers (Figure 2B) and consistent with the observed formation of dimers (15, Table 1). In the presence of 1 RecO/∼50- or ∼25 nt, the height of RecO particles was similar (Figure S3, Table 1). Since the dispersion of the data is negligent at higher protein concentrations (1 RecO/∼50 nt), at a low RecO:ssDNA ratios (1 RecO/∼50 nt) the ssDNA was not saturated, and both naked ssDNA and RecO–ssDNA complex could have still co-existed.
The theoretical volume of dimeric RecO was ∼96 nm3. The volume of RecO–ssDNA complexes revealed discrete peaks that agreed with the theoretical volume of RecO dimers and possibly two or three interacting dimers bound to ssDNA (Table 1). RecO bound to ssDNA ends, bridged the two non-complementary ends of a duplex (intramolecular) or the ends of different molecules (intermolecular bridging), by direct protein–protein interaction (Figure 2B and Table 2). Intermolecular bridging is also observed with eukaryotic Rad52 (29). Unlike eukaryotic Rad52, which binds and wraps ssDNA (30,31), RecO bridged two ssDNA molecules.
Table 2.
SsbA increased RecO-mediated bridging of ssDNA ends in a 3′-tailed non-homologous substrate
| Condition | Bounda | End bridged DNA molecules (in %) |
||||
|---|---|---|---|---|---|---|
| 1b | 2c | 3c | >3c | Total bridged (n)d | ||
| SsbA 1 nM | >99 | ND | ND | ND | ND | ND (153) |
| RecO 1 nM | 91.8 | 1.4 | 2.2 | 0.4 | <1 | 4.2 (174) |
| RecO 2.5 nM | 90.8 | 5.3 | 3.1 | 0.7 | <1 | 9.2 (151) |
| RecO 5 nM | 79.3 | 1.1 | 13.5 | 1.7 | 4.4 | 20.7 (177) |
| SsbA 1 nM, RecO 1 nM | 72.2 | 5.2 | 10.4 | 2.6 | 9.6 | 27.8 (117) |
| SsbA 1 nM, RecO 2.5 nM | 48.6 | 2.2 | 30.4 | 13.0 | 5.8 | 51.4 (138) |
| SsbA 1 nM, RecO 5 nM | 54.5 | 5.8 | 16.2 | 13.5 | 10.0 | 45.5 (115) |
aPercentage of ssDNA molecules bound to given protein(s).
bPercentage of bridged ssDNA ends in the same ssDNA molecule (intramolecular bridging).
cPercentage of intermolecular bridging involving two, three or more ssDNA ends.
dTotal bridged molecules (in %), and in parenthesis total number of molecules analyzed. ND, not detected.
SsbA increases RecO-mediated bridging of heterologous ssDNA molecules
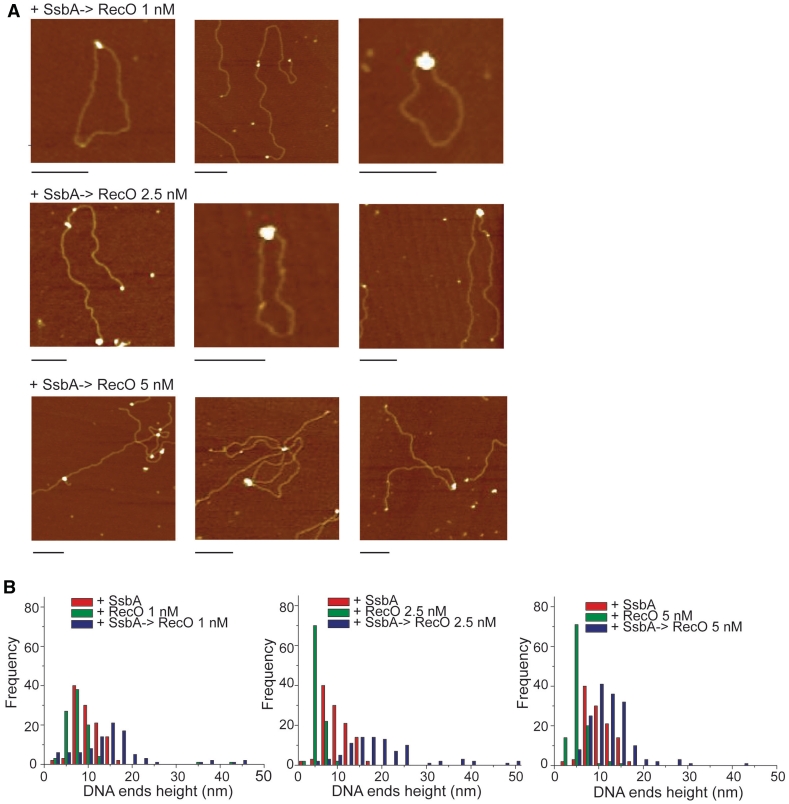
