Abstract
In Qβ RNA, sequestering the coat gene ribosome binding site in a putatively strong hairpin stem structure eliminated synthesis of coat protein and activated protein synthesis from the much weaker maturation gene initiation site, located 1300 nucleotides upstream. As the stability of a hairpin stem comprising the coat gene Shine–Dalgarno site was incrementally increased, there was a corresponding increase in translation of maturation protein. The effect of the downstream coat gene ribosome binding sequence on maturation gene expression appeared to have occurred only in cis and did not require an AUG start codon or initiation of coat protein synthesis. In all cases, no structural reorganization was predicted to occur within Qβ RNA. Our results suggest that protein synthesis from a relatively weak translational initiation site is greatly influenced by the presence or absence of a stronger ribosome binding site located elsewhere on the same RNA molecule. The data are consistent with a mechanism in which multiple ribosome binding sites compete in cis for translational initiations as a means of regulating protein synthesis on a polycistronic messenger RNA.
INTRODUCTION
Initiation of protein synthesis in prokaryotes has been extensively studied (1). Translation begins with the association between a 30S ribosomal subunit and the messenger RNA. In most cases, this interaction is dependent upon the hybridization between specific sequences in the 16S ribosomal RNA and the complementary Shine–Dalgarno region located upstream of the initiator codon on the RNA message (2). The initial interaction forms a reversible binary complex that can either dissociate into mRNA and 30S ribosome, or proceed into a nearly irreversible ternary complex that can initiate protein synthesis (3,4).
During the initiation phase of translation, several events occur that significantly contribute to the regulation of protein synthesis. For example, initiation factors drive the specificity of initiation, determine what kind of initiation codon is selected, and help to stabilize ribosome:RNA interactions (1). The extent of complementarity between the Shine–Dalgarno sequence of a cistron and the complementary region on the 16S ribosomal RNA has great impact on the efficiency of ribosome binding (5). Translation of a prokaryotic gene is sometimes dependent upon either the activity of a trans-acting protein factor, or the presence of trans-acting or antisense RNA that can bind the messenger RNA and inhibit translational initiation of another gene (6–9). On many polycistronic mRNAs, translation of one gene is dependent upon the coupled translation of an upstream gene sequence (1,10,11). The presence of secondary RNA structure at the initiation region of a gene can severely impede 30S ribosome association with the RNA (12,13). In the absence of coupled upstream translation, stable base pairing in a hairpin structure can often occlude ribosome binding.
Previously, we demonstrated a cis-acting mechanism in which one relatively strong translational initiation site on a polycistronic mRNA could modulate protein synthesis from a second gene present on the same molecule (8). In Qβ RNA, the presence of the translational initiation region for the coat gene inhibited translation of the maturation cistron, located nearly 1300-nt upstream. In contrast, when the coat initiation region was deleted, or masked by the presence of a trans-acting Qβ replicase protein, maturation protein was synthesized in significant quantities.
The effect of the coat gene initiation region on maturation gene expression was observed only in cis, but not when the two genes were on different mRNA molecules. It was further shown that the same coat gene initiation site also affected expression of the downstream replicase cistron in the absence of coupled coat gene translation. In all cases, there were no predicted alterations in the putative secondary structure of the Qβ RNA molecule. The results were consistent with a mechanism in which two ribosome binding sites present on the same RNA molecule could compete for translational initiations.
In this article, we demonstrate that if the Qβ coat gene initiation site is sequestered in hairpin structures of increasing stabilities, there is a concomitant increase in the levels of maturation protein synthesized from Qβ RNA transcripts. These data further support our previously proposed mechanism in which two ribosome binding sites on Qβ RNA compete in cis for translational initiations, and further imply that the extent of such competition is determined by the relative ribosome binding affinities between the two sites. Such a mechanism would necessarily have profound effects upon protein synthesis from polycistronic mRNAs.
MATERIALS AND METHODS
Bacterial strains
Escherichia coli MC1061 (14) was used for growth and maintenance of plasmids. Escherichia coli BL21(DE3) (15) carry the bacteriophage T7 RNA polymerase gene under the control of the E. coli lac operator. These cells were first transformed with the plasmid placIq (below), which overproduces lac repressor protein, then used for transformation and expression of inducible plasmid-generated proteins.
Materials
Restriction endonucleases, oligonucleotide linkers, T4 DNA polymerase and T4 DNA ligase were purchased from New England Biolabs, Inc., Beverly, MA, USA. Isopropyl β-d-thiogalacto-pyranoside (IPTG) was purchased from Sigma Chemical Co., St Louis, MO, USA. Oligonucleotide primers were obtained from Gene Link, Inc., Thornwood, NY, USA.
General procedures
The methods employed for plasmid constructions have been described (14). Plasmid DNAs were prepared using the Qiagen plasmid isolation kit; RNA was isolated using the Qiagen RNeasy kit; transcribed Qβ RNAs were subjected to the reverse transcriptase polymerase chain reaction using the Qiagen OneStep RT–PCR kit (Qiagen Inc., Chatsworth, CA, USA). The orientation and nucleotide sequence that resulted from plasmid constructions were confirmed by restriction enzyme and/or DNA sequence analysis. Transformed bacterial cultures were selected by growth in N-broth (16) supplemented with 50 µg/ml of the appropriate antibiotic. Procedures for electrotransformation have been described (17).
Plasmid constructions
The plasmid pBHQβ525, used as a mutagenesis target plasmid, was constructed by replacing a unique SalI/EcoRV fragment (nucleotides 5030–1471 of pBH95) with a 3.7 kb Qβ cDNA fragment (nucleotides 525–4227) obtained from the plasmid pQβm101 (18) following digestion with restriction endonucleases XhoI and HpaI.
To construct the plasmid placIq, a 2689 bp SphI/XmnI endonuclease restriction fragment containing the E. coli lac Iq gene was excised from the plasmid pET11c (New England Biolabs, Inc.). This DNA fragment was ligated into a 3 kDa SphI/FspI fragment from the plasmid pRep101 (19), replacing the Qβ replicase gene segment.
The plasmid pT7Qβ500 comprises the following sequences: nucleotides 1–4217 are the entire Qβ positive strand cDNA sequence; nucleotides 4218–4328 comprise a 100-bp poly-AT sequence followed by a PstI oligonucleotide linker; nucleotides 4329–4736 are a 408-bp PstI/PvuII fragment obtained from the plasmid pDL44 (20), containing a bacteriophage T7 transcription termination sequence; nucleotides 4737–7753 are the complement of a 3017-bp NheI/SspI fragment from pBR322 containing the amp gene and a modified ColE1 origin of DNA replication (19), but not including the ROP region (nucleotides 1283–2064 of pBR322); nucleotides 7754–8030 are a 277 bp NheI/PpuMI fragment from pT7MDV (21) containing the bacteriophage T7 RNA polymerase promoter directed into the first G residue of Qβ (+) cDNA. Plasmids that were used to generate variant Qβ RNA transcripts are derivatives of pT7Qβ500 that contain sequence variations indicated in Table 1.
Table 1.
Qβ genome mutations
| Mutation | Description | Phenotype |
|---|---|---|
| UAA1204 | out-of-frame stop at Qβ1204 | 41.9 kDa defective maturation protein (mat41.9) |
| UAA1245 | out-of-frame stop at Qβ1245 | 43.9 kDa defective maturation protein (mat43.9) |
| Δ1178/1365 | deletion of Qβ nt 1178–1365 | 41.9 kDa defective maturation protein, no coat SD |
| UAA1395 | stop codon at Qβ1395 | terminates 41.9 kDa Δ1178/1365 mutant |
| UAG3397 | stop codon at Qβ3397 | 38.4 kDa inactive replicase protein |
| heSD | −10.3 kcal/mol coat SD hairpin | inactive coat SD |
| heSD2a | −8.4 kcal/mol coat SD hairpin | inactive coat SD; no coat AUG |
| leSD | −2.7 kcal/mol coat SD hairpin | active coat SD; no coat AUG |
| ctSD[-3.9] | −3.9 kcal/mol coat SD hairpin | intermediate coat SD; no coat AUG |
| ctSD[-4.9] | −4.9 kcal/mol coat SD hairpin | intermediate coat SD; no coat AUG |
| ctSD[-5.4] | −5.4 kcal/mol coat SD hairpin | intermediate coat SD; no coat AUG |
| ctSD[-6.5] | −6.5 kcal/mol coat SD hairpin | intermediate coat SD; no coat AUG |
| ctSD[-7.2] | −7.2 kcal/mol coat SD hairpin | inactive coat SD; no coat AUG |
| ctSD[-8.4]a | −8.4 kcal/mol coat SD hairpin | inactive coat SD; no coat AUG |
| ctSD[-10.1] | −10.1 kcal/mol coat SD hairpin | inactive coat SD; no coat AUG |
actSD[-8.4] contains the same nucleotide mutations as heSD2.
