Abstract
For DNA replication to occur, chromatin must be remodeled. Yet, we know very little about which proteins alter nucleosome occupancy at origins and replication forks and for what aspects of replication they are required. Here, we demonstrate that the BRG1 catalytic subunit of mammalian SWI/SNF-related complexes co-localizes with origin recognition complexes, GINS complexes, and proliferating cell nuclear antigen at sites of DNA replication on extended chromatin fibers. The specific pattern of BRG1 occupancy suggests it does not participate in origin selection but is involved in the firing of origins and the process of replication elongation. This latter function is confirmed by the fact that Brg1 mutant mouse embryos and RNAi knockdown cells exhibit a 50% reduction in replication fork progression rates, which is associated with decreased cell proliferation. This novel function of BRG1 is consistent with its requirement during embryogenesis and its role as a tumor suppressor to maintain genome stability and prevent cancer.
INTRODUCTION
DNA replication occurs during S-phase of the cell cycle to duplicate each chromosome into two sister chromatids with a high degree of fidelity. As a prerequisite, a highly coordinated series of biochemical events must occur from early G1 to the G1-S-phase transition. During early G1, six ORC proteins assemble as origin recognition complexes (ORCs) at origins of replication throughout the genome at ∼25-kb intervals (1). In mammalian cells, these sites are presumably determined epigenetically because they are ubiquitous and do not share a consensus DNA sequence. Not all origins are competent to initiate replication, but many are licensed to do so when minichromosome maintenance (MCM) complex proteins 2–7 are loaded in an ORC1/Cdc6- and Cdt1-dependent manner to form pre-replicative complexes (pre-RCs) (2). At the G1-S-phase transition, CDC7 and CDK2 promote the recruitment of CDC45 and GINS complexes to a subset of pre-RCs, now considered pre-initiation complexes (pre-ICs), leading to activation of the MCM helicase, which collaborates with at least 20 additional cell-cycle proteins to initiate DNA replication in a bi-directional manner (3,4). Some initiation factors are also crucial for elongation. For example, proliferating cell nuclear antigen (PCNA) functions as a trimeric clamp that surrounds the DNA to increase DNA polymerase processivity and replication fork progression. All of the aforementioned steps culminate in replicons of ∼60–100 kb, which are unevenly distributed throughout the genome but emanate from every second or third origin on average (5,6). Finally, ∼10 neighboring replicons often coalesce as ∼1 Mb replication foci.
DNA replication is undoubtedly much more complicated than portrayed by current working models, as described above, because the replication machinery must interact with a nucleosomal template rather than naked DNA. For example, the molecular basis for selecting origins of replication in mammalian cells is not known because they do not share a common DNA sequence, but nucleosome phasing and covalent histone modifications are leading candidates. Histone acetylation is a particularly good candidate for the timing of DNA replication because licensed origins that fire early during S phase, such as gene-rich segments, tend to be hyperacetylated, whereas late-replicating sites such as heterochromatic regions are often hypoacetylated (7,8). This correlation is compelling for numerous genes that undergo X chromosome inactivation (XCI), genomic imprinting or allelic exclusion (9). At many of these loci, a specific origin that is in close proximity to a promoter will be hyperacetylated, replicate early, and be transcribed; in contrast, the identical DNA sequence on the homologous chromosome will be hypoacetylated, replicate late, and not be transcribed (10,11). This process appears to be complicated, involving Mbp intervals of DNA changing subnuclear position (12). It is also not clear whether transcription influences replication timing or vice versa in these cases (9,13). However, replication asynchrony is first observed during early embryonic development, which precedes the monoallelic expression that usually occurs much later in more differentiated cell types [e.g., imprinted genes in the placenta and central nervous system (CNS), odorant receptors in olfactory epithelium, IgH in B cells] (9). Therefore, the effect of histone acetylation on replication timing is apparently direct or at least not secondary to transcription.
Chromatin is also an impediment to DNA polymerases and must be remodeled for efficient replication fork progression (14). Following the removal of H1 linker histones, which allows 30-nm solenoid structures to unravel into 10-nm nucleosomal arrays, histone octamers are removed in a two-step process just ahead of the fork. H2A-H2B dimers are removed by ‘facilitates chromatin transcription’ (FACT) and then H3–H4 tetramers are removed by ASF1 (anti-silencing function 1) (14). Both of these histone chaperones directly (FACT) or indirectly (ASF1) interact with the MCMs, which might contribute to their recruitment to sites of DNA replication (14,15). In addition to acting as histone acceptors, FACT and ASF1 probably act as histone donors by interacting with CAF1 (chromatin assembly factor 1) to redeposit histone octamers onto sister chromatids immediately behind the replication fork (16). Both recycled parental histones and newly synthesized histones contribute to these newly formed nucleosomes (17,18).
