Abstract
Given that greater than 90% of the human genome is expressed, it is logical to assume that post-transcriptional regulatory mechanisms must be the primary means of controlling the flow of information from mRNA to protein. This report describes a robust approach that includes in silico, in vitro and in cellulo experiments permitting an in-depth evaluation of the impact of G-quadruplexes as translational repressors. Sequences including potential G-quadruplexes were selected within nine distinct genes encoding proteins involved in various biological processes. Their abilities to fold into G-quadruplex structures in vitro were evaluated using circular dichroism, thermal denaturation and the novel use of in-line probing. Six sequences were observed to fold into G-quadruplex structures in vitro, all of which exhibited translational inhibition in cellulo when linked to a reporter gene. Sequence analysis, direct mutagenesis and subsequent experiments were performed in order to define the rules governing the folding of G-quadruplexes. In addition, the impact of single-nucleotide polymorphism was shown to be important in the formation of G-quadruplexes located within the 5′-untranslated region of an mRNA. In light of these results, clearly the 5′-UTR G-quadruplexes represent a class of translational repressors that is broadly distributed in the cell.
INTRODUCTION
The life cycle of a messenger RNA (mRNA) species is full of diverse processing events and regulatory controls. For a long time, it has been believed that the primary means of regulating gene expression occurred at the transcription level. However, the discovery that over 90% of the genome is transcribed prompted the conclusion that post-transcriptional regulation is in fact the cornerstone for the regulation of gene expression (1). Post-transcriptional regulatory elements must be involved in order to direct the expression of specific subsets of genes within this large transcriptome. In terms of the mRNAs themselves, these regulatory elements can act at various steps in their life cycles, ranging from their processing events (e.g. capping, splicing and polyadenalytion) to their active transport, stability and translation (2). Several cellular factors are involved in these regulatory mechanisms. Some of them act as trans-acting regulatory elements. This is the case for the micro-RNAs, which generally interact with the 3′-untranslated regions (3′-UTR) of specific mRNAs, repressing their translation and/or decreasing their stabilities (3,4). There are also many cis-acting regulatory factors. In general, the latter are highly ordered RNA structures present in either the 5′- or 3′-UTRs. For example, the presence of a highly active hammerhead ribozyme in the 3′-UTRs of the rodent C-type lectin type II gene has been shown to reduce protein expression in mouse cells (5). Moreover, riboswitches, which are implicated in regulating gene expression, have been detected in the 5′-UTRs of a large variety of genes. Specifically, the binding of a metabolite to the aptamer domain has the effect of controlling the gene’s expression level, leading to either an increase or a decrease in the transcription and/or the translation levels. New riboswitches are frequently discovered, and both their complexities and diversities remain unappreciated (6). Clearly, the discovery and elucidation of post-transcriptional regulatory elements represent key components in achieving a good understanding of the molecular biology of the cell.
Guanine-rich nucleic acid sequences can fold into a non-canonical tetrahelical structure called a G-quadruplex. This structure involves the stacking of hydrogen-bonded G-tetrads, which, once stabilized by the chelation of monovalent metal ions such as potassium, represent an extremely stable four-stranded helical structure (7–9). Several bioinformatic studies have revealed both a significant level of conservation and an enrichment in potential G-quadruplex (PG4) sequences in various regulatory elements (e.g. telomeres, DNA promoters and both the 5′- and 3′-UTRs of mRNAs) within the genome, suggesting that they are involved in key biological processes (10–14). For example, the formation of G-quadruplexes at the eukaryotic telomeric sequences has been proposed to be associated with the telomeres’ maintenance by modulating their interactions with various proteins (15–17). The prevalence of PG4s within different functional classes of genes was determined using a computational approach (18). In addition, many G-quadruplexes have been found to be located in the promoters of various proto-oncogenes such as c-MYC, C-Kit, c-myb and KRAS (19–22). These PG4 sequences were suggested to be involved in the regulation of the transcriptional activity of these genes. Moreover, because the G-quadruplexes are directly linked to several key features in cancer cells, such as telomeres and oncogenes, great efforts have been made to try and find potential ligands that would act as anticancer agents. Some compounds that were shown to target DNA G-quadruplexes have already provided promising results, either by inhibiting the telomerase activity or by reducing oncogene expression (23).
While our knowledge of the DNA G-quadruplexes present in the human genome is increasing, our understanding of biologically relevant RNA G-quadruplexes remains limited. It is known that for a given sequence in vitro, an RNA G-quadruplex is usually more stable than its DNA counterpart (24). Moreover, unlike DNA, which is constrained mainly to a duplex form in the cell, RNA has no complementary strand limiting its structure. These two features make G-rich RNA sequences more susceptible to folding into a G-quadruplex structure in vivo. Several bioinformatic analyses searching for PG4 in the different regions of an mRNA have been reported (e.g. in the 5′-UTR, the 3′-UTR and the RNA processing sites) (12,25). Moreover, in some cases, RNA G-quadruplexes have been demonstrated to have functional roles (26–32). For instance, one G-quadruplex structure was shown to direct the discrimination of a proper target by the fragile X mental retardation protein, while another was reported to regulate an alternative splicing event, to name two examples (26,27). The original study showing a G-quadruplex structure acting as a translational repressor was performed in a cell-free system using the full-length NRAS 5′-UTR that includes such a structure (29). Subsequently, two other studies showed similar effects in cellulo using either a 27-nt Zic-1 RNA G-quadruplex, or a complete MT3-MPP 5′-UTR bearing a special purine-only RNA G-quadruplex (30,31). In each of these studies, only one RNA G-quadruplex was analyzed. More recently, the characterization of artificial cis-acting G-quadruplex repressors revealed an interesting correlation between the loop length and the number of G-tracks in terms of the translational inhibition level (32). Despite all of these studies, both the impact and the importance of the 5′-UTR G-quadruplex structures on the biology of the cell remain, most likely, underestimated. Here, we present a robust approach including in silico, in vitro and in cellulo experiments that permits a wider evaluation of the G-quadruplexes acting as translational repressors. Importantly, several G-quadruplex structures widely distributed within the transcriptome were studied and new rules governing the formation of the G-quadruplexes are reported. These rules permit the proposal of several regulatory mechanisms of G-quadruplex formation in an RNA strand.
MATERIALS AND METHODS
The sequences of all of the oligonucleotides used in this work are given in Table S1.
Bioinformatics
The 5′-UTR databases were derived from sequences taken from Transterm and UTRdb (33,34). These two databases contain spliced 5′-UTR sequences. PG4 sequences were identified using the above algorithm and the program RNAMotif (35). The results were subjected to various homemade Perl scripts and manually cured in order to obtain the PG4 databases presented in the Supplementary Data in an Excel file format. When a 5′-UTR PG4 was identified in a gene that generates more than one transcript with the same 5V-UTR, each transcript was treated individually and was counted as one more PG4. The gene ontology analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) web-accessible programs (36). The input data for the web-accessible program include the list of genes that included a PG4 in the complementary strand obtained from the human UTRfull database (Dataset S6). The single-nucleotide polymorphism (SNP) analysis was performed using a database of the SNPs present in various human mRNAs corresponding to NCBI dbSNP build 129, and the PG4 database obtained from the human UTRef database (Dataset S3). The presence of SNPs inside each PG4 sequence was examined using several homemade Perl scripts that compare the positions and the lengths of the PG4s to the positions of the SNPs present in the mRNAs. The list of SNPs found within the PG4 sequences was manually cured, and is presented in the Supplementary Data (Dataset S4).
RNA synthesis
All PG4 versions used for the in vitro experiments were synthesized by in vitro transcription using T7 RNA polymerase as described previously (37). Briefly, two overlapping oligonucleotides (2 µM each) were annealed, and double-stranded DNA was obtained by filling in the gaps using purified Pfu DNA polymerase in the presence of 5% dimethyl sulfoxide (DMSO). The double-stranded DNA was then ethanol-precipitated. The resulting DNA templates contained the T7 RNA promoter sequence followed by the PG4 sequence. After dissolution of the polymerase chain reaction (PCR) product in ultrapure water, runoff transcriptions were performed in a final volume of 100 µl using purified T7 RNA polymerase (10 µg) in the presence of RNase OUT (20 U, Invitrogen), pyrophosphatase (0.01 U, Roche Diagnostics) and 5 mM NTP in a buffer containing 80 mM HEPES-KOH, pH 7.5, 24 mM MgCl2, 2 mM spermidine and 40 mM DTT. The reactions were incubated for 2 h at 37°C. Upon completion, the reaction mixtures were treated with DNase RQ1 (Promega) at 37°C for 20 min. The RNA was then purified by phenol:chloroform extraction followed by ethanol precipitation. RNA products were fractionated by denaturing (8 M urea) 10% polyacrylamide gel electrophoresis (PAGE; 19:1 ratio of acrylamide to bisacrylamide) using 45 mM Tris-borate, pH 7.5/1 mM EDTA solution as running buffer. The RNAs were visualized by UV shadowing, and those corresponding to the correct sizes of the PG4s were excised from the gel and the transcripts eluted overnight at room temperature in buffer containing 1 mM EDTA, 0.1% SDS and 0.5 M ammonium acetate. The PG4s were then ethanol-precipitated, dried and dissolved in water. The concentrations were determined by spectrometry at 260 nm.