SD, Shine–Dalgarno; Δ, deletion.
The p2xQβ(+) plasmid system was previously described (8,22). This plasmid contains two copies of the Qβ cDNA genome, each with its own upstream T7 promoter/lac operator and downstream 5S T1T2 processing region. Having two genomes transcribed from the same plasmid ensures that both putative RNA genomes are synthesized in equal quantities within the host (18). For these experiments, each cDNA genome in the p2xQβ(+) plasmid was modified by site-directed mutagenesis to contain either an inactive Qβ coat gene Shine–Dalgarno site or an active Shine–Dalgarno site that is similar to the wild-type site. Upon induction with IPTG, this plasmid generates two intact Qβ RNA genomes simultaneously, each encoding a different set of mutations to distinguish the encoded proteins. To select against intra-plasmid recombination, the two cDNA genomes are separated by a chloramphenicol resistance gene on one side and an ampicillin resistance gene on the other.
The plasmid pT7QβMat(+) is a variation of pT7Qβ(+)500 with nucleotides 1406–4217 of the Qβ cDNA sequence deleted (23,24).
Site-directed mutagenesis
Mutations were incorporated into the Qβ cDNA sequence as described (25), using the target plasmid pBH95 (gift of Dr W.T. McAllister, University of Medicine and Dentistry of New Jersey). The target mutagenic primer was used to create point mutations in the cDNA sequence and was designed in such a way that it would anneal to the same strand as the selection primer. 25 pmol of the selection primer (25) and 25 pmol of the target primer were mixed with 200 ng of the template DNA and 2 μl of One-Phor-All buffer [100 mM Tris–acetate (pH 7.5), 500 mM potassium acetate (pH 7.5) and 100 mM Magnesium acetate] in a 20 μl reaction volume. The reaction mixture was incubated at 100°C for 5 min, chilled immediately on ice for another 5 min, followed by a 30 min incubation at room temperature. To the reaction mixture, 7 μl of nucleotide mix (2.86 mM dATP, 2.86 mM dCTP, 2.86 mM dGTP, 2.86 mM dTTP, 4.34 mM rATP, 1.43 × One-Phor-All buffer) and 3 μL of enzyme mix [FPLC pure T4 DNA polymerase (0.83–1.67 ku/ml], FPLC pure T4 DNA ligase (0.83–1.17 ku/ml) and T4 gene 32 protein (0.2–0.28 mg/ml) in aqueous buffer] were added. This mixture was incubated at 37°C for 1 h. The reaction was terminated by heating at 85°C for 15 min. The tubes were briefly chilled on ice. Electrophoresis was carried out using 2 μl of the reaction mixture and 40 μl of electrocompetent E. coli NMH22-mutS cells. The electrotransformants were selected on ampicillin (100 μg/ml) and kanamycin (100 μg/ml) plates.
Protein analysis
Escherichia coli BL21(DE3)/lacIq cells were electrotransformed, and grown in culture (16,17). Procedures for IPTG induction of protein synthesis, 14C-labeling of protein products, polyacrylamide gel electrophoresis (PAGE) and phosphorimage analysis have been described (8,26).
RESULTS
Generating Qβ RNA transcripts
Qβ RNA transcripts studied in these experiments were generated from three different plasmid sets. The first set comprises variations of the Qβ coliphage producing plasmid pT7Qβ(+)500 (Figure 1). This plasmid encodes the Qβ plus strand cDNA sequence under the control of a bacteriophage T7 promoter. Upon induction of Escherichia coli BL21(DE3) transformants with IPTG, stable full-length RNA transcripts are generated that contain the entire 4217 nucleotide Qβ plus strand sequence, followed by 100 adenosine residues and 3′ nucleotides from a bacteriophage T7 transcription terminator (20). The p2xQβ(+) plasmid system (below), containing two copies of the Qβ cDNA genome, has previously been described (8,22). In the presence of IPTG, p2xQβ(+) simultaneously generates equal quantities of two Qβ RNA transcripts, each comprising a different set of marker mutations to distinguish both the cDNA genomes and their encoded proteins. Finally, pT7Qβ(+)Mat (below) is a modified version of pT7Qβ(+)500 from which Qβ nucleotides 1406 through 4217 had been deleted. Transcription from this plasmid generates intact maturation gene mRNA and coat gene initiation sequences. For each of these plasmid sets, all Qβ cDNA genomes harbor a combination of specific mutations that result in a maturation protein defective for lysis function, and a replicase protein defective for both replication function and for the ability to serve as a trans-acting repressor of coat protein synthesis. In addition, all Qβ cDNA genomes contain either the wild-type or a mutated variation of the Qβ coat gene translational initiation region. Table 1 summarizes the specific Qβ cDNA mutations utilized in these experiments .
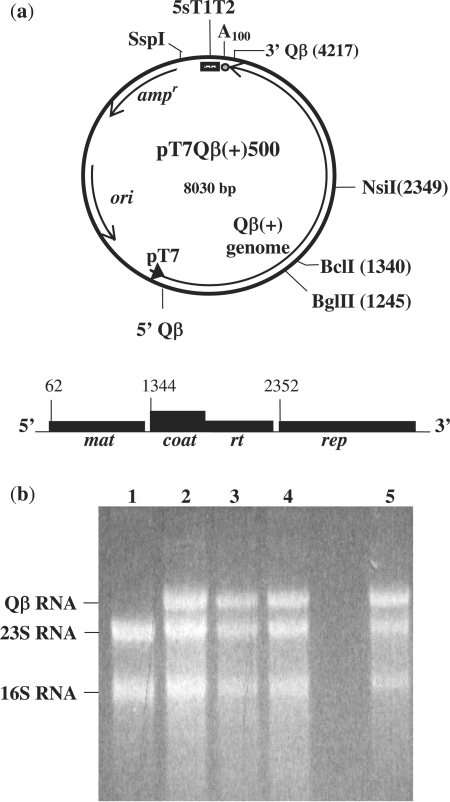
Figure 1.
(a) The pT7 plasmid system. In the plasmid pT7Qβ(+)500, a bacteriophage T7 promoter (dark arrow) is directed into 5′ Qβ(+) cDNA. Induction of E. coli BL21(DE3) host cells with IPTG results in RNA transcripts containing the entire 4217 nt Qβ(+) strand sequence, and a polyadenosine sequence (A100) 3′ to the last Qβ nucleotide. Transcripts terminate within a 3′ bacteriophage T7 5S T1T2 terminator region. The map of Qβ RNA shows the nucleotide positions of the translational initiation sites for the maturation (mat), coat/readthrough (coat/rt) and replicase (rep) cistrons. (b) Agarose gel analysis showing total RNA isolated from host cells in which pT7Qβ(+)500 plasmid derivatives were used to generate variant Qβ RNA transcripts: lane 1, uninduced; lane 2, T-502 RNA; lane 3, T-504 RNA; lane 4, T-503 RNA; and lane 5, T-500 RNA. For each, the Qβ RNA product represented 25–30% of the total RNA isolated relative to 16S and 23S RNAs. In each case, Qβ RNA transcripts were isolated and confirmed by RT–PCR.