To understand the mechanistic role of SsbA in RecO-mediated strand annealing, the interaction of RecO with SsbA and ssDNA was analyzed. Non-complementary 5′-resected duplex DNA was pre-incubated with SsbA (1 SsbA/∼50 nt) and then incubated with increasing concentrations of RecO (Figure 3). In the presence of SsbA, the height of protein–ssDNA particles, at ratios of 1 RecO/∼50- and ∼20 nt, were 13.4 and 16.7 nm, respectively. The 3′-tailed end could accommodate both proteins, as revealed by the increased volume of the complexes (Table 1). However, in the presence of 1 RecO/∼10 nt the volume of the nucleoprotein complexes were significantly smaller (10.4 nm, Table 1), suggesting that the SsbA–ssDNA complex was disrupted and, hence, explaining the decrease in the rate of annealing when excess RecO was added.
Figure 3.
RecO bridges SsbA–ssDNA complexes. (A) Selected images of nucleoprotein complexes on 5′-resected duplex DNA. (B) Histogram showing the height of the naked ssDNA and the quaternary (ssDNA–SsbA–RecO–ssDNA) or quinary (ssDNA–SsbA–RecO–SsbA–ssDNA) complexes. Scale bar = 200 nm.
SsbA bound to tailed ends failed to promote end-bridging (Figure 2A). However, RecO interaction with itself promoted intermolecular bridging that occurred with ∼10-fold higher efficiency when SsbA was present. RecO interaction with SsbA-bound to ssDNA led to the accumulation of transient quaternary (ssDNA–SsbA–RecO–ssDNA) or quinary (ssDNA–SsbA–RecO–SsbA–ssDNA) complexes (Figure 3, Table 2). From these results we deduce that: (i) the interaction of SsbA with ssDNA removes secondary DNA structures and inhibits spontaneous annealing; (ii) RecO interactions with itself and with SsbA-bound to ssDNA facilitates the formation of bridging complexes and (iii) RecO might promote dislodging of SsbA from ssDNA.
RecO destabilizes SsbA–ssDNA complexes
To address whether RecO destabilizes SsbA from the SsbA–ssDNA complex, filter binding assays were performed. RecO only forms transient complexes with poly[dT]-ssDNA that cannot be trapped in a filter (Figure 4, insert), whereas SsbA binds with high affinity to this ssDNA. The time-dependent decrease of SsbA–ssDNA complexes (1 SsbA tetramer/22 nt), upon addition of 20-fold excess of cold ssDNA and RecO, was used to calculate the half-life of the SsbA–ssDNA complex. The half-life of the SsbA–ssDNA complex was 18.3 ± 1.7 min.
In the presence of RecO, the half-life decreased ∼2-fold to 8.4 ± 0.6 min (Figure 4).
The half-life of the SsbSPP1·poly[dT]–ssDNA complex was short living (≤1 min) (data not shown). Since RecO did not interact with the non-cognate ssDNA binding protein, SsbSPP1 (15), and (ii) SsbSPP1-promoted DNA secondary structure removal did not facilitate RecO-mediated strand annealing, it is likely that RecO plays an active role in destabilizing the SsbA–ssDNA complexes rather than competing for ssDNA binding.