Circular dichroism spectroscopy
All circular dichroism (CD) experiments were performed using 4 µM of the relevant RNA sample dissolved in 50 mM Tris–HCl (pH 7.5) either in the absence of monovalent salt, or in the presence of 100 mM LiCl, NaCl or KCl. Prior to taking the CD measurement, each sample was heated to 70°C for 5 min and then slow-cooled to room temperature over a 1-h period. CD spectroscopy experiments were performed with a Jasco J-810 spectropolarimeter equipped with a Jasco Peltier temperature controller in a 1-ml quartz cell with a pathlength of 1 mm. CD scans, ranging from 220 to 320 nm, were recorded at 25°C at 50 nm min−1 with a 2-s response time, 0.1-nm pitch and 1-nm bandwidth. The means of at least three wavelength scans were compiled. Subtraction of the buffer was not required since control experiments in the absence of RNA showed negligible curves. CD melting curves were obtained by heating the samples from 25°C to 90°C at a controlled rate of 1°C min−1 and monitoring a 264-nm CD peak every 0.2 min. Melting temperature (Tm) values were calculated using ‘fraction folded’ (θ) versus temperature plots (38).
RNA labeling
In order to produce 5′-end-labeled PG4s, purified transcripts were dephosphorylated by adding 1 U of antartic phosphatase (New England BioLabs,) to 50 pmol of RNA and incubating the reaction mixture for 30 min at 37°C in a final volume of 10 µl containing 50 mM Bis-Propane (pH 6.0), 1 mM MgCl2, 0.1 mM ZnCl2 and RNase OUT (20 U, Invitrogen). The enzyme was inactivated by incubation for 5 min at 65°C. Dephosphorylated transcripts (5 pmol) were 5′-end-radiolabeled using 3 U of T4 polynucleotide kinase (Promega) for 1 h at 37°C in the presence of 3.2 pmol of [α-32P]ATP (6000 Ci/mmol; New England Nuclear). The reactions were stopped by adding formamide dye buffer (95% formamide, 10 mM EDTA, 0.025% bromophenol blue and 0.025% xylene cyanol), and the RNA molecules purified by 10% polyacrylamide gel electrophoresis. The bands of the correct sizes containing the 5′-end-labeled RNAs were excised and recovered as described above except that the detection was performed by autoradiography.
In-line probing
5′-end labelled RNA (50 000 c.p.m.), that is to say a trace amount of RNA (<1 nM), was heated at 70°C for 5 min and then slow-cooled to room temperature over 1 h in buffer containing 50 mM Tris–HCl (pH 7.5) and either no monovalent salt, or in the presence of 100 mM LiCl, NaCl or KCl in a final volume of 10 µl. Following this incubation, the final volume of each sample was adjusted to 100 µl such that the final concentrations were 50 mM Tris–HCl (pH 7.5), 20 mM MgCl2 and either no salt or 100 mM LiCl, NaCl or KCl. The reactions were then incubated for 40 h at room temperature, ethanol-precipitated and the RNAs dissolved in ice cold formamide dye loading buffer (95% formamide and 10 mM EDTA). For alkaline hydrolysis, 50 000 c.p.m. of 5′-end-labeled RNA (<1 nM) were dissolved in 5 µl of water, 1 µl of 1 N NaOH added and the reactions incubated for 1 min at room temperature prior to being quenched by the addition of 3 µl of 1 M Tris–HCl (pH 7.5). The RNA molecules were then ethanol-precipitated and dissolved in formamide dye loading buffer. An RNase T1 ladder was prepared using 50 000 c.p.m. of 5′-end-labeled RNA (<1 nM) dissolved in 10 µl of buffer containing 20 mM Tris–HCl (pH 7.5), 10 mM MgCl2 and 100 mM LiCl. The mixture were incubated for 2 min at 37°C in the presence of 0.6 U of RNase T1 (Roche Diagnostic), and was then quenched by the addition of 20 µl of formamide dye loading buffer. The radioactivity of the in-line probing samples and both ladders was calculated, and equal amounts in terms of counts per minute of all conditions and ladders of each candidate were fractionated on denaturing (8 M urea) 10% polyacrylamide gels.
Plasmid construction
The sequences of the 5′-UTRs were obtained from the NCBI database and correspond to the following Gene Identification (GI) for each candidate: EBAG9 (GI: 37694064), FZD2 (GI: 5922012), BARHL1 (GI: 31542183), NCAM2 (GI: 33519480), THRA (GI: 46255056), AASDHPPT (GI: 20357567) and TNFSF12 (GI: 23510442). The full-length 5′-UTRs of each candidate was reconstituted in vitro by the filling in of multiple overlapping oligonucleotides and various PCR steps (the specific sets of oligonucleotides used for each candidate are shown in Table S1). Wild-type and G/A-mutant 5′-UTR versions were synthesized for each candidate. In addition to the G/A-mutants, both C/A-mutants and CG/AA-mutants were synthesized for TNFSF12, and a C7 SNP 5′-UTR version was synthesized for AASDHPPT. The positions of all of the different mutations are the same as those used for the in vitro experiments. The list of oligonucleotides used for each candidate is shown in Table S1. The reconstituted 5′-UTRs were inserted in the NheI site in the pRL-TK plasmid vector (Promega). DNA sequencing of each candidate confirmed the insertion of the correct sequence.
Cell culture
HEK 293 cells (human embryonic kidney) were cultured in T-75 flasks (Sarstedt) in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1 mM sodium pyruvate and an antibiotic-antimycotic drug mixture (all purchased from Wisent) at 37°C in a 5% CO2 atmosphere in a humidified incubator.
Dual luciferase and quantitative real-time-PCR assays
HEK 293 cells (1.2 × 105) were seeded in 24-well plates. Twenty-four hours later, the cells were co-transfected with both the specific pRL-TK plasmid construction (renillia luciferase, Rluc) and the pGL3-control vector (firefly luciferase, Fluc) (Promega) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Twenty-four hours after transfection, 10% of the cells were used to measure the Rluc and Fluc activities using the Dual-luciferase Reporter Assay kit (Promega) according to the manufacturer’s protocol in a 5-ml test tube using a Berthold Lumat LB9501 luminometer (Berthold Technologies). For each lysate, the value of the Rluc was divided by the value of the Fluc. The ratios obtained for the G/A-mutant version were compared to those obtained with the wild type version of each candidate. Both the mean value and the standard deviation were calculated from at least three independent experiments for each candidate.
Total cellular RNA was extracted from the remaining cells using an Absolute RNA Microprep Kit (Stratagene) according to the manufacturer’s protocol that include a DNase treatment. Total RNA (200 ng) from each sample was reverse-transcribed using Transcriptor Reverse Transcriptase (Roche). The cDNA was subjected to quantitative real-time PCR using the FastStart Universal SYBR Green Master (Rox) mix (Roche) and a Rotor-GeneTM 3000 device (Corbett Research). The levels of Rluc and Fluc mRNAs were detected using the appropriate primers sets: forward primers Rluc 5′-(TGGGGTGCTTGTTTGGCATT)-3′ and Fluc 5′-(AAATGTCCGTTCGGTTGGCA)-3′ and reverse primers Rluc 5′-(TGGCAACATGGTTTCCACGA)-3′ and Fluc 5′-(ACTCCGATAAATAACGCGCCCA)-3′. The relative gene expression data were calculated using the ΔΔCT with the Fluc gene as internal control and the wild-type version as calibrator for each candidate (39).
RESULTS
Frequency of G-quadruplexes within 5′-UTR
In order to better understand the general role that G-quadruplexes play as translational repressors, a database of all potential G-quadruplexes located in the 5′-UTRs of the genes from 18 organisms, including humans, was constructed. We followed the same protocol as reported previously (12), except that 18 organisms were considered. The human 5′-UTR sequences were downloaded from both UTRdb and Transterm, while those from the other organisms (listed in Dataset S1) were downloaded only from Transterm (33,34). Potential G-quadruplex (PG4) sequences were identified using a previously available algorithm that searches for the sequence Gx-N1–7-Gx-N1–7-Gx-N1–7-Gx, where x ≥ 3 and N is any nucleotide (A,C,G or U) (40,41). These parameters were established by taking into account various results from in vitro studies on the G-quadruplex structure. With these guidelines, the 5′-UTR sequences were scanned in order to identify PG4s located on either the template or the complementary strand. The PG4s located on the template strands are composed of tracks of cytosines in the sequence database, while those located on the complementary strands correspond to tracks of guanosines and will be found in the mRNA. The primary analysis was focused on the 124 315 5′-UTRs obtained from the human UTRfull collection (Table 1 and Dataset S2). This yielded 9979 (8.0%) 5′-UTRs that contained at least one PG4 sequence. The numbers of 5V-UTRs with PG4s located in the template, versus those located in the complementary strand, was slightly different [6092 (4.9%) versus 5027 (4.0%) sequences, respectively]. In total, 17 844 PG4s were found in the 5′-UTRs, and are unequally distributed between the two strands. A significantly smaller number of potential G-quadruplex structures was observed in the complementary strand, that is to say in the mRNA, as compared to the template DNA strand (40.3% versus 59.7%, respectively), suggesting potential biological consequences. The same unequal strand distribution is observed in the four other species with >100 PG4s identified, supporting this statement (see Dataset S1). Moreover, a previous study reported the same bias for the distribution of the PG4s between the template and complementary strands (12). Another interesting observation was the higher PG4/5′-UTR ratio observed for the template strand, suggesting that the cell is better able to deal with consecutive G-quadruplexes in the template strand than in the mRNA (Table 1). However, some 5′-UTRs can contain up to five different PG4s in the complementary strand (e.g. ANKRD30B, CAV2 and CDKN2D; Dataset S2). The PG4 density in 5′-UTRs was estimated to be 0.292/kbase for the template strand, and 0.198/kbase for the complementary strand. In both cases, it represents a significant enrichment (4- to 5-fold) as compared to the reported PG4 density of the human genome (0.057 kb) using the same algorithm (12).