Sequestering the Qβ coat Shine–Dalgarno site activates the maturation gene
The initiation region of the Qβ coat gene contains a powerful ribosome binding site. Unlike the remainder of the Qβ RNA genome, the sequence in the vicinity of the coat Shine–Dalgarno region is poorly structured, allowing unlimited access to ribosomes (12,27,28). Following synthesis of RNA transcripts that carry a wild-type coat gene sequence, large quantities of coat protein are generated, whereas no other phage proteins can be detected (8). Previously, we have shown that if the coat gene initiation region is deleted from the genome, or if Qβ replicase protein is provided in trans to repress coat gene translation, significant levels of maturation protein are synthesized (8). We now demonstrate that if the coat Shine–Dalgarno region alone is sequestered within a relatively strong hairpin structure on intact Qβ RNA, synthesis of detectable coat protein is abolished, while significant amounts of maturation protein are generated. To determine the putative secondary structure surrounding the Qβ coat Shine–Dalgarno site, we utilized the RNA folding program MFOLD, version 3.0 (29). Mutations were then site-directed into the Qβ cDNA sequence of pT7Qβ(+)500 that were predicted to alter the secondary structure at the coat Shine–Dalgarno region, but not anywhere else in the genome.
Figure 2 illustrates the coat protein initiation regions for several variant Qβ RNA transcripts, their predicted local secondary structures, and proteins generated in vivo upon IPTG induction. Transcript T-500 contains the weakly structured wild-type coat initiation sequence, wtSD, which has a predicted negative free energy (ΔG0) of −3.2 kcal/mol (29). This RNA generates only coat protein and serves as a negative control for maturation gene expression. Transcript T-501 is a deletion mutant that lacks Qβ nucleotides 1178 through 1365, comprising all coat gene initiation sequences. This transcript generates substantial amounts of 41.9 kDa maturation protein, mat42, but no coat protein (8), and serves as a positive control for maturation gene expression.
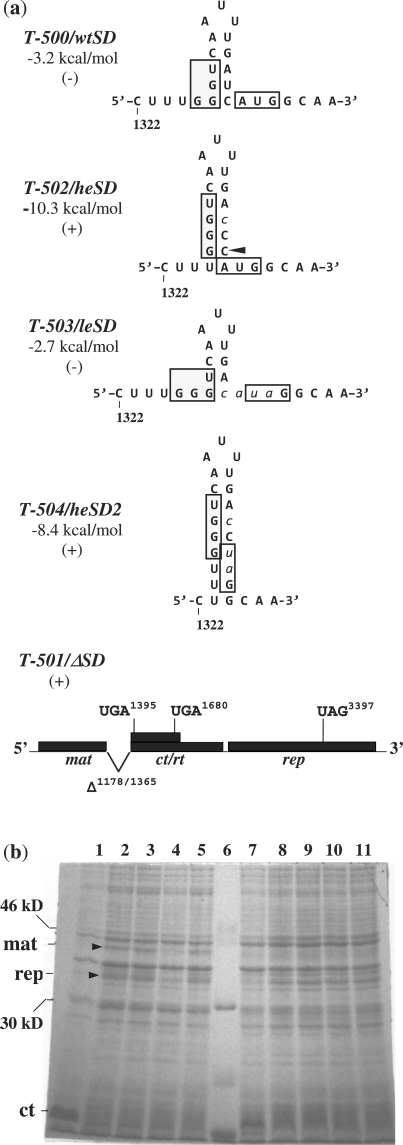
Figure 2.
(a) Mutations in the Qβ coat Shine–Dalgarno region. Nucleotides 1322–1346 are shown for variant Qβ RNA transcripts T-500, T-502, T-503 and T-504, and depict Qβ coat Shine–Dalgarno regions wtSD, heSD, leSD and heSD2, respectively. In transcript T-501, nucleotides 1178–1365, comprising the entire Qβ coat gene Shine–Dalgarno region, have been deleted from the Qβ genome. Predicted negative free energy (ΔG0) values are shown. Boxed nucleotides, coat gene Shine–Dalgarno region and location of the wild-type initiator codon; lower case italicized nucleotides, base substitutions; arrow, nucleotide insertion; +, maturation protein generated; −, no maturation protein generated; mat, maturation gene; rep, replicase gene; ct/rt, coat/readthrough gene. (b) PAGE analysis of proteins generated following growth of transformed E. coli BL21(DE3) in the presence (lanes 1–5) or absence (lanes 7–11) of IPTG and visualized by Coomassie blue staining. Lanes 1 and 7, T-500; lanes 2 and 8, T-501; lanes 3 and 9, T-502; lanes 4 and 10, T-503; lanes 5 and 11, T-504; lane 6, marker. Positions of Qβ maturation (mat), coat (ct) and replicase proteins are indicated.
Transcript T-502 differs from T-500 by only two point mutations in the coat gene initiation region. These mutations are predicted to create the hairpin stem structure heSD, which encompasses the entire coat gene Shine–Dalgarno sequence. The predicted ΔG0 of this hairpin stem is −10.3 kcal/mol, 7.1 kcal/mol stronger than that of the wtSD sequence (29). It has been estimated that for a hairpin structure that harbors a ribosome binding site, a change in ΔG0 of −1.5 kcal/mol at 37°C corresponds to a tenfold reduction in the rate of translational initiations from that site (12). Using this calculation, the heSD structure in T-502 would be expected to reduce translational initiations of the coat gene by a factor of 10(7.1/1.5), nearly 50 000 times. Although this reduction reflects an immeasurable difference, we know that pT7Qβ(+)502 never yields any detectable coat protein. Note that the stable heSD duplex is the only structural alteration predicted for the T-502 transcript. The remainder of the Qβ RNA molecule is predicted to retain its native conformation (29).
Transcripts T-500, T-501 and T-502 were synthesized from their respective pT7Qβ(+)500 plasmids upon IPTG induction of transformed E. coli. Figure 2b shows Qβ protein products generated from these variant RNAs. Transcript T-500, carrying the wtSD coat initiation region, generated only coat protein (lane 1), whereas T-501, with no Qβ coat gene initiation site, yielded both a 41.9 kDa maturation protein and a 38.4 kDa truncated replicase protein (lane 2). As expected, transcript T-502, carrying the coat gene initiation site in the strong heSD hairpin stem structure, produced no detectable coat protein (lane 3). Instead, maturation and replicase proteins were synthesized at levels comparable to that from the control transcript T-501. Presumably, because the strong Qβ coat gene initiation site was occluded by stable base-pair associations in the heSD hairpin, its availability for 30S ribosome binding was eliminated such that no coat protein could be produced (30,31). The corresponding activation of the maturation cistron is consistent with our previous observations in which maturation gene translation was activated in response to coat gene repression by Qβ replicase that was supplied in trans (8). The data collectively support the possibility of a mechanism by which blocking ribosome binding at the coat gene Shine–Dalgarno sequence would enable local ribosomes to recognize and initiate translation at the much weaker upstream maturation gene start site late in phage infection when maturation protein is needed to lyse the host.
An AUG initiator codon is not required for long-range inhibition
For the strong Qβ Shine–Dalgarno site to inhibit translation from the weaker maturation gene start site, neither an AUG initiator codon nor coat protein synthesis are required. For these experiments, we utilized RNA transcripts T-503 and T-504 (Figure 2a) in which the AUG initiator codons for the respective coat genes had been altered. In T-503, the coat Shine–Dalgarno sequence is located within the poorly structured leSD region, similar to that of the wild-type sequence (29). Such a weakly structured Shine–Dalgarno site would be expected to access E. coli 30S ribosomes easily. Alternatively, the coat Shine–Dalgarno region in T-504 was sequestered within a putatively strong hairpin structure, heSD2, with a predicted ΔG0 of −8.4 kcal/mol. Note that although the stabilities of their respective coat hairpin stems are extremely different, T-503 and T-504 differ from one another by only one nucleotide. Neither RNA transcript contains an AUG initiator codon for the coat gene, and so it is possible that only binary complexes will form between the initiation region and 30S ribosomes (32). However, T-503 also contains an out-of-frame AUA codon at the −1 position, and AUA has been shown to serve as a functional start codon for the coat proteins of related phages (33,34).