Identification and characterization of chromatin-modifying factors that participate in DNA replication will greatly increase our understanding of how this process occurs efficiently in the condensed chromatin environment of the interphase nucleus. SWI/SNF-related complexes are good candidates because of their well-characterized role in nucleosome remodeling (19–22). These 1–2 MDa complexes are recruited to specific sites in the genome and confer DNA-dependent ATPase activity to break DNA–histone contacts and alter nucleosome conformation and/or position (23). For example, when recruited to promoters by sequence-specific transcription factors, SWI/SNF-related complexes can slide or evict nucleosomes away from transcription start sites to enable RNA polymerase II to initiate transcription. Although the vast majority of SWI/SNF-related studies have focused on transcriptional regulation, this does not exclude them from playing a role in DNA replication. For instance, FACT and ASF1 also function in transcription, and transcriptional data have been drawn upon to provide insight into their function in replication (14). More importantly, SWI/SNF was first discovered in Saccharomyces cerevisiae where it is required for efficient replication of at least one autonomous replication sequence (24), which is equivalent to an origin of replication, and swi/snf mutant strains grow slowly (20–22). In addition, remodels structure of chromatin (RSC) complexes are similar to SWI/SNF in their biochemical composition and activity but are 10-fold more abundant and essential for yeast viability (20–22).
Similarly, mammalian SWI/SNF-related complexes are essential based on null mutations of the BRG1 (brahma-related gene 1, also known as SMARCA4) catalytic ATPase or two other core subunits (BAF155 and SNF5/BAF47) as well as one non-core subunit (BAF250) that each confer early embryonic lethality in mice (19,22,25,26). To circumvent this lethality, which precludes a functional assessment of their role in transcription and replication, we previously used ethylnitrosourea (ENU) to generate a Brg1 hypomorphic mutation in the mouse. This Brg1ENU1 mutant allele encodes a BRG1 protein with an E1083G substitution in the catalytic ATPase domain that is stable and assembles into SWI/SNF-related complexes (27). However, it has diminished chromatin-remodeling properties, and Brg1null/ENU1 mutants die at midgestation due to severe anemia. Utilizing these mutants, we have previously defined the role of BRG1 in chromatin remodeling/looping and transcription of the α and β globin loci (27–30) and now report a novel function for BRG1 in DNA replication.
MATERIALS AND METHODS
Mice, BrdU and histology
All mouse experiments were performed using protocols approved by the Institutional Animal Care and Use Committees (IACUC) of UNC and in accordance with federal guidelines. Timed matings were initiated, and BrdU was injected i.p. (0.1 mg per gram body weight) 12 days after detection of a copulation plug. Embryos were removed 2–4 h after the injection, fixed in 4% paraformaldehyde, and processed for the production of paraffin sections. Sections (5–8 µm) were stained with a BrdU staining Kit (Zymed) or stained with hematoxylin and eosin (H&E).
Flow cytometry/sorting
Flow cytometry was performed as previously described (27). Briefly, fetal livers (FLs) were triturated in phosphate-buffered saline (PBS) containing 2% fetal bovine serum (FBS) using 1-cc syringes fitted with needles having progressively smaller apertures (18, 22 and 25 gauges). Approximately 2 × 106 cells were incubated with anti-KIT-FITC, anti-TER119-PE and Annexin V-APC (each from Pharmingen) on wet ice in the dark for 1 h. Cells were washed several times and analyzed using FACScan and MoFlow instruments (Becton Dickinson). For each experiment, wild-type cells were also incubated with each antibody alone as well as no antibody or Annexin V to serve as compensation controls.
Reverse transcription quantitative polymerase chain reaction
RNA was prepared using Trizol reagent (Invitrogen) and reverse-transcribed using a mixture of random hexamers and Oligo d(T) (BioRad iScript) according to standard procedures. Validated TaqMan assays (Applied Biosystems) were used with TaqMan gene expression master mix (Applied Biosystems) on an ABI7300 instrument under default cycling conditions (95°C 15 s followed by 60°C 1 min for 45 cycles). Gapdh was used as a normalization control. Relative expression levels were determined using the ΔΔCt method.