Table 1.
Incidence of potential G-quadruplexes in a human 5′-UTR database
| Template strand | Complementary strand | Total | |
|---|---|---|---|
| Nb. of 5′UTR | – | – | 124 315 |
| Nb of 5′UTR with PG4 (%) | 6092 (4.9) | 5024 (4.0) | 9979 (8.0) |
| 5′UTR with 1 PG4 (%) | 3313 (54.4) | 3399 (67.7 | 5133 (51.4) |
| 5′UTR with more than 1 PG4 (%) | 2779 (45.6) | 1625 (32.3) | 4846 (48.6) |
| Nb. PG4 | 10 646 | 7198 | 17 844 |
| % of PG4 | 59.7 | 40.3 | – |
| Ratio PG4/5′UTR | 1.75 | 1.43 | 1.79 |
| PG4 density | 0.292/kb | 0.198/kb | 0.490/kb |
Ability of the selected candidates to fold into G-quadruplex structures in vitro
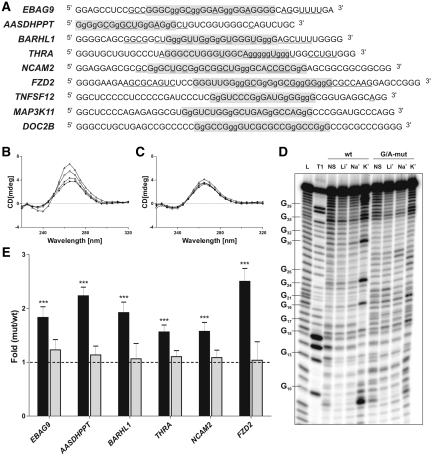
With the goal of evaluating the impact of the G-quadruplex structure on the transcriptome, several candidates were selected with which to continue this study. Specifically, nine 5′-UTRs bearing a PG4 on their complementary strand were chosen based on the bioinformatic analysis (Table 2). The main criterion of selection was that these candidates’ mRNAs had to encode proteins important for various cellular pathways; therefore, they constituted a good representation of gene heterogeneity. The first step was the demonstration of whether or not the candidate’s PG4 sequences adopted a G-quadruplex structure in vitro. Three different biochemical methods were used with each candidate, providing a reliable evaluation of the situation. The experiments were performed using transcripts that exceed the PG4 sequence requirement (i.e. they are longer) in order to better reflect the biological context of each 5′-UTR instead of only considering the guanosine tracks. However, it was not possible to use the complete 5′-UTR sequence, due to both technical constraints and difficulties in analyzing the results. Consequently, in all in vitro experiments the sequence of a PG4 was flanked by ∼15 nt both upstream and downstream, and began with at least two consecutive guanosines as these are required for efficient in vitro transcription (see Figure 1A for the detailed sequences). Moreover, a G/A-mutant created by mutating several guanosines into adenosines in such a way that it prevents the formation of a G-quadruplex structure was also synthesized for each candidate (Figure 1A). These mutants were used as negative controls.
Table 2.
Gene ontology of the nine candidate genes
| Gene | Function | Process |
|---|---|---|
| EBAG9 | Apoptotic protease activator activity | Apoptosis/regulation of cell growth |
| FZD2 | G-protein-coupled receptor activity | G-protein-coupled receptor protein signaling pathway |
| BARHL1 | Protein binding/transcription factor activity | Midbrain development/neuron migration |
| NCAM2 | Protein binding | Cell adhesion/neuron adhesion |
| THRA | Thyroid hormone receptor activity | Hormone-mediated signaling |
| AASDHPPT | Transferase activity | Macromolecule biosynthetic process |
| TNFSF12 | Cytokine activity | Apoptosis/cell differentiation |
| MAP3K11 | JUN kinase kinase kinase activity | Regulation of JNK cascade/cell proliferation |
| DOC2B | Calcium ion binding/transporter activity | Transport |
Figure 1.
Identification of G-quadruplex structures and translational repressors. (A) Sequences of the wild-type PG4s of the nine selected candidates. Sequences of the original potential G-quadruplexes are shaded in gray. The lowercase guanosines (g) correspond to those changed to adenosines in the G/A-mutants. Nucleotides that were cleaved significantly more in the presence of KCl, as compared to in the presence of LiCl, in the in-line probing experiments as determined by the quantification with the SAFA software, are underlined. (B, C) Circular dichroism spectra for the EBAG9 PG4 candidate using 4 µM of either the wild type (B) or the G/A-mutant (C) sequences and either in the absence of salt (close circles) or in the presence of 100 mM LiCl (closed triangles), NaCl (open circles) or KCl (open triangles). (D) Autoradiogram of a 10% denaturing (8M urea) polyacrylamide gel of the in-line probing of the 5′-labeled EBAG9 wild-type and G/A-mutant PG4 versions performed either in the absence of salt (NS), or in the presence of 100 mM LiCl, NaCl or KCl. The lanes designated L and T1 are an alkaline hydrolysis and a ribonuclease T1 (RNase T1) mapping of the wild-type version, respectively. Representative guanosine residues are indicated on the left of the gel. (E) Gene expression levels of the different constructs used at the protein level using either the luciferase assay (black bars) or the mRNA level as determined by RT-qPCR (gray bars). The x-axis identifies the candidates and the y-axis the fold difference that corresponds to the value obtained for the G/A-mutant version divided by that obtained for the wild type version for each candidate. The RT-qPCR values were obtained using the ΔΔCT method with the Fluc gene as internal control and the wild-type version as calibrator. Error bars were calculated using a minimum of three independent experiments. ***P < 0.001.
The first method used for detecting G-quadruplex formation was analysis by CD. This is a classical technique that detects G-quadruplex structures possessing the typical spectrum caused by the topology of the four-stranded helical structure (i.e. parallel or anti-parallel). Due to the nature of its sugar, an RNA G-quadruplex structure is compelled to adopt a parallel form. More specifically, the ribose residues prefer the puckering C3′-endo conformation. This in turn favors that the glycosidic bond of every guanosine involved in the core of G-tetrads be in the anti orientation (42). The formation of a parallel G-quadruplex structure provokes the appearance of a negative peak at 240 nm and a positive one at 264 nm (43). It is important to focus on the transition of both characteristic peaks, when comparing the spectra recorded under two different conditions, in order to propose that the RNA molecule forms a G-quadruplex. The analysis cannot rely on a single spectrum, because other RNA structural features exist that can give a positive peak around 260 nm, as this would lead to a potential false positive G-quadruplex signature. The CD spectra for each candidate were initially recorded either in the absence of salt, or in the presence of 100 mM LiCl, two conditions that do not support the formation of G-quadruplex structures. The presence of Li+ is the most reliable control in order to identify the ‘intrinsic’ or initial structure of the RNA molecule because it provides the same ionic force as Na+ or K+, but it cannot support the formation of a G-quadruplex structure. Subsequently, the experiments were repeated in the presence of 100 mM of either NaCl or KCl, two conditions that should favor the formation of G-quadruplex structures. Panel B of Figure 1 shows the recorded CD spectra for the EBAG9-derived transcripts as an example of a result typical of one positive for G-quadruplex formation, while the panel 1C illustrates the corresponding G/A-mutant. Clearly, there is a significant transition to a higher positive peak at 264 nm, and a negative one at 240 nm, when using the wild-type version in the presence of KCl. No corresponding transition was observed for the G/A-mutant. Six out of nine candidates exhibited CD spectra with G-quadruplex signatures. Specifically, the BARHL1 and NCAM2 PG4 sequences appear to fold into G-quadruplex structures in the presence of either KCl or NaCl, while the EBAG9, FZD2, THRA and AASDHPPT sequences adopt this structure solely in the presence of KCl. Conversely, the TNFSF12, MAP3K11 and DOC2B PG4 sequences did not show any evidence of a G-quadruplex signature, regardless of the nature of the salt present in the buffer. Similarly, the G/A-mutants never exhibited a significant transition characteristic of the formation of a G-quadruplex structure.