Figure 2b shows the proteins generated from transformed host cells following induction with IPTG. Transcript T-503, containing the weakly structured leSD coat Shine–Dalgarno sequence, resulted in no maturation protein even though no coat protein was made (lane 4). Alternatively, transcript T-504, containing the strongly duplexed heSD2 coat gene initiation region, resulted in significant synthesis of maturation protein as well as replicase protein (lane 5). These results are consistent with the idea that when the Qβ coat gene initiation site is exposed in a poorly structured conformation, putative binary complex association between the coat gene Shine–Dalgarno sequence and a 30S ribosome might be sufficient to prevent expression from the upstream maturation gene as well as from the downstream replicase gene.
Strength of the coat Shine–Dalgarno site directly affects maturation gene expression
To test the effect of the coat gene initiation site solely on maturation gene translation, we constructed truncated RNA genomes that contained only the maturation cistron and the downstream coat Shine–Dalgarno region, but no replicase cistron. Progressively strengthening the hairpin stem structure surrounding the Qβ coat gene ribosome binding site resulted in a concomitant increase in synthesis of maturation protein from the upstream cistron. We used site-directed mutagenesis to create hairpin stems of varying strengths in the region of the Qβ coat gene Shine–Dalgarno site. Figure 3 shows the encoded T-ctSD series of RNA transcripts with their putative secondary structures. The predicted negative free energy (ΔG0) for each of the mutated hairpin sequences varied from −10.1 kcal/mol, the most stable structure, to −3.9 kcal/mol, the least stable (29). Note that for each variant transcript, no alternative structures were predicted to form at any other location within the RNA.
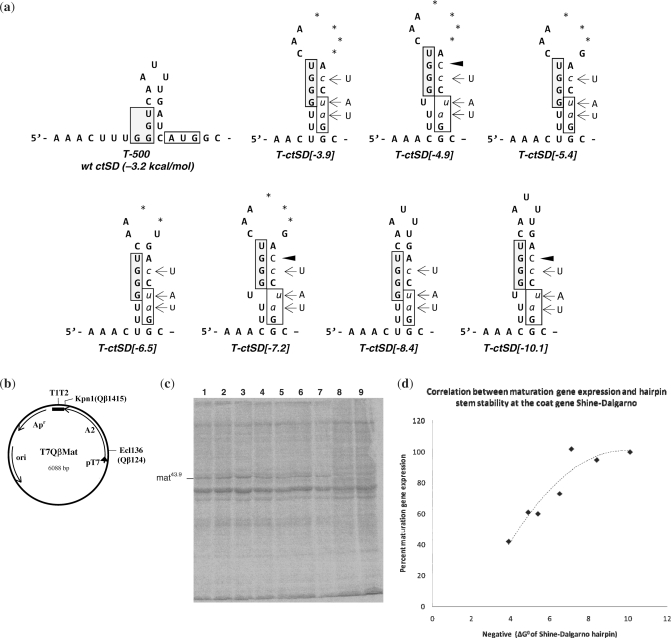
Figure 3.
(a) Putative secondary structures of the variant Qβ coat gene Shine–Dalgarno regions. Numbers in brackets indicate the predicted negative free energy (ΔG0) values. Wild-type nucleotides that were replaced are shown to the side; lower case italicized nucleotides, base substitutions; dark arrow, nucleotide insertion; boxed nucleotides, coat gene Shine–Dalgarno region and location of the wild-type coat gene initiator codon; *, nucleotide deletion; +, maturation protein generated; −, no maturation protein generated. (b) circular map of the plasmid pT7QβMat. This plasmid contains the entire Qβ maturation gene (A2) and coat Shine–Dalgarno sequence under the control of the T7 promoter (pT7). Transcripts terminate within a 3′ bacteriophage T7 5S T1T2 terminator region. Apr, ampicillin gene; ori, origin of replication. (c) PAGE analysis showing 14C-labeled proteins synthesized following growth of transformed E. coli BL21(DE3) in the presence of IPTG: lane 1, T-ctSD[-10.1]; lane 2, T-ctSD[-8.4]; lane 3, T-ctSD[-7.2]; lane 4, T-ctSD[-6.5]; lane 5, T-ctSD[-5.4], lane 6, T-ctSD[-4.9]; lane 7, T-ctSD[-3.9]. Lanes 8 and 9 are proteins from uninduced cells transformed with the parent plasmid pT7QβMat. (Note that the ctSD[-8.4] mutation in transcript T-ctSD[-8.4] is the same as the heSD2 mutation.) (d) The ratio of maturation protein synthesized relative to total protein expressed for each of the above Qβ coat gene Shine–Dalgarno mutants. Values were determined from phosphorimage analysis of 14C-labeled proteins generated after 30 min of induction with IPTG. For each, the percent of maturation protein synthesized was determined relative to the maximum amount (100%) that was synthesized from the T-502/heSD2 transcript (Figure 2).
Each mutant coat gene Shine-Dalgarno sequence was incorporated into a modified version of the pT7Qβ(+)500 plasmid, pT7Qβ(+)Mat, from which Qβ nucleotides 1406 through 4220 had been deleted (Figure 3b). Expression from this plasmid generates truncated Qβ mRNA transcripts that contain the entire maturation gene as well as the coat gene initiation sequences (24). Upon IPTG induction of cells transformed with the T-ctSD variants, maturation protein was synthesized at varying levels (Figure 3c). For each transformant, we utilized phosphorimage analysis to quantitate the ratio of maturation protein synthesized relative to total cellular protein. The relative amount of maturation protein synthesized from variant T-ctSD[-10.1] (lane 1) was the same as what we can obtain from the heSD control transcript T-502 (above) and was therefore normalized to 100% of the maximum that could be made within our system. For each of the other variant transcripts, T-ctSD[-8.4], T-ctSD[-7.2], T-ctSD[-6.5], T-ctSD[-5.4], T-ctSD[-4.9], and T-ctSD[-3.9] (lanes 2–7, respectively), we determined the amount of maturation protein synthesized as a percentage relative to that synthesized from variant T-ctSD[-10.1]. Figure 3d shows the results of our analysis. As the putative stability of the Qβ coat Shine–Dalgarno hairpin was increased, the amount of maturation protein synthesized progressively increased from <50% for transcript T-ctSD[-3.9] to 100% for transcripts T-ctSD[-7.2], T-ctSD[-8.4] and T-ctSD[-10.1]. Clearly, there is a correlation between the single-strandedness of the Qβ coat gene initiation region and expression of the distal upstream maturation gene. Apparently, once the coat gene Shine–Dalgarno is occluded in a duplexed hairpin of about −7 kcal/mol, the inhibitory effect of this site is abolished. Consequently, at this point, the maturation gene appears to be maximally activated. These data provide additional evidence that the coat gene Shine–Dalgarno region is able to modulate maturation gene translation via competition for translational initiations.
Note that the increase in maturation protein production that we observed here was not proportional to the decrease in coat protein initiations. We would not expect the activation of the maturation gene to quantitatively equal the inactivation of the coat gene. There was, however, a parallel trend between the increase in stability of the hairpin structure at the coat gene Shine–Dalgarno site and the increase in maturation protein to a maximum that could be reached. We propose that the suppression of the Qβ coat gene initiation site is necessary to allow synthesis of only enough maturation protein necessary to lyse the host late in phage infection.
A strong site inhibits a weak site in cis
Previously, we have shown that a deletion of the coat gene initiation site from Qβ RNA activates translation from the upstream maturation gene when present on the same RNA genome, but has no effect on maturation protein synthesis from a second genome (8). The following experiment shows that when the coat gene initiation site is present but sequestered in a strong hairpin stem structure, the same result is observed. That is, in Qβ RNA, occlusion of the coat gene initiation site within a strong hairpin stem structure will enable expression of an upstream maturation gene in cis, but has no effect in trans on maturation gene translation from a second Qβ RNA genome present in the same cell.