RecO performs homology search and strand annealing
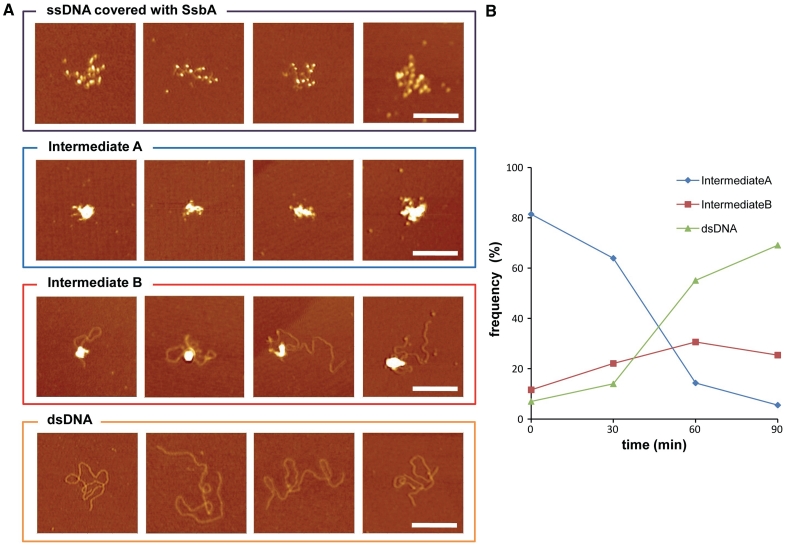
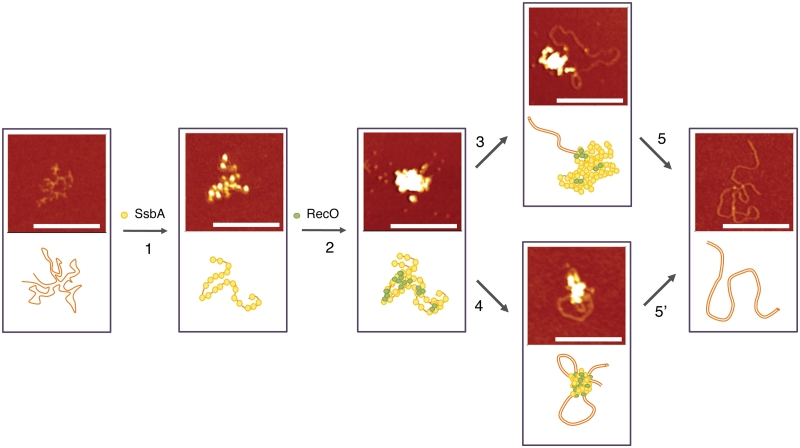
RecO-mediated strand annealing between two long homologous DNA strands was monitored by AFM. SsbA-bound to 2958-nt long ssDNA removed secondary structures, but failed to bridge complementary strands (Figure 5). SsbA-bound ssDNA markedly increased the frequency of bead-shaped RecO (1 RecO/∼20 nt) interactions with SsbA (1 SsbA/∼50 nt) and with itself, leading to the accumulation of transient quaternary or quinary intermediates bridged structures (Figures 5A and 6, steps 1 and 2). Optimal annealing occurred in the presence of both proteins. At type A intermediate (bridged structures) RecO facilitated SsbA partial dislodging from ssDNA and bridging of two ‘naked’ homologous segment. Consequent RecO facilitation of transient random interactions until a complementary region is found, results in the propagation of strand annealing of complementary strands (Figures 5B and 6, steps 3 and 4), with SsbA removing secondary structures. Indeed, RecO promoted interaction between homologous segments facilitates local Watson–Crick-type base pairing, with the conversion of bridged structures (intermediate A) into bridged structures with paired segments (intermediate B) and type B intermediates into duplex DNA (Figures 5 and 6, steps 3 and 5′). This stabilization can result in RecO-mediated propagation of annealing of complementary strands. Once the DNA becomes base-paired, RecO and SsbA are released from the duplex, consistent with the lower affinity of RecO for dsDNA and with the incapability of SsbA to bind dsDNA (Figures 5A and 6, steps 5 and 5′).
Figure 5.
RecO bridges SsbA–ssDNA and promotes strand annealing. (A) Heat-denatured 2958-nt long ssDNA was pre-incubated with SsbA (10 nM, 1 SsbA/50 nt) in buffer C. RecO (25 nM, 1 RecO/20 nt) was added and the reaction incubated for a variable time. Scale bar = 200 nm. (B) Time course of the reaction. Samples were taken at 30 (n = 43), 60 (n = 86), 90 (n = 49) and 120 min (n = 55) and the proportion of type A and B intermediates and final product of the recombination reaction were quantified.