With the goal of confirming that some of the PG4 sequences do indeed fold into G-quadruplexes, thermal denaturation studies were then performed. The formation of a G-quadruplex in the presence of an appropriate cation (e.g. Na+ or K+) should lead to an increased stability (i.e. a higher Tm) of the RNA molecule as compared to one containing a structure involving only Watson–Crick base pairs (44). The six PG4 sequences that, according to CD analysis, folded into G-quadruplex structures were examined in this way. The presence of LiCl provokes only a small increase in the Tm as compared to the value obtained in the absence of salt (Table 3). This stabilization of the RNA structure is due to the counterion effect of the cations that reduces the repulsion of the negative charge of the phosphate backbone. Conversely, the presence of KCl in the solution led to a significant increase in the Tm of each PG4 sequence (Table 3). In fact, all of the RNA structures were incompletely denatured, even at 90°C. Clearly, this experiment provides additional physical evidence supporting the conclusion that these six PG4 sequences adopt a G-quadruplex structure at a physiological KCl concentration (i.e. 100 mM). Finally, an increase in the Tm value was also observed in the presence of NaCl, but only for the BARHL1 and NCAM2 PG4-derived sequences, in agreement with the CD analysis. The G/A-mutants exhibited no increase in their Tm values under all conditions, indicating that their structures were not altered by the addition of a monovalent cation such as Na+ or K+ (Table 3). However, relatively high Tm values were obtained for some of the G/A-mutants that we cannot explain so far. Thus, the thermal denaturation and CD analyses were in perfect agreement: if a PG4 sequence folded into a G-quadruplex in the presence of either Na+ or K+, it was detectable with both methods.
Table 3.
Thermal denaturation analysis
| 5′ UTR | No salt | LiCl | NaCl | KCl |
|---|---|---|---|---|
| EBAG9 | ||||
| wt | n.a. | n.a. | 51.4 ± 1.1 | >90 |
| mut | 58.0 ± 1.3 | 72.7 ± 2.1 | 72.2 ± 0.9 | 71.2 ± 1.1 |
| FZD2 | ||||
| wt | 65.1 ± 1.8 | 77.0 ± 1.9 | 79.3 ± 1.7 | >90 |
| mut | 58.8 ± 2.3 | 70.5 ± 0.1 | 68.0 ± 1.5 | 64.0 ± 0.1 |
| BARHL1 | ||||
| wt | 40.1 ± 1.0 | 44.2 ± 1.9 | 70.6 ± 3.1 | >90 |
| mut | 48.2 ± 0.6 | 64.7 ± 1.7 | 61.8 ± 2.7 | 61.2 ± 1.6 |
| NCAM2 | ||||
| wt | 67.6 ± 4.0 | 78.4 ± 1.4 | > 90 | > 90 |
| mut | 68.9 ± 1.7 | 70.4 ± 1.4 | 73.8 ± 2.8 | 72.1 ± 0.2 |
| THRA | ||||
| wt | 67.8 ± 1.4 | 76.0 ± 0.8 | 75.4 ± 1.1 | >90 |
| mut | 82.3 ± 1.1 | 82.4 ± 1.2 | 82.6 ± 2.2 | 81.5 ± 1.0 |
| AASDHPPT | ||||
| wt | 65.5 ± 1.8 | 77.7 ± 0.6 | 75.1 ± 4.7 | >90 |
| mut | 52.4 ± 0.7 | 59.2 ± 1.3 | 61.5 ± 0.2 | 59.1 ± 0.5 |
| snp | 59.9 ± 2.2 | 75.5 ± 3.7 | 75.2 ± 1.7 | 73.7 ± 0.9 |
| TNFSF12 C/A | ||||
| wt | 46.4 ± 3.1 | 60.5 ± 0.2 | 56.9 ± 0.9 | 79.8 ± 0.6 |
| muta | 49.0 ± 1.2 | 59.4 ± 1.9 | 62.5 ± 4.2 | 57.8 ± 1.9 |
| DOC2B C/A | ||||
| wt | 59.8 ± 2.0 | 65.6 ± 0.5 | 68.4 ± 0.1 | 74.1 ± 0.5 |
| mutb | 56.9 ± 0.4 | 64.3 ± 1.9 | 64.5 ± 0.1 | 63.3 ± 2.6 |
| MAP3K11 C/A | ||||
| wt | 59.1 ± 0.5 | 68.1 ± 1.7 | 67.8 ± 0.4 | 73.9 ± 0.3 |
| mutc | 54.0 ± 0.7 | 63.6 ± 0.1 | 64.1 ± 1.4 | 59.4 ± 2.0 |
n.a. The magnitude of the curves did not permit the determination of accurate Tm values.
aCorresponds to the TNFSF12 CG/AA-mutant version.
bCorresponds to the DOC2B CG/AA-mutant version.
cCorresponds to the MAP3K11 CG/AA-mutant version.
CD and thermal denaturation analyses are typical methods used to study G-quadruplex structures. However, because of their requirement for a relatively large amount of RNA (i.e. in the low micromolar range), they do not permit discrimination between the formation of a unimolecular, a bimolecular or a tetramolecular G-quadruplex structure. In the context of a G-quadruplex present in the 5′-UTR of an mRNA, the unimolecular topology is most likely; however, it is not impossible that several mRNAs may interact together through the formation of either a bimolecular or a tetramolecular structure. In order to address this question, an in-line probing was performed on all of the PG4 wild-type and G/A-mutant versions. Trace amounts of 5′-32P-radiolabeled transcripts (<1 nM) were incubated for 40 h in a slightly basic buffer (pH 8.3) that included a relatively high magnesium concentration (20 mM MgCl2), and either in the absence or the presence of monovalent cations (Li+, K+ or Na+). During the incubation, the presence of the magnesium led to the cleavage of the phosphodiester backbone of the single-stranded nucleotides often found at the periphery of the RNA structure (45). If a PG4 sequence adopts a unimolecular G-quadruplex structure, the nucleotides in the loops should bulge out of the RNA’s structure and therefore be susceptible to in-line attack by the magnesium ions. A typical example of an autoradiogram for an in-line attack experiment is illustrated for the EBAG9 PG4-derived sequence in panel D of Figure 1. Clearly, an important difference in the intensity of the banding patterns was observed at several positions of the wild-type PG4 in the presence of 100 mM KCl when compared to all other conditions. Specifically, there was a drastic increase in the intensity of the bands representing the nucleotides located between the guanosine tracks (e.g. C20, A25 and A31), and those corresponding to the loops of the PG4. In addition, the inability of the G/A-mutants to fold into a G-quadruplex structure was confirmed, regardless of the PG4 candidate. In order to provide a reliable evaluation, a quantitative analysis was performed. Briefly, at least two gels for each candidate were exposed to a phosphor screen and revealed by phosphor imaging using a Storm apparatus coupled with the SAFA software for the quantitative analysis (46). The intensity of each band in the K+ lane was divided by that of the corresponding band in the Li+ lane. A nucleotide was considered significantly more accessible when this ratio was higher than an arbitrarily fixed threshold of 2. A summary of all of the accessible nucleotides is shown in panel A of Figure 1. The nucleotides for which the accessibility was significantly modified by the addition of KCl are underlined. These nucleotides were always found to be located between the G-tracts, as well as in the vicinity of the PG4 (which should become single-stranded upon the formation of the G-quadruplex). These results validate the hypothesis that the G-quadruplex structures identified in vitro are able to fold according to a unimolecular topology. The conditions used in this experiment (i.e. trace amount of RNA, <1 nM) should not support the formation of intermolecular G-quadruplexes. It is important to note that, in order to validate this technique, two important controls have been performed in conjunction to the in-line probing experiment for the EBAG9 PG4-derived sequence. First, the impact of a high concentration of magnesium (10 mM) on the G-quadruplex’s formation was tested by CD experiments and it does not interfere with the ability to form a G-quadruplex structure (data not shown). Second, DMS probing experiment was performed in parallel to the in-line probing and many guanines in the tracks identified by bioinformatic were protected only in the presence of 100 mM KCl (data not shown). Finally, the in-line probing data were in perfect agreement with those obtained from both the CD and the thermal denaturation analyses. The same set of six candidates identified in the previous experiments gave a positive G-quadruplex signature in the in-line probing experiments performed in the presence of KCl, while the other three did not. Moreover, only the BARHL1 and NCAM2 PG4-derived sequences appeared to fold into a G-quadruplex structure in the presence of NaCl.
In summary, the three different methods used provided consistent data for the set of PG4 candidates tested (see Table 4 for a summary). The PG4 sequences from the EBAG9, FZD2, BARHL1, NCAM2, THRA and AASDHPPT 5′-UTRs fold into G-quadruplex structures in vitro at a physiological concentration of KCl, while their G/A-mutant versions do not. The TNFSF12-, MAP3K11- and DOC2B-derived sequences do not fold into G-quadruplex structures under these conditions.
Table 4.
Summary of the in vitro and in cellulo analysis of the candidates in terms of their ability to adopt a G-quadruplex structure
| 5′ UTR |
In vitro |
In cellulo (fold) | ||
|---|---|---|---|---|
| CD | In line probing | Tm | ||
| EBAG9 | Yes | Yes | Yes | 1.83 |
| FZD2 | Yes | Yes | Yes | 2.50 |
| BARHL1 | Yes | Yes | Yes | 1.92 |
| NCAM2 | Yes | Yes | Yes | 1.57 |
| THRA | Yes | Yes | Yes | 1.56 |
| AASDHPPT | Yes | Yes | Yes | 2.24a/1.48b |
| TNFSF12 | No | No | – | 1.25 |
| TNFSF12 C/A | Yes | Yes | Yes | 0.38c/1.29d |
| MAP3K11 | No | No | – | – |
| MAP3K11 C/A | Yes | Yes | Yes | – |
| DOC2B | No | No | – | – |
| DOC2B C/A | Yes | Yes | Yes | – |
aFold difference in protein expression for the AASDHPPT G/A-mutant versus the wt version.
bFold difference in protein expression for the AASDHPPT C7 SNP versus the wt version.
cFold difference in protein expression for the TNFSF12 C/A-mutant versus the wt version.
dFold difference in protein expression for the TNFSF12 CG/AA-mutant versus the wt version.