For these experiments, we utilized the p2xQβ(+) two-genome plasmid system (Figure 4a). This plasmid contains two cDNA copies of Qβ RNA as direct repeats, each with its own upstream T7 promoter/lac operator and downstream 5S T1T2 processing region (8,22). Having both genomes present on the same plasmid enables both encoded Qβ RNA genomes to be transcribed in equal amounts within the host (18). To distinguish the encoded maturation proteins, each cDNA genome contained one of two frame-shift mutations in the maturation gene cDNA sequence (Figure 4b): either a UAA1204 mutation, which results in a truncated 41.9 kDa maturation protein mat41.9, or a UAG1245 mutation, which results in a truncated 43.9 kDa maturation protein, mat43.9 (Table 1). Both variant maturation proteins are defective for lysis function. Each cDNA genome also contained a UAG3397 mutation (Table 1), which results in a truncated replicase protein, defective for replicase function and incapable of repressing the coat gene initiation site (8). Finally, each cDNA genome harbored either the heSD2 coat gene mutation from transcript T-504, or the leSD coat gene mutation from transcript T-503, neither of which contains the coat gene AUG initiator codon, thereby eliminating the possibility for coat protein synthesis (Figure 2).
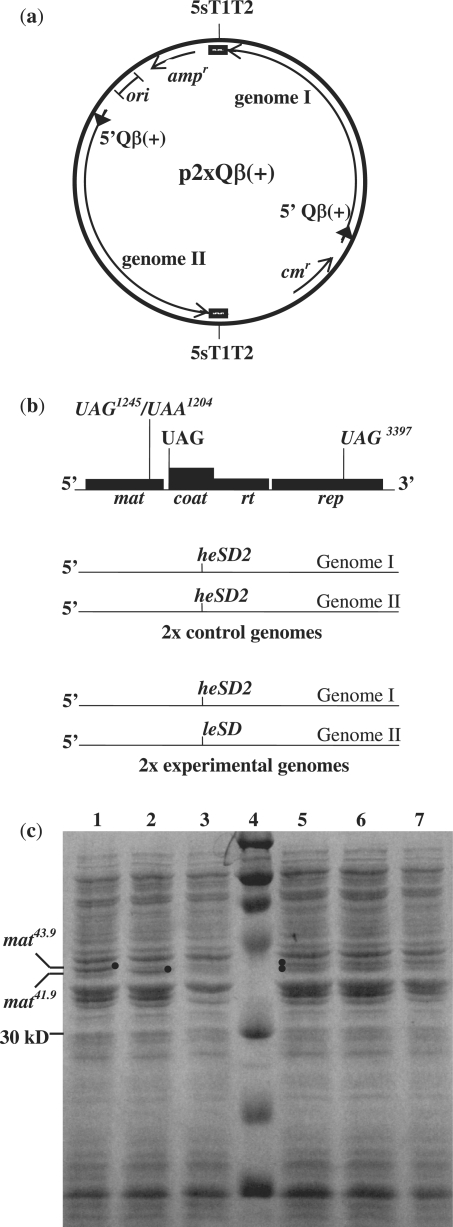
Figure 4.
Cis and trans effects of the Qβ coat initiation site on Qβ maturation protein synthesis. (a) The two message plasmid p2xQβ(+) contains two cDNA copies of the entire Qβ(+) genome, each with its own T7 promoter/lac operator element (block arrow) and bacteriophage T7 5S T1T2 terminator region. Upon transcription with T7 RNA polymerase, two full-length Qβ RNA genomes (I and II) are generated in equivalent amounts. To select against intra-plasmid recombination, the two Qβ genomes are separated by a chloramphenicol resistance gene on one side and an ampicillin resistance gene on the other. (b) The map of the Qβ RNA genome shows the positions of various mutations incorporated into the cDNA of genomes I and II. Each Qβ genome contains either a UAA1204 mutation, which yields a 41.9 kDa maturation protein mat41.9, or a UAG1245 mutation, which yields a 43.9 kDa maturation protein, mat43.9. Each genome also contains UAG3397 mutation, which results in a truncated replicase protein that cannot repress the coat gene, and thus cannot activate the maturation gene (Table 1). Both the heSD2 and the leSD coat gene mutations eliminate the coat gene initiation codon, replacing it with a UAG termination codon. In control p2xQβ(+) plasmids, both genome I and genome II harbor the heSD2 coat gene mutation. In the experimental p2xQβ(+) plasmid, only genome I encodes the heSD2 mutation, while genome II encodes the leSD coat gene mutation. (c) PAGE analysis of proteins generated following growth of transformed E. coli BL21(DE3) in the presence of IPTG and visualized by Coomassie blue staining. Lane 1, pT7Qβ(+)504 (heSD2) encoding mat43.9; lane 2, pT7Qβ(+)504 encoding mat41.9; lane 3, pT7Qβ(+)503 (leSD2) encoding mat41.9; lane 4, marker; lane 5, p2xQβ(+)[504/504]a, encoding mat43.9 on genome I and mat41.9 on genome II; lane 6, p2xQβ(+)[504/504]b, encoding mat41.9 on genome I and mat43.9 on genome II; lane 7, p2xQβ(+)504/503, encoding mat43.9 on genome I and mat41.9 on genome II.
To test the cis and trans effects of the Qβ coat gene Shine-Dalgarno site on maturation gene synthesis, different p2xQβ(+) plasmid sets were constructed, each containing a different set of mutations. In two different control plasmids, p2xQβ(+)[504/504]a and p2xQβ(+)[504/504]b, both genome I and genome II encoded the T-504 transcript, which contains the heSD2 mutation that putatively sequesters the coat gene Shine–Dalgarno site in a stable hairpin stem (Figure 4b). As demonstrated above, transcript T-504 was unable to generate Qβ coat protein, but yielded large amounts of maturation protein when generated from the single genome pT7Qβ(+)500 plasmid (Figure 4c, lanes 1 and 2; Figure 2b). In the first two-genome control plasmid, p2xQβ(+)[504/504]a, genome I encoded mat43.9, and genome II encoded mat41.9. In the second control plasmid, p2xQβ(+)[504/504]b, the maturation gene mutations were reversed: genome I encoded mat41.9, and genome II encoded mat43.9. Following induction with IPTG, each of these control plasmids yielded comparable amounts of both mat43.9 and mat41.9 proteins (Figure 4c, lanes 5 and 6).
The experimental two-genome plasmid was designed to test the effect of the Qβ coat gene Shine–Dalgarno site on maturation protein synthesis in trans. In the plasmid p2xQβ(+)[504/503], only genome I contained the heSD2 coat gene hairpin stem mutation, and encoded the mat43.9 maturation gene. Genome II, however, encoded the T-503 transcript carrying the leSD mutation, and harbored the mat41.9 gene. As demonstrated earlier, the leSD mutation has relatively weak secondary structure at the coat gene initiation site and completely inhibits translation from the upstream maturation gene when generated from the single genome plasmid pT7Qβ(+)503 (Figure 4c, lane 3; Figure 2b). Following IPTG induction of p2xQβ(+)[504/503], only mat43.9 was synthesized from genome I, but no detectable mat41.9 protein was synthesized from genome II (Figure 4b, lane 7). These results demonstrate that when the strong coat gene Shine–Dalgarno site is occluded within a strong hairpin stem structure, protein synthesis at the upstream maturation gene is activated in cis, but there is no detectable in trans effect on maturation protein synthesis from a second Qβ RNA genome present in the same cell. Seemingly, the presence alone of a second ribosome binding site on the same RNA molecule has a significant effect on the synthesis of Qβ maturation protein. These results are consistent with our previous data in which we demonstrated that activation of the Qβ maturation cistron is cis to the RNA genome on which the coat gene initiation site had been deleted, and that the presence of coat protein on a second genome had no effect on the maturation cistron in trans (8).
Note that when maturation proteins are generated from two-genome plasmids, they appear to be present in reduced amounts relative to the level of maturation protein expressed from either of the single genome plasmids (lanes 1 and 2). We have observed this before with the two-genome plasmid system (8,18). A possible explanation is that transcription by T7 RNA polymerase is limited, resulting in reduced amounts of RNA generated from each Qβ genome. Alternatively, the larger size of the two-genome plasmid might yield a lower copy number than that of the single genome plasmids. Although maturation proteins are generated in reduced quantities, the simultaneous transcription from the same plasmid consistently yields two RNA genomes in equal proportions (18).
DISCUSSION
Our results demonstrate that when the Qβ coat gene Shine–Dalgarno sequence was occluded by stable hairpin stem structure, translational initiation from the upstream maturation cistron was activated in cis. The secondary structure at the coat gene Shine–Dalgarno site had no effect on the translation of a maturation gene present on a different RNA molecule. There was a direct correlation between the stability of a hairpin stem structure that sequestered the coat gene initiation site and the degree of maturation gene activation. Furthermore, the inhibitory effect of the coat gene initiation site on maturation gene synthesis did not require either an AUG initiator codon or initiation of coat protein synthesis.