Figure 6.
RecO mediates strand annealing of SsbA-coated ssDNAs. ssDNA was pre-incubated with SsbA (10 nM, 1 SsbA/50 nt, step 1) in buffer C. RecO (25 nM, 1 RecO/20 nt, step 2) was added and the reaction incubated for a viable time (steps 3 to 5′). In steps 3 and 4, a partial RecO-mediated dislodging of SsbA leads to RecO promoted homology search and strand annealing. In steps 5 and 5′, RecO-mediated strand annealing leads to the naked dsDNA product. The samples were taken at different times. Schematic representation of the images is shown below. Scale bar = 200 nm.
DISCUSSION
The RecO protein plays a central role in various HR processes. In this paper, we show that SsbA or SsbSPP1 bound to ssDNA inhibits the spontaneous renaturation of complementary DNA strands. RecO-mediated annealing increased ∼80-fold when the ssDNA was coated with SsbA, irrespective of the Mg2+ presence (Figures 1 and S1). SsbA facilitates RecO-mediated strand annealing through the accumulation of non-productive ternary complexes, by protein–protein and protein–ssDNA interactions. The short-living SsbSPP1–ssDNA complex, however, inhibited RecO-mediated annealing of complementary ssDNA strands (Figures 1 and S1). Indeed, deletion of the C-terminal ‘acidic tail’ of SSBEco or SsbA suppresses the interaction with RecOEco or RecO, respectively, and blocks the action of the RecO modulator (7,32; data not shown).
It has been widely reported that a subset of ‘recombinase mediators’ (namely bacterial RecO/RecOEco and eukaryotic Rad52) promote the annealing of complementary strands coated with a species-specific single-stranded binding protein, which by themselves inhibit spontaneous annealing, in the absence of any energy cofactor (11,13,19,21,33–35, this study). However, there are marked differences in the way this is achieved: (i) RecO (this study) or Rad52 (33) binds preferentially ssDNA, independently of the presence of Mg2+ ions, with at least 20-fold higher affinity and/or stability than to dsDNA(13); RecOEco, on the other hand, binds ssDNA and dsDNA with similar efficiency, but only in the presence of Mg2+ (13); (ii) RecOEco-mediated strand annealing strictly requires Mg2+ ions (13); and (iii) the rate and extent of RecOEco- or Rad52-mediated DNA strand annealing is significantly reduced in the presence of their cognate ssDNA-binding proteins, SSBEco or RPA, respectively (11,30,31).
To gain an insight into the mechanistic role of RecO-mediated strand annealing we have used AFM to visualize the accumulation of non-productive (Figure 3) or productive (Figure 5) recombination intermediates. RecO forms discrete globular shaped structures, larger than the monomer volume, on ssDNA. This is consistent with the observation that RecO forms dimers in protein cross-linking experiments (15), and the putative involvement of RecODra C-terminal end and the OB-fold in the protein self-interaction, despite it crystallizes as a monomer (21).
Unlike RecO (Figure 3), Rad52 forms homo-oligomers of seven or more subunits with the ssDNA wrapping around (35,36). Furthermore, the single-strand annealing proteins (SSAP) as RecTEco or the recombinase RecAEco, in the ATPγS bound form, are organized as ring-shaped structures and can assemble as helical filaments when they are in presence of ssDNA (37,38). Pairing proteins of the SSAP and the RecA families promote unidirectional polymerization on ssDNA and strand annealing, in the presence of a divalent metal ion or a divalent metal ion and a nucleotide cofactor, respectively. Both RecAEco·ATP·Mg2+ and RecTEco·Mg2+, when complexed with ssDNA, accumulate large networks of co-aggregates where homology search occurs (38–41). Co-aggregation was shown to facilitate the establishment of homologous contacts and the determining factor for recognition of homology was proposed to be the unstacking of bases that allow base flipping and switching at A:T-rich regions (38–42). A plausible explanation on how RecO-mediated DNA strand annealing occurs would be: (i) SsbA bound to ssDNA recruits RecO to form a ternary complex (ssDNA–SsbA–RecO), but such protein–protein interaction per se might not promote SsbA dislodging from ssDNA; (ii) RecO interaction with SsbA–ssDNA and with itself leads to the formation of bridged structures, with RecO decreasing the half-life of the SsbA–ssDNA complexes; and (iii) RecO once bound to naked ssDNA distorts the structure of ssDNA and prevents SsbA binding or relief it from ssDNA (42). Alternatively, RecO by interaction with SsbA facilitates the spontaneous SsbA sliding along ssDNA (43), favoring its displacement from ssDNA. The annealing process is based on transient and random interactions between naked ssDNA segments facilitated by RecO and inhibited by SsbA, with RecO facilitating homology search. Indeed, the ssDNA bound to RecOTth has structural properties similar to that of ssDNA bound to RecAEco·ATP·Mg2+ or RecTEco·Mg2+, suggesting that disruption of base stacking and base flipping are conserved mechanisms (42,43). RecO-mediated annealing of complementary strands is produced in the absence of metal ions or large networks of co-aggregates (Figures 5 and 6). It is likely that these functionally unrelated proteins mediate DNA strand annealing by a different modus operandi. In addition, homology search occurs in the presence of both proteins (Figure 6, steps 3 and 4) with SsbA facilitating RecO-mediated bridged structures. On the contrary, SSBEco limits RecAEco·ATP·Mg2+ or RecTEco·Mg2+ filamentation onto the DNA, hence preventing homology search (7).