Ability of the identified G-quadruplexes to repress translation in cellulo
Subsequently, the characterization of the G-quadruplexes identified in vitro was performed by verifying their potential effects on translation in cellulo. Both the full-length wild type and the G/A-mutant 5′-UTRs of the candidates folding into G-quadruplexes in vitro were cloned upstream of a luciferase reporter gene (Rluc) (see ‘Materials and Methods’ section). HEK293 cells were then co-transfected with either the wild type or the G/A-mutated Rluc construction and a Fluc reporter gene, thereby permitting the normalization of the transfection efficiency. Cells were harvested 24 h post-transfection and lysed. The resulting lysates were used in luciferase activity assays in order to estimate the quantity of luciferase protein synthesized. The Rluc activity was normalized with the Fluc activity for each sample. A ratio of luciferase activities was calculated by dividing the value determined for the G/A-mutant 5′-UTRs by that of the corresponding wild-type 5′-UTR. This analysis yielded an estimation of the relative differences in luciferase protein resulting from the abolition of the G-quadruplex structure in each case. For example, the EBAG9 G/A-mutated 5′-UTR construct (in which only six guanosines out of a total of 235 nt were substituted for adenosines) produced a 1.8-fold greater level of luciferase activity than did its corresponding wild-type counterpart (Figure 1E). The six G-quadruplex structures studied yielded estimated differences ranging from an increase of 1.56- to 2.50-fold in terms of the quantity of luciferase protein. In other words, the formation of the G-quadruplex structure significantly decreased the level of luciferase expression in all cases.
RNA was also extracted from the above cells and RT-qPCR experiments performed in order to verify if the differences in gene expression occurred at the transcriptional or the post-transcriptional level. This analysis provided an evaluation of the quantity of mRNA produced by each construct. The same normalization methodology was used with both the Fluc gene and the G/A-mutant version. Both the wild-type and the G/A-mutant versions of the EBAG9 5′-UTR produced the same Rluc mRNA level, namely ∼1 (Figure 1E). Similar data were obtained for all of the other candidates. Because the mRNA levels did not vary between the wild-type and the G/A-mutant versions, regardless of the candidate examined, this indicated that the formation of the G-quadruplex structure has a post-transcriptional effect.
The use of a different cell line yielded similar results at both the protein and the RNA levels (i.e. MCF-7 cells, data not shown). Similar experiments, but in which the reporter and normalizer genes were inverted (i.e. 5′-UTR inserted upstream of the Fluc gene), were also carried out and virtually identical data were obtained (data not shown). Together, these results show that all of the PG4 sequences able to fold into G-quadruplexes in vitro repressed the expression levels of two different reporter genes in cellulo, and did so in two different cell lines. Moreover, this repression occurs post-transcriptionally, most likely by repressing the translation level of the mRNA species in question.
Transforming negative candidates into positive candidates
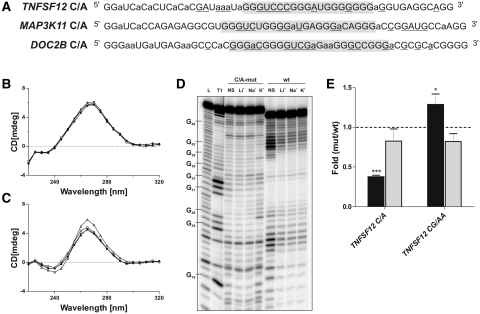
Three of the nine candidates identified by the bioinformatic analysis were shown to be unable to fold into G-quadruplex structures in the presence of KCl (i.e. TNFSF12, MAP3K11 and DOC2B). Since these sequences possessed the requirement of four consecutive guanosine tracts, we wondered why they did not adopt a G-quadruplex structure. Initially, the primary sequences of all of the PG4 sequences used for the in vitro experiments were compared. The first observation made was that these three sequences include significantly more cytosines than did those that folded into G-quadruplexes (Table 5). In fact, the only interesting correlation observed was that a high G/C ratio appeared to be associated with the ability of the PG4 sequence to fold into a G-quadruplex structure (Table 5). The presence of a larger number of cytosines obviously lowers the G/C ratio. The relatively high level of cytosines most likely increases the ability of a given sequence to form stable stem structures resulting from GC Watson–Crick base pair formation. In order to verify this hypothesis, the stabilities, in terms of Gibbs free energy (ΔG) of the predicted secondary structures adopted by all of the PG4 sequences, were estimated using several bioinformatic programs (i.e. mfold, KineFold and MC-Fold) (47–50). The predicted structures of the TNFSF12, MAP3K11 and DOC2B sequences all had lower ΔG values, as compared to the others, regardless of the software used, indicating that they represent the most stable structures (Table 5). In these predicted structures, several of the guanosines required for the G-quadruplex formation where in fact involved in GC Watson–Crick base pairs. This has the effect of stabilizing the rod-like predicted secondary structure (data not shown). It is well known that the rod-like secondary structure is formed relatively rapidly, while G-quadruplex formation usually requires a fairly long period of time (44,51,52). We hypothesized that the presence of a relatively stable secondary structure may prevent the formation of a G-quadruplex. If this is indeed the case, the reduction of the stability of the initial secondary structure should favor the formation of an alternative one that includes a G-quadruplex. Consequently, mutants in which several randomly chosen cytosines were substituted for adenosines were synthesized (i.e. C/A-mutants) (Figure 2A). The number of substitutions was calculated so as to yield a final G/C ratio equal to that of the lowest G/C ratio of the positive candidates, specifically the 2.2 of NCAM2 (Table 5). Moreover, mutants in which important guanosines of the PG4 were mutated to adenosines, in addition to the C/A mutations, were also generated (CG/AA-mutants). In order to verify if these mutants were able to fold into G-quadruplex structures, they were subjected to the in vitro experiments described earlier.
Table 5.
Primary sequence and secondary structure analysis of in vitro PG4s
| Gene | Length (nt) | Nb G | %G | Nb C | %C | %GC | %AT | Ratio G/C | Mfold (kcal/mol) | KineFOLD (kcal/mol) | MC-Fold (kcal/mol) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| EBAG9 | 45 | 26 | 57.8 | 9 | 18.4 | 77.8 | 22.2 | 2.9 | −13.9 | −14.2 | −37.4 |
| FZD2 | 60 | 36 | 60.0 | 13 | 21.7 | 80.0 | 20.0 | 3.0 | −16.8 | −18.6 | −49.2 |
| BARHL1 | 46 | 30 | 65.2 | 5 | 10.9 | 76.1 | 23.9 | 6.0 | −8.5 | −8.6 | −30 |
| NCAM2 | 54 | 33 | 61.1 | 15 | 27.8 | 88.9 | 11.1 | 2.2 | −22.4 | −21.1 | −48.0 |
| THRA | 49 | 28 | 57.1 | 9 | 18.4 | 75.5 | 24.5 | 3.1 | −19.7 | −19.1 | −38.4 |
| AASDHPPT | 38 | 22 | 57.9 | 8 | 21.1 | 79 | 21.0 | 2.8 | −13.7 | −14.2 | −34.2 |
| TNFSF12 | 54 | 23 | 42.6 | 20 | 37.0 | 79.6 | 20.4 | 1.2 | −30.4 | −26.1 | −55.2 |
| TNFSF12 C/A | 54 | 23 | 42.6 | 10 | 18.5 | 61.1 | 38.9 | 2.3 | −10.2 | −13.1 | −42.5 |
| MAP3K11 | 55 | 26 | 47.3 | 16 | 29.1 | 76.4 | 23.6 | 1.6 | −23.8 | −23.1 | −54.0 |
| MAP3K11 C/A | 55 | 26 | 47.3 | 10 | 18.2 | 65.5 | 34.5 | 2.6 | −12.3 | −14.5 | −47.8 |
| DOC2B | 56 | 28 | 50.0 | 24 | 42.9 | 92.9 | 7.1 | 1.2 | −31.6 | −27.4 | −57.6 |
| DOC2B C/A | 56 | 28 | 50.0 | 12 | 21.4 | 71.4 | 28.57 | 2.3 | −9.7 | −10.0 | −43.5 |
Figure 2.