The data are in agreement with previous observations (8). During infection, the Qβ maturation gene is generally kept silent by extensive long-range secondary structure (27,28). In fact, the thermodynamic stability of the entire 5′ domain of Qβ RNA is such that there are essentially no viable alternative competing structures predicted (27). The single-strandedness of the coat gene initiation region renders it an extremely strong ribosome binding site relative to the much weaker maturation gene initiation site (12,13,27,28). However, despite the long-range structural interaction in the 5′ domain, when Qβ replicase binds the coat gene initiation site in trans to repress coat protein synthesis, the maturation gene becomes activated (8). Because maturation protein mediates host cell lysis (35), this mechanism is likely an efficient means of generating increased amounts of maturation protein when it is required late in infection.
In our experiments, when the coat gene Shine–Dalgarno site was sequestered in a stable hairpin structure to repress coat protein synthesis, both the replicase and maturation genes were activated. Presumably under these conditions, ribosomes were physically inhibited from binding to the coat gene Shine–Dalgarno site. Previously, we proposed a mechanism in which multiple translational initiation sites that have different ribosome binding strengths will compete for association with a single ribosome in cis at any moment in time (8). It is likely that, in the current experiments using the maturation gene plasmids, the progressive decrease in the single-strandedness at the coat gene initiation site caused a corresponding alteration in the relative ribosome binding affinities between the coat and maturation gene sites. Consequently, the probability that a ribosome would bind the less accessible upstream maturation gene site was increased. We propose the possibility that the Qβ coat and maturation gene ribosome binding sites compete in cis for ribosome binding as a means of regulating differential protein synthesis.
Intramolecular competition for ribosome binding could be a general mechanism which would allow a very weak ribosome binding site on any polycistronic RNA to become activated whenever a stronger site within the same molecule is rendered incapable of accessing ribosomes. Indeed, we have demonstrated that the Qβ replicase gene is activated when the coat gene initiation site is either blocked or eliminated (8), indicating that replicase expression does not need to be coupled to coat gene expression as previously thought (8,10,18,22). We have further noticed that if we eliminate the coat gene ribosome binding sequence from Qβ cDNA and incorporate a strong heterologous ribosome binding site from the bacteriophage T7 gene10 (36,37) 2.3-kb downstream of the maturation gene initiation site, maturation protein can be synthesized from the encoded Qβ RNA transcripts (unpublished results). Based on our observations, we suggest that there might not be anything inherent in the Qβ phage RNA coat gene initiation region that specifically leads to translational repression of the maturation gene. Instead, translational inhibition of one cistron by the presence of a strong distal ribosome binding site is likely a general mechanism that might apply to any prokaryotic polycistronic messenger RNA.
It should be mentioned that wild-type Qβ bacteriophage RNA has three ribosome binding sites. Whereas the coat gene Shine–Dalgarno site has the strongest affinity for ribosomes, the maturation gene site has the weakest. Consider that during active Qβ phage infection, coat protein needs to be made early and in large quantities. At first, translation through the coat gene region opens the replicase gene initiation site and allows translation of the replicase protein (10,11,18). Later in infection, excess replicase protein binds and represses translation of the coat cistron, and excess coat protein binds and represses translation of the replicase cistron (38). Elimination of both coat and replicase Shine–Dalgarno sites leaves only the maturation gene inititation site for ribosomes to access.
Such a mechanism in which a ribosome will bind at one particular translational initiation site and not another on the same mRNA would necessarily rely on a number of factors. These include the fixed distance between two ribosome binding sites on the same molecule, the dynamic equilibrium association of a 30S ribosome at a Shine–Dalgarno site, and the differential binding affinities of competing Shine–Dalgarno sites for a 30S ribosome. We will consider each of these factors below.
Fixed distance between two ribosome binding sites
Because the distance between two ribosome binding sites on the same RNA is a constant, these sites can be considered to be at high concentration relative to one another, and independent of the cellular message RNA concentration. However, when two sites lie on separate molecules, their concentration is a function of the cellular mRNA concentration. Consequently, the relative concentration of any two ribosome binding sites with respect to a single ribosome is dependent upon whether or not the two sites are present on the same molecule. Consider a volume of a cell in which free mRNAs are equally distributed, and mRNA concentration is a function of the number of RNA molecules present. Under these conditions, unbound ribosomes would be distributed proportionally among free mRNAs. If each RNA molecule contained only one Shine–Dalgarno sequence, and these had equal ribosome binding affinities, then ribosome association and translation would be proportional to the concentration of messenger RNAs.
Alternatively, when two different Shine–Dalgarno sites are present in cis on the same polycistronic message, the situation is very different. The concentration of the intramolecular sites relative to one another is now independent of cell volume. Instead, it is a constant that is determined by a fixed distance between the two sites. This distance would be determined both by the number of nucleotides between the two sites, and by RNA structure that can bring the two sites into closer proximity. Hence, the concentration of two ribosome binding sites relative to one another can be extremely high compared with that of available ribosomes. As such, the local concentration of unbound 30S ribosomes would always be limiting with respect to these two sites, regardless of the ribosome or mRNA concentration in the cell. The immediate reaction then becomes that of two ribosome binding sites ‘competing’ for association with only one ribosome. Note that this competition model considers available ribosomes only, and not those already involved in translational elongation throughout the cell.
Dynamic equilibrium of a 30S ribosome complex
It is generally accepted that because ribosomes in a cell are usually present in excess of message RNA molecules, all accessible ribosome binding sites can be saturated. However, binary complex association is a dynamic reversible equilibrium process (31), and so the 30S ribosome is never permanently bound at any one Shine–Dalgarno site. Following the association at a Shine–Dalgarno locus, the 30S ribosomal subunit will proceed in one of two ways: either it will translocate to the initiator codon and undergo protein synthesis, thereby eliminating itself from the pool of unbound ribosomes; or it will dissociate from the RNA. As such, Shine–Dalgarno sites would never be completely saturated at any given moment, but would be continually accessible for 30S ribosome binding.
Differential affinities of ribosome binding sites
Several factors contribute to the ribosome binding affinity of a Shine–Dalgarno region. Among these are: the degree of complementarity between a Shine–Dalgarno region and 16S ribosomal RNA (2,39); the secondary structure that comprises the Shine–Dalgarno region (30,31); the presence of either a nearby ribosomal protein S1 binding site or an enhancer site on the mRNA (40–42); the presence of putative standby sites for 30S ribosomes close to a Shine–Dalgarno sequence (13); and the putative interaction of a trans-acting protein or RNA that can bind a message RNA to block ribosome access (8,38). Hence, when two Shine–Dalgarno sites are present on the same polycistronic message, the probability that a single ribosome will associate with one or the other site is determined by their relative affinities for a 30S ribosome. The greater the difference in binding affinities, the more dramatic would be the competition between the two sites.
General implications of intramolecular competition between two ribosome sites
Our findings suggest that all polycistronic translational systems might be affected to some degree by competition in cis between multiple ribosome entry sites. Since competition within a single RNA molecule is putatively independent of both mRNA and ribosome concentration, all mRNAs carry the potential for this type of translational regulation. The more extreme the differences are in ribosome binding affinities among multiple sites on an RNA message, the more profound would be the regulatory effect. Competition in cis would not only affect the translational balance between multiple cistrons within a polycistronic mRNA, but also the possibility exists that pseudo-ribosome entry sites affect the efficiency of translational initiations at one or more genes on any given messenger RNA. Although association between a 30S ribosome and a messenger RNA depends upon a number of factors (see above), the 30S:mRNA association does not require an AUG initiator codon. As such, a translational initiation site might be rendered inactive simply because it is inhibited by a second, more competitive pseudo-ribosome binding site present within the same RNA molecule. Indeed, even ribosome binding sites that appear to be silent due to long-range secondary interactions are capable of accessing ribosomes in the absence of stronger competing sites. For example, the Qβ maturation gene was once believed to be completely inactive because of long-range secondary structure, but appears to be expressed to a maximum in the absence of the stronger downstream coat gene initiation site (8). We have previously proposed the possibility that for a large folded RNA domain, the kinetics of folding and re-folding can be very slow, thus allowing occasional exposure of a translational initiation site (8,22).