Plasmids and transfections/RNA interference
The methodology for knocking down BRG1 in MiaPaCa2 cells and BRM in LNCaP have already been described (31,32). For the D98OR HeLa knockdown cells, which have BRG1 and BRM reductions, we first established a stable cell line (D98OR Brg1i-11) with BRG1 knocked down using an identical procedure as for the MiaPaCa2 cells (31).
All transfections were performed using Lipofectamine 2000TM (Invitrogen) per the manufacturer’s instructions on cells that had been equally plated in 100 mm culture plates for stable transfections. Cells were plated in antibiotic-free media and allowed to attain 70–80% confluency in a 37°C/5% CO2-supplemented/100% humidity incubator prior to transfection. pHTPBrmi and pcDNA3 (Invitrogen, CMV driven with G418 resistance gene) were transfected into D98orBrg1-11 using a total of 40 ng of DNA in a respective 5 : 1 ratio. Transfectant material was replaced after 24 h with fresh RPMI 1640 supplemented with 10% FBS. After 24 h, the cells were released by trypsin-treatment and equally divided among 8–100-mm culture plates. For selection, puromycin (2 µg/ml) and G418 (1 µg/ml) was applied and maintained for 10–21 days after which single colony clones were captured, isolated in 24-well plates, and allow to mature. Protein was harvested and western blots run to verify the status of the target proteins as described (31).
Nucleotide analog labeling of DNA
For chromatin fiber studies, EdU was added to NHF1 and SW13 cultures at a final concentration of 30 µM. For FL cells, pregnant mice were injected i.p. with 420 µl of 10 mM EdU. FLs were removed and placed on ice between 15 and 25 min after injection. For DNA fiber studies, pregnant mice were injected i.p. with 20 mM IdU and after 11 min they were then injected i.p. with 100 mM CldU. FLs were removed and placed on ice 5 min after injection. For the tissue culture studies, D98OR or Mia PaCa2 cells were incubated with IdU at a final concentration of 50 µM for 10 min, washed with PBS twice and then incubated with CldU at a final concentration of 100 µM for 20 min. Afterwards, the cells were washed and prepared for DNA fiber analysis.
DNA fiber studies
For the preparation of the DNA fiber spreads upon slides, 2 μl of a 200 cell/μl cell suspension were spread on a SILANEPREPTM slide (Sigma-Aldrich, S4651), close and parallel to the label. The sample was allowed to evaporate until almost, but not completely dry and then overlaid with 10 μl of spreading buffer [0.5% SDS in 200 mM Tris–HCl (pH 7.4), 50 mM EDTA]. After ∼10 min the slide was tilted at ∼15° to allow the cell lysate to slowly move down the slide, and the resulting DNA spreads were air-dried, fixed in 3:1 methanol/acetic acid for 2 min, air-dried overnight, then stored at −20°C for at least 24 h. The staining of the IdU and CldU tracks (red and green, respectively) in the DNA fibers were performed as described previously (33–36). Microscopy was carried out using an Olympus FV500 confocal microscope using the sequential scanning mode.
Statistical analyses
In the experiments comparing replication track lengths, statistical significance was determined using Student’s two-tailed t-test.
Extended chromatin fiber studies
Extended chromatin fibers were prepared and immunofluorescence was performed essentially as previously described (37). Briefly, cells were pelleted and resuspended in warm hypotonic buffer (0.075 M KCl) at 37°C for 20 min. After hypotonic buffer treatment, approximately 8000 cells were cytospun onto Superfrost Plus slides (Fisher Scientific). After removal of excess fluid, 20 µl of a lysis buffer (25 mM Tris, pH 7.5, 0.5 M NaCl, 0.75% Triton X-100, and 0.2 M urea) that contains 4′,6-diamidino-2-phenylindole (DAPI; Sigma, 0.2 mg/ml) was added and the liquid immediately covered with a 22 × 22 mm square coverslip. Slides were incubated overnight protected from light. DAPI staining allowed for the visualization of chromatin fibers and slides were chosen for further processing based on the quality of chromatin fiber extension.