Rescue of G-quadruplex structures in vitro and in cellulo. (A) Sequences of the C/A-mutant PG4 versions for each of the three candidates that did not initially fold into G-quadruplex structures. The original PG4 sequences are highlighted in gray. Lowercase adenosines (a) were cytosines in the wild-type changed to adenosines in the C/A-mutant versions. Underlined nucleotides correspond to those that were cleaved significantly more in the presence of KCl, as compared to in the presence of LiCl in the in-line probing experiment after quantification with the SAFA software. (B, C) Circular dichroism spectra for the TNFSF12 candidate using 4 µM of either the wild-type (B) or the C/A-mutant versions (C) and either in the absence of salt (close circles) or in the presence of 100 mM LiCl (close triangles), NaCl (open circles) or KCl (open triangles). (D) Autoradiogram of a 10% denaturing (8 M urea) polyacrylamide gel of in-line probing of the 5′-end labeled TNFSF12 C/A-mutant and wild-type PG4 versions performed either in the absence of salt (NS) or in the presence of 100 mM LiCl, NaCl or KCl. The lanes designated L and T1 are an alkaline hydrolysis and an RNase T1 mapping of the C/A-mutant version, respectively. Representative guanosine residues are indicated on the left of the gel. (E) Gene expression levels of the different constructs used at either the protein level, determined using luciferase assays (black bars), or the mRNA level, determined using RT-qPCR (gray bars). The x-axis identifies the mutated version of the TNFSF12 candidates, and the y-axis the fold difference corresponding to the value obtained for either the C/A-mutant or the CG/AA-mutant version divided by the value obtained for the wild-type version of TNFSF12. The RT-qPCR values were obtained using the ΔΔCT method with the Fluc gene as internal control and the wild-type version as calibrator. Error bars were calculated using a minimum of three independent experiments. *p < 0.05 and ***P < 0.001.
First, CD spectra experiments were performed on all of the C/A-mutants. The C/A-mutant of TNFSF12 produced a shift to the G-quadruplex characteristic spectrum in the presence of KCl, while no change was observed for the corresponding wild type sequence (Figure 2B and C). Similar results were obtained for the C/A-mutants of both MAP3K11 and DOC2B (Table 4). Second, thermal denaturation experiments corroborated the CD results. Specifically, all of the C/A-mutants showed a significant increase in the Tm value in the presence of KCl, while those of the CG/AA-mutants remained relatively constant. The increase in stability observed in the presence of KCl is a good indication that G-quadruplexes structures were adopted by the C/A-mutants. Finally, the in-line probing experiments also added to the physical evidence that the C/A-mutants fold into G-quadruplexes, while their wild-type counterparts adopt rod-like structures involving GC Watson–Crick base pairs. For example, the wild-type TNFSF12 sequence’s probing gel showed that the cytosine-rich sequence located from positions 5 to 13 most likely interacted with the guanosine-rich sequence located in positions 31–39 (Figure 2D). This helical region includes seven GC, one AU and one GU base pairs out of a total of 18 nt from both strands. Therefore, it appears reasonable to suggest that its formation impaired the G-quadruplex formation. In the case of the corresponding C/A-mutant, stronger bands corresponding to the loop appeared in the presence of KCl only between the guanosine tracts, and in the middle of the seven guanosine long tract (Figure 2D). As observed before, this pattern is characteristic of a G-quadruplex structure. Similar data were also obtained for the MAP3K11 and DOC2B candidates. Clearly, the in-line probing experiments support the initial hypothesis that the stable secondary structures formed by these sequences prevent the formation of the G-quadruplexes. Together, these approaches make a strong case for explaining why, even though the TNFSF12, MAP3K11 and DOC2B sequences possess all of the basic requirements for the adoption of a G-quadruplex structure in the presence of the KCl, they instead fold into a stable secondary structure containing a relatively long double-stranded helical domain. However, it should be noted that the introduction of mutations that destabilize this initial secondary structure favors the formation of the corresponding G-quadruplex structures.
Subsequently, whether or not this G-quadruplex rescue (i.e. the C/A-mutants) had the ability to repress translation in cellulo was investigated. The appropriate plasmid constructions (i.e. full-length wild-type, C/A-mutant and CG/AA-mutant 5′UTR versions for TNFSF12) were cloned upstream of the Rluc reporter gene, transfected into HEK293 cells and the gene expression analyzed at both the protein and mRNA levels as described previously. Astonishing decreases in the amounts of protein synthesized for the C/A-mutant, as compared to those for the wild-type version, were observed (Figure 2E and Table 4). Specifically, in the case of the C/A-mutant of TNFSF12, a 2.6-fold less level of protein was detected. The CG/AA-mutant showed a small increase of 1.29-fold, at the protein level, giving a net repression estimated to be 3.3-fold by the TNFSF12 G-quadruplex. In all the cases, the mRNA level remained the same (Figure 2E). Thus, these results confirmed that by modifying the initial secondary structure, it was possible to modulate the formation of the G-quadruplex in vitro as well as in cellulo.
SNPs in 5′-UTR G-quadruplexes
According to the results presented here, it appears reasonable to suggest that G-quadruplexes located in the 5′-UTRs of mRNAs act as translational repressors of several genes in human cells. Therefore, it is logical to wonder if variability exists in these repressors, and, if yes, can this variability change the level of repression between individuals. A bioinformatic search was performed in order to identify SNPs within the human PG4 sequences from the UTRef collection of the UTRdb database (Dataset S3). A total of 327 SNPs were found in 271 different PG4 sequences with a distribution of 184 SNPs in 155 PG4 sequences located in the template strand, and 143 SNPs in 116 PG4 sequences located in the complementary strand (see Dataset S4). Thus, 5.0% of all PG4 sequences included at least one SNP. The PG4 with the highest number of SNPs was found in the 5′-UTR of the dihydrofolate reductase mRNA at position 35 (RefSeq: NM_000791). It contains a total of eight different SNPs.
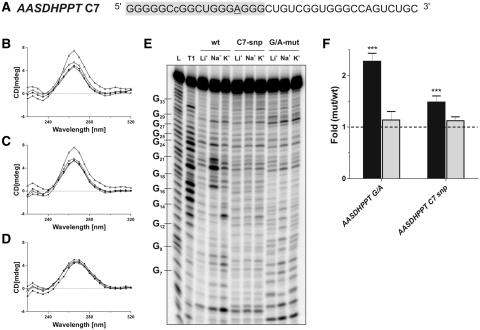
Interestingly, an SNP was identified in one of our initial candidates, namely AASDHPPT. It consisted of a substitution for the guanosine located in position 7 by a cytosine (Figure 3A). This guanosine was previously shown to be important for the formation of the G-quadruplex structure by the in-line probing analysis (Figure 1A). In order to investigate if this single substitution found in some individuals can affect not only the formation of the G-quadruplex structure, but also its ability to repress translation, the same set of in vitro and in cellulo experiments as described previously were performed. CD spectra analysis showed that both the wild type (G7) and SNP (C7) PG4 versions exhibited a G-quadruplex signature in the presence of KCl, while the G/A-mutant did not (Figure 3B–D). Conversely, the thermal denaturation experiment showed no increase in the Tm value in the presence of KCl for the SNP (C7), suggesting that the G-quadruplex structure was not adopted (Table 3). Similarly, the in-line probing gel displayed no specific structural rearrangement in the presence of KCl for the SNP (C7) version, result comparable to that observed for the G/A-mutant. It is noteworthy that the nucleotides located in the PG4 loops tended to become more accessible in the wild type (G7) sequence under these conditions (Figures 1A and 3E). These banding patterns demonstrated that, in the presence of trace amounts of RNA, the wild-type (G7) version can fold into a G-quadruplex structure in vitro in the presence of KCl while both the SNP (C7) and G/A-mutant versions did not. The discrepancy observed with the CD spectra may result from the large amount of RNA required by this method (i.e. micromolar quantities). This result suggests that CD analysis may provide misleading results.
Figure 3.
Effects of a SNP in a 5′-UTR G-quadruplex. (A) Sequence of the C7 SNP PG4 version of the AASDHPPT candidate. The gray box indicates the sequence of the original PG4 identified by the algorithm. The lowercase cytosine (c) corresponds to the guanosine changed to a cytosine in the C7 SNP version. The underlined nucleotides correspond to the nucleotides that were cleaved significantly more in the presence of KCl, as compared to in the presence of LiCl, in the in-line probing experiment after quantification with the SAFA software. (B–D) Circular dichroism spectra for the AASDHPPT candidate using 4 µM of either the wild-type (B), the C7 SNP (C) or the G/A-mutant versions (D) and either in the absence of salt (close circles) or in the presence of 100 mM LiCl (close triangles), NaCl (open circles) or KCl (open triangles). (E) Autoradiogram of a 10% denaturing (8 M urea) polyacrylamide gel of the in-line probing of 5′labeled wild-type, C7 SNP and G/A-mutant PG4 versions of AASDHPPT performed either in the absence of salt (NS) or in the presence of 100 mM LiCl, NaCl or KCl. The lane designated L and T1 are an alkaline hydrolysis and an RNase T1 mapping of the wild-type version, respectively. Representative guanosine residues are indicated on the left of the gel. (F) Gene expression levels of the different constructs used at the protein level, as determined using luciferase assays (black bars), or at the mRNA level, as determined using RT-qPCR (gray bars). The x-axis identifies the mutated version of AASDHPPT, and the y-axis the fold difference corresponding to the value obtained for either the G/A-mutant or C7 SNP version divided by the value obtained for the wild-type version. The RT-qPCR values were obtained using the ΔΔCT method with the Fluc gene as internal control and the wild-type version as calibrator. Error bars were calculated using a minimum of three independent experiments. ***P < 0.001.