The effect of a putative competing ribosome entry site on distal gene translation might be modulated by several factors, such as: coupled translation with a second gene; interaction with either a trans-acting protein factor, or with antisense RNA (9); processing of an RNA into two or more separate mRNA molecules; or the formation of alternate RNA conformations. Consequently, the proposed mechanism of competition in cis would enable a single polycistronic mRNA to exist as one of two or more different functional messenger RNAs, each capable of translating a different proportion of the same encoded proteins. Such a process would provide a sophisticated means of translational auto-regulation not necessarily confined to the RNA coliphages. Since RNA phage genomes are highly adapted to utilizing the host translational apparatus, it is possible that other bacterial messenger RNA systems usefully employ a similar regulatory mechanism. It has been shown that prokaryotic polycistronic mRNAs can generate different proteins in quantities that vary over three orders of magnitude (3,43,44). Consequently, it is crucial to understand how alternative regulatory mechanisms govern the differential synthesis of multiple protein products from these RNA messages.
Many intriguing systems exist in which potential ribosome entry site competition might influence prokaryotic translation in cis. For example, there are intragenic ribosome entry sites that have been shown to affect gene expression (45,46). Mechanisms also exist that are responsible for masking independent initiation of translation (47–49). Translational competition has been shown to occur between one or more cistrons that are fused to a reporter gene within an RNA message (50). Translation of some cellular genes might be selectively enhanced by trans-acting repressor proteins, e.g. the T4 RegA repressor (51). In the E. coli rpmI-rplT operon encoding ribosomal proteins L35 and L20, a kinetic model is suggested in which the L20 repressor protein competes with 30S ribosomes for binding at the operator region to regulate translation (52). In addition, studies in eukaryotic systems suggest that translational regulation of human fibroblast growth factor might be affected by competition between a cap-dependent translational mechanism and an internal ribosome entry site-dependent mechanism (53,54).
In conclusion, we suggest that the following points should be considered with respect to translational control mechanisms: (i) all ribosome entry sites on a single messenger RNA can compete in cis for a single 30S ribosome; (ii) these ribosome entry sites are never saturated with ribosomes at any instant in time; (iii) competition in cis can occur when the cellular messenger RNA concentration is extremely low relative to the local concentration of two ribosome binding sites on the same messenger RNA; and (iv) reference genes that are inserted into a messenger RNA molecule might significantly influence translational initiations that occur at a distal experimental gene. Each of these points should be carefully considered when conducting experiments with cis-acting reporter genes and truncated message RNA molecules. It might be necessary to carry out such studies using intact mRNA molecules in the presence of any trans-acting RNA binding proteins, or antisense RNA transcripts that could influence gene expression.
FUNDING
Research investment initiative award through the Dean’s office of the State University of New York, Health Science Center at Brooklyn; Interim Grant from SUNY downstate. Funding for open access charge: Personal.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 2.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity nonsense triplets and ribosome binding sites. Proc. Natl Acad. Sci. USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gold L, Stormo G. Translational initiation. In: Neidhart FC, editor. Cellular and Molecular Biology of Escherichia coli and Salmonella typhimurium. Washington, DC: American Society for Microbiology; 1987. pp. 1302–1307. [Google Scholar]
- 4.McCarthy JE, Gualerzi C. Translational control of prokaryotic gene expression. Trends Genet. 1990;3:78–85. doi: 10.1016/0168-9525(90)90098-q. [DOI] [PubMed] [Google Scholar]
- 5.Ringquist S, MacDonald M, Gibson T, Gold L. Nature of the ribosomal mRNA track: analysis of ribosome-binding sites containing different sequences and secondary structures. Biochemistry. 1993;32:10254–10262. doi: 10.1021/bi00089a048. [DOI] [PubMed] [Google Scholar]
- 6.Inokuchi Y, Hirashima A. Interference with viral infection by RNA replicase deleted at the carboxy-terminal region. J. Biochem. 1990;108:3–58. doi: 10.1093/oxfordjournals.jbchem.a123162. [DOI] [PubMed] [Google Scholar]
- 7.Atkins JF, Gesteland RF. A case for trans translation. Nature. 1996;379:9–70. doi: 10.1038/379769a0. [DOI] [PubMed] [Google Scholar]
- 8.Priano C, Arora R, Jayant L, Mills DR. Translational activation in coliphage Qβ: on a polycistronic messenger RNA, repression of one gene can activate translation of another. J. Mol. Biol. 1997;271:299–310. doi: 10.1006/jmbi.1997.1194. [DOI] [PubMed] [Google Scholar]
- 9.Wassarman KM, Zhang A, Storz G. Small RNAs in Escherichia coli. Trends Microbiol. 1999;7:37––45. doi: 10.1016/s0966-842x(98)01379-1. [DOI] [PubMed] [Google Scholar]
- 10.Ball LA, Kaesberg P. A polarity gradient in the expression of the replicase gene of RNA bacteriophage Qβ. J. Mol. Biol. 1973;74:547–562. doi: 10.1016/0022-2836(73)90046-6. [DOI] [PubMed] [Google Scholar]
- 11.Skripkin E, Jacobson AB. A two-dimensional model at the nucleotide level for the central hairpin of coliphage Qβ RNA. J. Mol. Biol. 1993;233:261–269. doi: 10.1006/jmbi.1993.1503. [DOI] [PubMed] [Google Scholar]
- 12.de Smit MH, van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc. Natl Acad. Sci. USA. 1990;87:7668–7672. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Smit MH, van Duin J. Translational standby sites: how ribosomes may deal with the rapid folding kinetics of mRNA. J. Mol. Biol. 2003;331:737–743. doi: 10.1016/s0022-2836(03)00809-x. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 15.Studier FW, Moffat BA. Use of bacteriophage T7 RNA polymerase to direct selective high level expression of cloned genes. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 16.Mills DR, Priano C, Merz P, Binderow BD. Qß RNA bacteriophage: mapping cis-acting elements within an RNA genome. J. Virol. 1990;64:3872–3881. doi: 10.1128/jvi.64.8.3872-3881.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with Fission Yeast: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- 18.Arora R. 1995. Structural and functional analysis of the readthrough cistron of the RNA coliphage Qβ: its role in the viral life cycle. PhD Dissertation, State University of New York at Brooklyn, NY. [Google Scholar]
- 19.Mills DR, Priano C, DiMauro P, Binderow BD. Qß replicase: mapping the functional domains of an RNA dependent RNA polymerase. J. Mol. Biol. 1989;205:751–764. doi: 10.1016/0022-2836(89)90319-7. [DOI] [PubMed] [Google Scholar]
- 20.Lyakhov DL, He B, Zhang X, Studier FW, Dunn JJ, McAllister WT. Mutant bacteriophage T7 RNA polymerases with altered termination properties. J. Mol. Biol. 1997;269:28–40. doi: 10.1006/jmbi.1997.1015. [DOI] [PubMed] [Google Scholar]
- 21.Axelrod VD, Brown E, Priano C, Mills DR. Coliphage Qβ RNA replication: RNA catalytic for single strand release. Virology. 1991;184:595–608. doi: 10.1016/0042-6822(91)90430-j. [DOI] [PubMed] [Google Scholar]