These and previous results suggest that RecO has three activities coordinated by SsbA: (i) it recruits RecA onto SsbA-coated ssDNA (15,23); (ii) it modulates the extent of RecA-mediated DNA strand exchange (15); and (iii) it bridges SsbA-coated ssDNA molecules, which, when complementary, promote annealing (this study). The first two activities work at the pre- and synaptic stages respectively and are essential for RecA-dependent DSB repair during HR (15,23). On the other hand, RecO-mediated strand annealing is critical for RecA filament extension and strand exchange during RecA-mediated recombination (postsynaptic stage) (15,23), in a similar manner as reported for eukaryotic Rad52 (44,45). RecO is also important in RecA-independent genetic recombination. We have previously reported the involvement of DNA recombination and RecO during natural transformation (19,20). We propose that RecO-mediated annealing facilitation of plasmid establishment in natural competent cells is key to successful transformation and, hence, it indirectly contributes to the lateral acquisition of plasmid-borne multi-drug- resistance genes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Ministerio de Ciencia e Innovación (MICINN) (grants BFU2009-07167 and CSD2007-00010); Comunidad de Madrid (grant S-2009MAT-1507 to J.C.A.); Grant-in-aid for Priority Area from the MEXT of Japan (to K.T.) and JSPS (to Y.S.). Funding for open access charge: Spanish Ministry of Science and Innovation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Ayora and C. E. César for helpful comments on the manuscript. Y.S. is a Research Fellow of the Japan Society for the Promotion of Science (JSPS). T.Y. is PhD fellow of the International Fellowship Programme of La Caixa Foundation (La Caixa/CNB).
REFERENCES
- 1.Cox MM. Motoring along with the bacterial RecA protein. Nat. Rev. Mol. Cell. Biol. 2007;8:127–138. doi: 10.1038/nrm2099. [DOI] [PubMed] [Google Scholar]
- 2.Galletto R, Kowalczykowski SC. RecA. Curr. Biol. 2007;17:R395–R397. doi: 10.1016/j.cub.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 3.McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 4.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 5.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell. Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 6.Beernink HT, Morrical SW. RMPs: recombination/replication mediator proteins. Trends Biochem. Sci. 1999;24:385–389. doi: 10.1016/s0968-0004(99)01451-6. [DOI] [PubMed] [Google Scholar]
- 7.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Umezu K, Chi NW, Kolodner RD. Biochemical interaction of the Escherichia coli RecF, RecO, and RecR proteins with RecA protein and single-stranded DNA binding protein. Proc. Natl Acad. Sci. USA. 1993;90:3875–3879. doi: 10.1073/pnas.90.9.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shan Q, Bork JM, Webb BL, Inman RB, Cox MM. RecA protein filaments: end-dependent dissociation from ssDNA and stabilization by RecO and RecR proteins. J. Mol. Biol. 1997;265:519–540. doi: 10.1006/jmbi.1996.0748. [DOI] [PubMed] [Google Scholar]
- 10.Hobbs MD, Sakai A, Cox MM. SSB protein limits RecOR binding onto single-stranded DNA. J. Biol. Chem. 2007;282:11058–11067. doi: 10.1074/jbc.M611007200. [DOI] [PubMed] [Google Scholar]