Subsequently, the full-length wild type, SNP (C7) and G/A-mutant versions of the AASDHPPT 5′-UTR sequence were cloned upstream of the Rluc reporter gene. After the transfection of HEK293 cells, the levels of gene expression were monitored at both the protein and the mRNA levels by comparing either the G/A-mutant to the wild type (G7), or the SNP (C7) to the wild type (G7). Increases of 2.24- and 1.48-fold at the protein level were observed for the G/A-mutant and SNP (C7) sequences, respectively. At the mRNA level, no variation was observed in both cases (Figure 3F and Table 4), clearly showing that both the G/A-mutant and SNP (C7) versions were able to disrupt, or at least weaken sufficiently, the G-quadruplex structure leading to an increase in the translation of the downstream gene.
DISCUSSION
The importance of the G-quadruplexes found in RNA molecules, in terms of the life cycle of the cell, remains to be appreciated. In fact, the field appears to be only in its infancy. Some researchers are investigating the physical rules that surround RNA G-quadruplex structures, while others try to find an associated biological role. The bioinformatic search reported here, as well as a previously reported one (12), demonstrate that sequences potentially capable of forming G-quadruplexes are located in thousands of 5′-UTRs. This observation led to the formulation of the hypothesis that G-quadruplexes are in fact translational repressors that are involved in various pathways within the cell. When analyzing the 5024 different human mRNAs possessing at least one PG4 in their 5′-UTR retrieved in this work, we found not only their presence noteworthy, but also the fact that they were found in a broad variety of genes in terms of gene ontology. For example, PG4 sequences were enriched in many of the mRNAs encoding the proteins involved in transcription regulation, mRNA transcription, protein modification, G-protein-mediated signaling, cation transport and developmental processes, to name only a few examples (with P-values of 6.4 × 10−19, 2.2 × 10−14, 4.3 × 10−14, 7.8 × 10−10, 4.5 × 10−9 and 2.7 × 10−8, respectively; see Dataset S5). However, it is important to consider that this type of bioinformatic search would undoubtedly overestimate the real prevalence and impact of G-quadruplex structures in the 5′-UTRs of the transciptome because it is based solely on sequence criteria. This point is well illustrated here by the fact that only six out of the nine PG4 candidates tested did in fact fold into a G-quadruplex structure (according to the in vitro experiments performed). However, if the resulting percentage of true G-quadruplex is indicative of all possibilities (67%), it suggests that there still are several thousand G-quadruplexes located exclusively in 5′-UTRs.
In order to obtain a reliable evaluation of the importance of the presence of G-quadruplexes in the 5′-UTRs of mRNAs, we selected nine PG4 sequences retrieved in the mRNAs encoding proteins belonging to various cellular pathways. Of these, classical methods such as CD analysis and thermal denaturation (using a version smaller than the active 5′-UTR) provided consistent data indicating that six of the sequences were in fact folding into G-quadruplex structures (Figure 1 and Table 4). Specifically, the CD spectroscopy led to the observation of a G-quadruplex characteristic spectrum transition, while the thermal denaturation permitted the observation of higher Tm values in the presence of Na+ or K+, as compared to that expected for structures based on Watson–Crick base pairs. In order to provide additional physical support, in-line probing experiments were also performed. To our knowledge, this represents the first time that this technique was extensively used to analyze G-quadruplex structures, although it is routinely used to characterize other biologically relevant RNA structures such as riboswitches (45). In-line probing is simple to perform, and does not require important RNA concentrations as compared to the other usual techniques. In addition, it is an efficient, reproducible and reliable method for studying RNA structure. The use of only trace amounts of RNA (<1 nM) should favor the formation of unimolecular structures while reducing the probability of intermolecular ones. This study provides data in agreement with the physico-chemical approaches, as well as permitting the determination of specific physical features such as the positions and the nucleotides of the loops of the G-quadruplex structures. All of the G-quadruplexes identified by in-line probing, possessed three loops intercalated by four guanosine tracks, whose results support the formation of a unimolecular G-quadruplex. However, this does not discard the possibilities that under certain conditions these G-quadruplex structures may involve more than one RNA molecule. Moreover, the in-line probing gels permitted the determination of the nature of the inhibitory secondary structure formed by the three candidates that initially could not fold into a G-quadruplex. Clearly, in-line probing appears as a method of choice and compared to conventional enzymatic and chemical footprinting experiments: it does not require either special chemicals or biochemical reagents (e.g. high-grade DMS and specific ribonucleases) or specific characteristics of the sequence (i.e. nature and accessibility of the nucleotides). Finally, it was striking to observe that the six G-quadruplexes identified in vitro repressed translation in cellulo in the context of their full-length 5′-UTR.
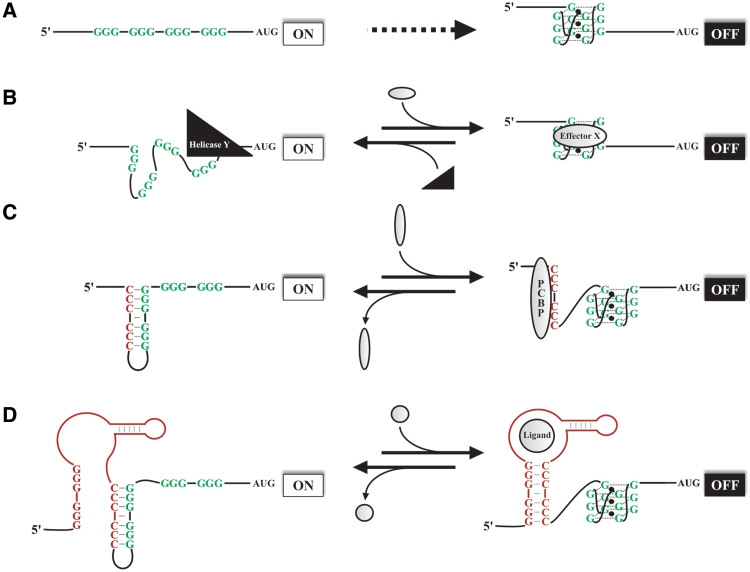
Taken together, the data from the in vitro and in cellulo experiments showed that the G-quadruplex structures are, indeed, very important in 5′-UTR sequences, specifically because of their ability to repress translation. In light of these results, they appear to be a key component in translational regulation (Figure 4A). Probably the easiest way to illustrate this is shown in Figure 4A where the simple presence of a guanosine-rich sequence in a 5′-UTR, in conjunction with the appropriate thermodynamic parameters, is sufficient to form a G-quadruplex structure. An interesting subsequent question to ask would be how this structure can be modulated in both space and time in the cell. G-quadruplex structures can certainly be the targets of specific proteins that decrease the minimal energy required for their formation; however, specific helicases have also been reported to unwind these stable RNA structures (Figure 4B) (26,53,54). Alternatively, the three candidates that did not fold into G-quadruplex structures bring another level of complexity to the situation. Clearly, it is not simply because a 5′-UTR contains a PG4 sequence that it forms a G-quadruplex in the presence of K+. The nature of the nucleotides located in the vicinity of the PG4 sequence is important in determining whether or not a G-quadruplex structure is adopted. In some cases, the G-quadruplex structure is not favored over stable secondary structure based solely on Watson–Crick base pairs. The presence of cytosine tracks appears to be detrimental to G-quadruplex formation, as they interact and form stable stem secondary structures with the guanosine tracks. In this case, the driving forces involved in the G-quadruplex folding pathway will be too weak to promote its formation. Nevertheless, this characteristic could be implicated in the regulation of the formation of new G-quadruplexes. For instance, it increases the range of proteins potentially involved in these mechanisms to include poly(C)-binding proteins, or stem–loop RNA helicases, which could disrupt the inhibitory secondary structure thereby allowing the G-quadruplex formation to proceed (Figure 4C). The GC stem secondary structures could also be disturbed by the RNA itself. As is observed for riboswitches, an RNA aptamer present either upstream or downstream of the G-quadruplex could change the local structure of the RNA upon the binding of a metabolite, thereby leading to the removal of the inhibitory GC stem (Figure 4D). The G-quadruplex would act as the expression platform of such a riboswitch. With the RNA G-quadruplexes field growing rapidly, the discovery of more RNA G-quadruplex regulators will become essential to accurately defining their different roles.
Figure 4.
Proposed models for the regulation by 5′-UTR G-quadruplexes. Representations of different means of regulation by 5′-UTR G-quadruplexes. The general symbols are as follows: green G, the guanosines involved in the G-quadruplex structure; AUGs, the initiation codon of the open reading frames; white (ON) and black (OFF) rectangles, reflect the translation status; and, small black spheres, represent the monovalent cation needed for the G-quadruplex formation (most likely K+). (A) Simplest model for the G-quadruplex formation based solely on favorable thermodynamic parameters. (B) Modulation of the G-quadruplex formation by proteins interacting directly with the G-quadruplex structure. The black triangles represent a helicase with the specific activity of unwinding G-quadruplex structures. Gray horizontal ovals represent a protein that binds G-quadruplex structures and either promotes its formation, and/or stabilizes the final structure. (C) The red C corresponds to the cytosine tracks that can interact, by the formation of Watson–Crick base pairs, with some of the guanosine tracks involved in the G-quadruplex structure. The gray vertical ovals represent a poly(C)-binding protein (PCBP) that is able to bind the cytosine tracks and disrupt the initial inhibitory secondary structure, thereby allowing the formation of the G-quadruplex. (D) The red parts correspond to an RNA aptamer able to bind a specific ligand (gray spheres). The binding of the ligand promotes the final folding step of the aptamer and the formation of a new stem involving both the cytosine and guanosine tracks of the aptamer. This rearrangement removes the initial inhibitory secondary structure and allows the formation of the G-quadruplex.