Primary antibodies and dilutions used: BRG1 (G-7) diluted 1 : 50, ORC1 (N-17) diluted 1 : 100, PCNA (FL-261) diluted 1 : 200, all from Santa Cruz Biotechnology Inc., Santa Cruz CA; ORC2 diluted 1 : 200, Stressgen Bioreagents Victoria BC; PSF2 (1 : 2000) from Abcam Inc., Cambridge MA; acetyl-H3 (1 : 100), Upstate Cell Signaling Solutions, Temecula, CA. Anti-BRG1 (J1) was diluted 1 : 100 was a generous gift from Drs. W. Wang and J. Crabtree. Secondary antibodies were diluted 1 : 200 and were Alexa Fluor conjugates purchased from Invitrogen. All incubations were conducted in a moist chamber at room temperature. Microscopy was carried out using an Olympus FV500 confocal microscope using the sequential scanning mode. Photomicrographic images were analyzed using Image J software (Rasband, W.S., ImageJ, National Institutes of Health, Bethesda, Maryland, USA, 1997-2004) with UCSD (http://rsb.info.nih.gov/ij/plugins/ucsd.html) and McMaster Photobionics (Colocalization Highlighter; Pierre Bourdoncle, Institut Jacques Monod, Service Imagerie) plugins. Results were analyzed and graphed using Microsoft Excel 2007.
Co-IP experiment
Immunoprecipitations were performed on E12.5 FL cells using anti-BRG1 (J1) as previously described (30). Western blots were performed using a TOPBP1 antibody (Signal Transduction Laboratories) as described (38).
RESULTS
BRG1 regulates cell proliferation during development
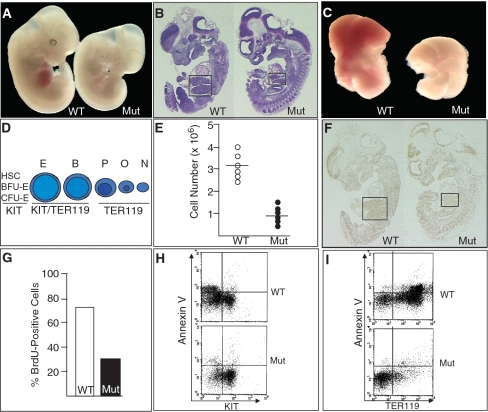
The overall body size of Brg1null/ENU1 mutants at embryonic day (E) 12.5 is smaller than wild-type controls (Figure 1A). Mutant FLs are particularly small based on gross anatomical examination as well as H&E staining of parasagittal sections (Figure 1A and B). Dissection of mutant FLs highlights their pallor and decreased size (Figure 1C). We previously demonstrated that the pallor is a result of reduced α and β globin transcription and fewer hemoglobin-containing cells due to a partial developmental block at the transition from basophilic erythroblasts to polychromatic erythroblasts (Figure 1D). To quantify the smaller size of mutant FLs, we performed cell counts and determined that they have a ∼3-fold reduction in cell number (Figure 1E).
Figure 1.
BRG1 regulates cell proliferation in erythroid cells of the fetal liver. (A) Photograph comparing wild-type (WT) and mutant (Mut) E12.5 embryos. (B) Parasagittal sections from WT and Mut E12.5 embryos stained with H&E. The sections are in the optimal focal plane for visualizing the FLs, which are outlined by boxes. (C) Photograph of FLs removed from E12.5 embryos. (D) Schematic of the erythroid lineage in the FL from undifferentiated (left) to differentiated (right). HSC, hematopoietic stem cell; BFU-E, burst forming unit-erythroid; CFU-E, colony forming unit-erythroid; E, proerythroblast; B, basophilic erythroblast; P, polychromatic erythroblast; O, orthochromatic erythroblast; N, non-nucleated reticulocyte. Beneath each cell type is the expression status of the cell-surface markers KIT (a marker of hematopoietic progenitors) and TER119 (a pan-erythoid marker). (E) Hemocytometer-based cell counts of WT and Mut FLs. (F) BrdU incorporation of WT and Mut E12.5 embryos using sections adjacent to those shown in (B). FLs are outlined by boxes. (G) Percentage of BrdU-positive cells from WT and Mut E12.5 FLs. (H) Flow cytometry of E12.5 FLs showing the Annexin V status of KIT-positive and –negative cells. (I) Flow cytometry of E12.5 FLs showing the Annexin V status of TER119-postive and -negative cells.