- 11.Kantake N, Madiraju MV, Sugiyama T, Kowalczykowski SC. Escherichia coli RecO protein anneals ssDNA complexed with its cognate ssDNA-binding protein: A common step in genetic recombination. Proc. Natl Acad. Sci. USA. 2002;99:15327–15332. doi: 10.1073/pnas.252633399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luisi-DeLuca C. Homologous pairing of single-stranded DNA and superhelical double-stranded DNA catalyzed by RecO protein from Escherichia coli. J. Bacteriol. 1995;177:566–572. doi: 10.1128/jb.177.3.566-572.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luisi-DeLuca C, Kolodner R. Purification and characterization of the Escherichia coli RecO protein. Renaturation of complementary single-stranded DNA molecules catalyzed by the RecO protein. J. Mol. Biol. 1994;236:124–138. doi: 10.1006/jmbi.1994.1123. [DOI] [PubMed] [Google Scholar]
- 14.New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 15.Manfredi C, Carrasco B, Ayora S, Alonso JC. Bacillus subtilis RecO nucleates RecA onto SsbA-coated single-stranded DNA. J. Biol. Chem. 2008;283:24837–24847. doi: 10.1074/jbc.M802002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 17.Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 18.Kidane D, Sanchez H, Alonso JC, Graumann PL. Visualization of DNA double-strand break repair in live bacteria reveals dynamic recruitment of Bacillus subtilis RecF, RecO and RecN proteins to distinct sites on the nucleoids. Mol. Microbiol. 2004;52:1627–1639. doi: 10.1111/j.1365-2958.2004.04102.x. [DOI] [PubMed] [Google Scholar]
- 19.Kidane D, Carrasco B, Manfredi C, Rothmaier K, Ayora S, Tadesse S, Alonso JC, Graumann PL. Evidence for different pathways during horizontal gene transfer in competent Bacillus subtilis cells. PLoS Genet. 2009;5:e1000630. doi: 10.1371/journal.pgen.1000630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez S, Kobayashi Y, Ogasawara N, Alonso JC. Analysis of the Bacillus subtilis recO gene: RecO forms part of the RecFLOR function. Mol. Gen. Genet. 1999;261:567–573. doi: 10.1007/s004380051002. [DOI] [PubMed] [Google Scholar]
- 21.Makharashvili N, Koroleva O, Bera S, Grandgenett DP, Korolev S. A novel structure of DNA repair protein RecO from Deinococcus radiodurans. Structure. 2004;12:1881–1889. doi: 10.1016/j.str.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 22.Leiros I, Timmins J, Hall DR, McSweeney S. Crystal structure and DNA-binding analysis of RecO from Deinococcus radiodurans. EMBO J. 2005;24:906–918. doi: 10.1038/sj.emboj.7600582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carrasco B, Manfredi C, Ayora S, Alonso JC. Bacillus subtilis SsbA and dATP regulate RecA nucleation onto single-stranded DNA. DNA Repair (Amst) 2008;7:990–996. doi: 10.1016/j.dnarep.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 24.Carrasco B, Ayora S, Lurz R, Alonso JC. Bacillus subtilis RecU Holliday-junction resolvase modulates RecA activities. Nucleic Acids Res. 2005;33:3942–3952. doi: 10.1093/nar/gki713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pratto F, Suzuki Y, Takeyasu K, Alonso JC. Single-molecule analysis of protein·DNA complexes formed during partition of newly replicated plasmid molecules in Streptococcus pyogenes. J. Biol. Chem. 2009;284:30298–30306. doi: 10.1074/jbc.M109.035410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alonso JC, Stiege AC, Dobrinski B, Lurz R. Purification and properties of the RecR protein from Bacillus subtilis 168. J. Biol. Chem. 1993;268:1424–1429. [PubMed] [Google Scholar]
- 27.Barrett SD. Image SXM v 1.90. 2010 http://www.liv.ac.uk/∼sdb/Research/ (April 2010, date last accessed) [Google Scholar]