SNP occurs when a single nucleotide in the genome differs between the members of a species. SNPs are also involved in several diseases (e.g. cancer and Alzheimer’s), and can be related to how a person will react to a specific treatment (55). After having analyzed the impact of the flanking sequences of the G-quadruplex on their formation, the effect of a change in the sequence of the G-quadruplex itself was investigated. From all the bioinformatic results presented here, the database of SNPs inside 5′-UTR PG4s (Dataset S4) clearly represents the most important novelty in the field concerning in silico information available and should be of general interest for researchers working in many fields. At least one SNP was found in 116 different 5′-UTR PG4s located on the complementary strand. Several of the corresponding genes are known to be implicated in various diseases (e.g. the RAD51 (NM_002875) and CAV2 (NM_001233) genes in cancer). The presence of the SNP within the AASDHPPT 5′-UTR, which was used as a model PG4 sequence, abolishes the G-quadruplex structure formation in vitro and increases the translation of a reporter gene in cellulo. These results suggest that two individuals could have a different expression for a given gene due to the difference in their PG4 SNP. Thus, SNPs located in 5′-UTR G-quadruplexes might be involved either in the predisposition, or in the appearance of, various diseases and cancers by altering the gene expression background of a specific individual. However, the bioinformatic approach used most likely identifies only the SNPs that lead to the abolition of a G-quadruplex because it searched for the presence of an SNP inside an already discovered PG4. Moreover, it has been shown that the sequences adopting a non B-DNA structure (e.g. G-quadruplexes) possessed higher levels of polymorphism (56). Consequently, there is a higher error frequency in these sequences during the DNA replication. This fact adds to the importance of exploring the presence of SNPs in G-quadruplexes. In summary, a deeper analysis of SNPs in G-quadruplex structures remains essential, but this study provides an original proof-of-principle of their relevancy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Canadian Institutes of Health Research (CIHR, grant number MOP-44022 to J.-P.P.); the Université de Sherbrooke and the CIHR (grant number PRG-80169 to the RNA group). J.D.B. was the recipient of pre-doctoral fellowships from both the CIHR and the Fonds de Recherche en Santé du Québec (FRSQ). J.-P.P. holds the Canada Research Chair in Genomics and Catalytic RNA and is member of the Centre de Recherche Clinique Étienne-Lebel. Funding for open access charge: The Canadian Institutes of Health Research (CIHR, grant number MOP-44022 to J.-P.P.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge the technical assistance of Patrice Coulombe. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
REFERENCES
- 1.ENCODE Project Consortium. Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halbeisen RE, Galgano A, Scherrer T, Gerber AP. Post-transcriptional gene regulation: from genome-wide studies to principles. Cell. Mol. Life Sci. 2008;65:798–813. doi: 10.1007/s00018-007-7447-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;13:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martick M, Horan LH, Noller HF, Scott WG. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature. 2008;454:899–902. doi: 10.1038/nature07117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 2009;78:305–34. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neidle S, Balasubramanian S. Quadruplex Nucleic Acids. Cambridge: RSC Publishing; 2006. [Google Scholar]
- 8.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huppert JL. Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chem. Soc. Rev. 2008;37:1375–1384. doi: 10.1039/b702491f. [DOI] [PubMed] [Google Scholar]
- 10.Verma A, Halder K, Halder R, Yadav VK, Rawal P, Thakur RK, Mohd F, Sharma A, Chowdhury S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 2008;51:5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- 11.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:403–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du Z, Zhao Y, Li N. Genome-wide colonization of gene regulatory elements by G4 DNA motifs. Nucleic Acids Res. 2009;37:6784–6798. doi: 10.1093/nar/gkp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008;36:1321–1333. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 16.Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl Acad. Sci. USA. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell. Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palumbo SL, Memmott RM, Uribe DJ, Krotova-Khan Y, Hurley LH, Ebbinghaus SW. A novel G-quadruplex-forming GGA repeat region in the c-myb promoter is a critical regulator of promoter activity. Nucleic Acids Res. 2008;36:1755–1769. doi: 10.1093/nar/gkm1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paramasivam M, Membrino A, Cogoi S, Fukuda H, Nakagama H, Xodo LE. Protein hnRNP A1 and its derivative Up1 unfold quadruplex DNA in the human KRAS promoter: implications for transcription. Nucleic Acids Res. 2009;37:2841–2453. doi: 10.1093/nar/gkp138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saccà B, Lacroix L, Mergny JL. The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 2005;33:1182–1192. doi: 10.1093/nar/gki257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kostadinov R, Malhotra N, Viotti M, Shine R, D’Antonio L, Bagga P. GRSDB: a database of quadruplex forming G-rich sequences in alternatively processed mammalian pre-mRNA sequences. Nucleic Acids Res. 2006;34:D119–D124. doi: 10.1093/nar/gkj073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 27.Gomez D, Lemarteleur T, Lacroix L, Mailliet P, Mergny JL, Riou JF. Telomerase downregulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Res. 2004;32:371–39. doi: 10.1093/nar/gkh181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonnal S, Schaeffer C, Créancier L, Clamens S, Moine H, Prats AC, Vagner S. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J. Biol. Chem. 2003;278:39330–39336. doi: 10.1074/jbc.M305580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arora A, Dutkiewicz M, Scaria V, Hariharan M, Maiti S, Kurreck J. Inhibition of translation in living eukaryotics cells by an RNA G-quadruplex motif. RNA. 2008;14:1290–1296. doi: 10.1261/rna.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris MJ, Basu S. An unusually stable G-quadruplex within the 5′-UTR of the MT3 matrix metalloproteinase mRNA represses translation in eukaryotic cells. Biochemistry. 2009;48:5313–5319. doi: 10.1021/bi900498z. [DOI] [PubMed] [Google Scholar]
- 32.Halder K, Wieland M, Hartig JS. Predictable suppression of gene expression by 5′-UTR-based RNA quadruplexes. Nucleic Acids Res. 2009;37:6811–6817. doi: 10.1093/nar/gkp696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mignone F, Grillo G, Licciulli F, Iacono M, Liuni S, Kersey PJ, Duarte J, Saccone C, Pesole G. UTRdb and UTRsite: a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2005;33:D141–D146. doi: 10.1093/nar/gki021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobs GH, Chen A, Stevens SG, Stockwell PA, Black MA, Tate WP, Brown CM. Transterm: a database to aid the analysis of regulatory sequences in mRNAs. Nucleic Acids Res. 2009;37:D72–76. doi: 10.1093/nar/gkn763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macke T, Ecker D, Gutell R, Gautheret D, Case DA, Sampath R. RNAMotif – A new RNA secondary structure definition and discovery algorithm. Nucleic Acids Res. 2001;29:4724–4735. doi: 10.1093/nar/29.22.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 37.Beaudoin JD, Perreault JP. Potassium ions modulate a G-quadruplex-ribozyme’s activity. RNA. 2008;14:1018–1025. doi: 10.1261/rna.963908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mergny JL, Lacroix L. UV Melting of G-Quadruplexes. Curr. Protoc. Nucleic Acid Chem. 2009 doi: 10.1002/0471142700.nc1701s37. Chapter 17: Unit 17.1. [DOI] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang CF, Shafer RH. Engineering the quadruplex fold: nucleoside conformation determines both folding topology and molecularity in guanine quadruplexes. J. Am. Chem. Soc. 2006;128:5966–5973. doi: 10.1021/ja0603958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paramasivan S, Rujan I, Bolton PH. Circular dichroism of quadruplex DNAs: applications to structure, cation effects and ligand binding. Methods. 2007;43:324–331. doi: 10.1016/j.ymeth.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 44.Lane AN, Chaires JB, Gray RD, Trent JO. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods Mol. Biol. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- 46.Laederach A, Das R, Vicens Q, Pearlman SM, Brenowitz M, Herschlag D, Altman RB. Semiautomated and rapid quantification of nucleic acid footprinting and structure mapping experiments. Nat. Protoc. 2008;3:1395–1401. doi: 10.1038/nprot.2008.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 48.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xayaphoummine A, Bucher T, Isambert H. Kinefold web server for RNA/DNA folding path and structure prediction including pseudoknots and knots. Nucleic Acid Res. 2005;33:605–610. doi: 10.1093/nar/gki447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 51.Onoa B, Tinoco I., Jr RNA folding and unfolding. Curr. Opin. Struct. Biol. 2004;14:374–379. doi: 10.1016/j.sbi.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 52.Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu Y, Shin-ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Creacy SD, Routh ED, Iwamoto F, Nagamine Y, Akman SA, Vaughn JP. G4 resolvase 1 binds both DNA and RNA tetramolecular quadruplex with high affinity and is the major source of tetramolecular quadruplex G4-DNA and G4-RNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2008;283:34626–34634. doi: 10.1074/jbc.M806277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shastry BS. SNPs in disease gene mapping, medicinal drug development and evolution. J. Hum. Genet. 2007;52:871–880. doi: 10.1007/s10038-007-0200-z. [DOI] [PubMed] [Google Scholar]
- 56.Wells RD. Non-B DNA conformations, mutagenesis and disease. Trends Biochem. Sci. 2007;32:271–278. doi: 10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.