- 22.Jacobson AB, Arora R, Zuker MM, Priano C, Lin CH, Mills DR. Structural plasticity in RNA and its role in the regulation of protein translation in coliphage Qβ. J. Mol. Biol. 1998;274:589–600. doi: 10.1006/jmbi.1997.1472. [DOI] [PubMed] [Google Scholar]
- 23.Priano C, Arora R, Butke J, Mills DR. A complete plasmid-based complementation system for RNA coliphage Qβ: three proteins of bacteriophages Qβ (GroupIII) and SP (group IV) can be interchanged. J. Mol. Biol. 1995;249:283–297. doi: 10.1006/jmbi.1995.0297. [DOI] [PubMed] [Google Scholar]
- 24.Jayant L. 2002. Different ribosome binding sites on a single mRNA molecule influence the translation of each other in cis. PhD Dissertation, State University of New York at Brooklyn, NY. [Google Scholar]
- 25.He B, Rong M, Lyakhov DL, Gartenstein H, Diaz G, Castagna R, McAllister WT, Durbin RK. Rapid mutagenesis and purification of phage RNA polymerases. Protein Express. Purif. 1997;9:142–151. doi: 10.1006/prep.1996.0663. [DOI] [PubMed] [Google Scholar]
- 26.Arora R, Priano C, Jacobson AB, Mills DR. Cis-acting elements within an RNA coliphage genome: fold as you please, but fold you must!! J. Mol. Biol. 1996;258:433–446. doi: 10.1006/jmbi.1996.0260. [DOI] [PubMed] [Google Scholar]
- 27.Jacobson AB, Zuker M. Structural analysis by energy dot plot of a large mRNA. J. Mol. Biol. 1993;233:261–269. doi: 10.1006/jmbi.1993.1504. [DOI] [PubMed] [Google Scholar]
- 28.Beekwilder J. 1996. Secondary structure of the RNA genome of bacteriophage Qβ. PhD Dissertation, Leiden University, The Netherlands. [Google Scholar]
- 29.Zuker M, Mathews DH, Turner DH. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In: Barciszewski J, Clark B.FC, editors. RNA Biochemistry and Biotechnology. NATOASI Series, Kluwer Academic Publishers, Dordrecht, The Netherlands; 1999. pp. 11–43. [Google Scholar]
- 30.Hall MN, Gabay J, Debarbouille M, Schwartz M. A role for mRNA secondary structure in the control of translation initiation. Nature. 1982;295:616–618. doi: 10.1038/295616a0. [DOI] [PubMed] [Google Scholar]
- 31.de Smit MH, van Duin J. Control of translation by mRNA secondary structure in Escherichia coli: a quantitative analysis of literature data. J. Mol. Biol. 1994;244:144–150. doi: 10.1006/jmbi.1994.1714. [DOI] [PubMed] [Google Scholar]
- 32.Hartz D, McPheeters DS, Green L, Gold L. Detection of Escherichia coli ribosome binding at translational initiation sites in the absence of tRNA. J. Mol. Biol. 1991;218:99–105. doi: 10.1016/0022-2836(91)90876-8. [DOI] [PubMed] [Google Scholar]
- 33.Köpke AK, Leggatt JS. Initiation of translation at an AUA codon for an archaebacterial protein gene expressed in. E. coli. Nucleic Acids Res. 1991;19:5169–5172. doi: 10.1093/nar/19.19.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romero A, Garcia P. Initiation of translation at AUC, AUA and AUU codons in Escherichia coli. FEMS Microbiol. Lett. 1991;68:325–330. doi: 10.1016/0378-1097(91)90377-m. [DOI] [PubMed] [Google Scholar]
- 35.Winter RB, Gold L. Over-production of bacteriophage Qβ maturation (A2) protein leads to cell lysis. Cell. 1983;33:877–885. doi: 10.1016/0092-8674(83)90030-2. [DOI] [PubMed] [Google Scholar]
- 36.Rosenberg AH, Lade BN, Chui D.-S, Lin S.-W, Dunn JJ, Studier FW. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56:125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- 37.Studier FW, Rosenberg AH, Dunn JJ, Dubendorf JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 38.Weber H, Billeter MA, Kahane S, Weissmann C, Hindley J, Porter A. Molecular basis for repressor activity of Qβ replicase. Nature New Biol. 1972;237:166–170. doi: 10.1038/newbio237166a0. [DOI] [PubMed] [Google Scholar]
- 39.Backendorf C, Overbeek GP, van Boom JH, Van der Marel G, Veeneman G, van Duin J. Recognition of the 16S RNA in ribosome messenger recognition. Eur. J. Biochem. 1980;110:599–604. doi: 10.1111/j.1432-1033.1980.tb04904.x. [DOI] [PubMed] [Google Scholar]
- 40.Goelz S, Steitz JA. Escherichia coli ribosomal protein S1 recognizes two sites in bacteriophage Qβ RNA. J. Biol. Chem. 1977;252:15177–15179. [PubMed] [Google Scholar]
- 41.Boni IV, Isaeva DM, Musychenko ML, Tszareva NV. Ribosome messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 1991;19:155–162. doi: 10.1093/nar/19.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farwell MA, Roberts MW, Rabinowitz JC. The effect of ribosomal protein S1 from Escherichia coli and Micrococcus luteus on protein synthesis in vitro by E. coli and Bacillus subtilis. Mol. Microbiol. 1992;6:3375–3383. doi: 10.1111/j.1365-2958.1992.tb02205.x. [DOI] [PubMed] [Google Scholar]
- 43.Kozak M, Nathans D. Translation of the genome of a ribonucleic acid bacteriophage. Bacteriol. Rev. 1972;36:109–134. doi: 10.1128/br.36.1.109-134.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ray PN, Pearson Ml. Functional inactivation of bacteriophage λ morphogenetic gene mRNA. Nature. 1975;253:647–650. doi: 10.1038/253647a0. [DOI] [PubMed] [Google Scholar]
- 45.Ivanov I, Alexandrova R, Dragulev B, Saraffova A, Abouttaidar MG. Effect of tandemly repeated AGG triplets on the translation of CAT-mRNA in. E. coli. FEBS Lett. 1992;307:173–176. doi: 10.1016/0014-5793(92)80761-5. [DOI] [PubMed] [Google Scholar]
- 46.Matten SR, Schneider TD, Ringquist S, Brusilow WA. Identification of an intragenic ribosome binding site that affects expression of the uncB gene (atp) of the E. coli proton-translocating ATPase (unc) operon. J. Bacteriol. 1998;180:3940–3945. doi: 10.1128/jb.180.15.3940-3945.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sor F, Bolotin-Fukuhara M, Nomura M. Mutational alterations of translational coupling in the L11 ribosomal protein operon of Escherichia coli. J. Bacteriol. 1987;169:3495–3507. doi: 10.1128/jb.169.8.3495-3507.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pediconi D, Spurio R, La Teana A, Jemiolo D, Gualerzi CO, Pon CL. Translational regulation of infC operon in Bacillus stearothermophilus. Biochem. Cell Biol. 1995;73:1071–1078. doi: 10.1139/o95-115. [DOI] [PubMed] [Google Scholar]
- 49.Supply P, Magdalene J, Locht C. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol. Microbiol. 1997;26:991–1003. doi: 10.1046/j.1365-2958.1997.6361999.x. [DOI] [PubMed] [Google Scholar]
- 50.McCarthy JEG, Schairer HV, Sebald W. Translational initiation frequency of ATP genes from E. coli: identification of an intercistronic sequence that enhances translation. EMBO J. 1985;4:519–526. doi: 10.1002/j.1460-2075.1985.tb03659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller ES, Karam J, Dawson M, Trojanowska M, Gauss P, Gold L. Translational repression: biological activity of plasmid-encoded bacteriophage T4 RegA protein. J. Mol. Biol. 1987;194:397–410. doi: 10.1016/0022-2836(87)90670-x. [DOI] [PubMed] [Google Scholar]
- 52.Haentjens-Sitri J, Allemand F, Springer M, Chiaruttini C. A competition mechanism regulates the translation of the Escherichia coli operon encoding ribosomal proteins L35 and L20. J. Mol. Biol. 2008;375:612–625. doi: 10.1016/j.jmb.2007.10.058. [DOI] [PubMed] [Google Scholar]
- 53.Vagner S, Gensac M, Maret A, Bayard F, Amalric F, Prats H, Prats A. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol. Cell. Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arnaud E, Touriol C, Boutonnet C, Gensac M, Vagner S, Prats H, Prats A. A new 34-kilodalton isoform of human fibroblast growth factor 2 is cap dependently synthesized by using a non-AUG start codon and behaves as a survival factor. Mol. Cell Biol. 1999;19:505–514. doi: 10.1128/mcb.19.1.505. [DOI] [PMC free article] [PubMed] [Google Scholar]