- 28.Hamon L, Pastre D, Dupaigne P, Le Breton C, Le Cam E, Pietrement O. High-resolution AFM imaging of single-stranded DNA-binding (SSB) protein–DNA complexes. Nucleic Acids Res. 2007;35:e58. doi: 10.1093/nar/gkm147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ristic D, Modesti M, Kanaar R, Wyman C. Rad52 and Ku bind to different DNA structures produced early in double-strand break repair. Nucleic Acids Res. 2003;31:5229–5237. doi: 10.1093/nar/gkg729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grimme JM, Honda M, Wright R, Okuno Y, Rothenberg E, Mazin AV, Ha T, Spies M. Human Rad52 binds and wraps single-stranded DNA and mediates annealing via two hRad52-ssDNA complexes. Nucleic Acids Res. 2010;38:2917–2930. doi: 10.1093/nar/gkp1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singleton MR, Wentzell LM, Liu Y, West SC, Wigley DB. Structure of the single-strand annealing domain of human RAD52 protein. Proc. Natl Acad. Sci. USA. 2002;99:13492–13497. doi: 10.1073/pnas.212449899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lecointe F, Serena C, Velten M, Costes A, McGovern S, Meile JC, Errington J, Ehrlich SD, Noirot P, Polard P. Anticipating chromosomal replication fork arrest: SSB targets repair DNA helicases to active forks. EMBO J. 2007;26:4239–4251. doi: 10.1038/sj.emboj.7601848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl Acad. Sci. USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugiyama T, New JH, Kowalczykowski SC. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl Acad. Sci. USA. 1998;95:6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shinohara A, Shinohara M, Ohta T, Matsuda S, Ogawa T. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells. 1998;3:145–156. doi: 10.1046/j.1365-2443.1998.00176.x. [DOI] [PubMed] [Google Scholar]
- 36.Stasiak AZ, Larquet E, Stasiak A, Muller S, Engel A, Van Dyck E, West SC, Egelman EH. The human Rad52 protein exists as a heptameric ring. Curr. Biol. 2000;10:337–340. doi: 10.1016/s0960-9822(00)00385-7. [DOI] [PubMed] [Google Scholar]
- 37.Yu X, VanLoock MS, Yang S, Reese JT, Egelman EH. What is the structure of the RecA-DNA filament? Curr. Protein Pept. Sci. 2004;5:73–79. doi: 10.2174/1389203043486883. [DOI] [PubMed] [Google Scholar]
- 38.Erler A, Wegmann S, Elie-Caille C, Bradshaw CR, Maresca M, Seidel R, Habermann B, Muller DJ, Stewart AF. Conformational adaptability of Redbeta during DNA annealing and implications for its structural relationship with Rad52. J. Mol. Biol. 2009;391:586–598. doi: 10.1016/j.jmb.2009.06.030. [DOI] [PubMed] [Google Scholar]
- 39.Folta-Stogniew E, O'Malley S, Gupta R, Anderson KS, Radding CM. Exchange of DNA base pairs that coincides with recognition of homology promoted by E. coli RecA protein. Mol. Cell. 2004;15:965–975. doi: 10.1016/j.molcel.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 40.Noirot P, Gupta RC, Radding CM, Kolodner RD. Hallmarks of homology recognition by RecA-like recombinases are exhibited by the unrelated Escherichia coli RecT protein. EMBO J. 2003;22:324–334. doi: 10.1093/emboj/cdg027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–484. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 42.Masuda T, Ito Y, Terada T, Shibata T, Mikawa T. A non-canonical DNA structure enables homologous recombination in various genetic systems. J. Biol. Chem. 2009;284:30230–30239. doi: 10.1074/jbc.M109.043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roy R, Kozlov AG, Lohman TM, Ha T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature. 2009;461:1092–1097. doi: 10.1038/nature08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nimonkar AV, Sica RA, Kowalczykowski SC. Rad52 promotes second-end DNA capture in double-stranded break repair to form complement-stabilized joint molecules. Proc. Natl Acad. Sci. USA. 2009;106:3077–3082. doi: 10.1073/pnas.0813247106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugiyama T, Kantake N, Wu Y, Kowalczykowski SC. Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture. EMBO J. 2006;25:5539–5548. doi: 10.1038/sj.emboj.7601412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.