The decreased number of mutant FL cells could be due to decreased cell proliferation and/or increased apoptosis so we performed bromodeoxyuridine (BrdU) incorporation and Annexin V assays, respectively. BrdU incorporation was markedly reduced in mutant FLs (Figure 1F), and quantification at high magnification revealed a 2- to 3-fold decrease (Figure 1G), which is consistent with the decreased cell number. In contrast, no difference was observed for Annexin V staining using flow cytometry (Figure 1H and I). These experiments utilized two cell-surface markers: 1, KIT, a stem/progenitor cell marker of multiple hematopoietic lineages (including erythroid); 2, TER119, a pan-erythroid marker. Due to the partial developmental block, mutants have a normal number of KIT-positive cells (Figure 1H, right side) but a decreased number of KIT-negative cells (Figure 1H, left side). The KIT-negative cells are primarily TER119-positive polychromatic erythroblasts, orthochromatic erythroblasts and reticulocytes (Figure 1D) and are underrepresented in mutants (Figure 1I, right side). However, Annexin V is not elevated in either KIT- or TER119-positive mutant cells, indicating that the mutant cells were not undergoing apoptosis more frequently than normal (Figure 1H and I). In addition, we did not detect a difference in trypan blue vital dye staining (data not shown), which indicated the mutant cells were not undergoing necrosis. These results indicate that BRG1 regulates cell proliferation but not cell death in the FL.
The Brg1 mutant phenotype is most severe in the erythroid lineage because Brm is not expressed and cannot compensate
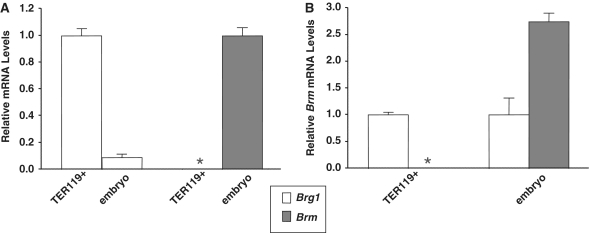
The cell proliferation defect in Brg1 mutants is more severe in the FL than other cell types of the E12.5 embryo. One explanation is that erythroid cells of the FL proliferate more rapidly than most or all other tissues at this stage of development. A second explanation, which is not mutually exclusive, relates to functional redundancy. We reasoned that Brm, which encodes an alternative catalytic subunit 75% identical to BRG1 with similar nucleosome remodeling properties, might functionally compensate in most embryonic cell types but not in the FL. To test this hypothesis, we compared Brg1 and Brm mRNA levels by reverse transcription quantitative polymerase chain reaction (RT-qPCR). Brg1 is expressed at high levels in TER119 flow-sorted erythroid cells of the FL with mRNA levels ∼10-fold greater than the rest of the embryo (Figure 2A). In contrast, Brm is not expressed in TER119 cells but is strongly expressed in the rest of the embryo (Figure 2A). In fact, Brm mRNA levels are nearly 3-fold higher than Brg1 in the whole embryo (Figure 2B). A survey of adult tissues also showed that Brm expression is as high or higher than Brg1 in each organ that was analyzed (Supplementary Figure 1). These results indicate that erythroid cells of the FL are unusual because Brm is not expressed, and, as a corollary, suggest that these cells exhibit a particularly severe phenotype in Brg1 mutants because Brm cannot compensate.
Figure 2.
Brm is not expressed in erythroid cells of the fetal liver. (A) RT-qPCR analysis of Brg1 and Brm mRNA levels normalized to Gapdh levels in wild-type flow-sorted erythroid cells (TER119+) and whole mouse embryos minus their fetal livers (embryo) at E12.5. The white histograms on the left show Brg1 expression of embryos relative to TER119+ samples. The gray asterisk on the right indicates Brm mRNA levels are below the limit of detection in TER119+ samples and is compared to Brm expression in embryoss (gray histogram). Each histogram represents the mean ± SE for two (TER119+) or three (embryo) independent experiments. (B) Brm mRNA levels relative to Brg1 mRNA levels based on data presented in (A). For TER119+ (left) and embryo samples (right), Brg1 mRNA levels (white histograms) are set at 1.0 and Brm mRNA levels (gray asterisk/histogram) are shown in proportion. The asterisk indicates Brm was analyzed for TER119 samples but mRNA was not detected.
BRG1 is required for efficient replication fork progression
BRG1 activates transcription of cyclin-dependent kinase inhibitors (p16INK4A and p21CIP1/WAF1) and physically associates with retinoblastoma (RB) to inhibit cell-cycle progression in vitro (39–46). However, it is not known whether this mechanism is significant in vivo. For example, based on our previous gene expression profiling experiments on flow-sorted erythroid cells from wild-type versus mutant FLs, we did not identify any genes in the RB pathway or other cell-cycle regulatory factors as candidates to explain the cell proliferation phenotype described above (27). In addition, the gene expression profiles of Brg1 mutant mammary tumors, which are heterogeneous in nature, do not exhibit altered expression of cell-cycle genes in the RB pathway (47).
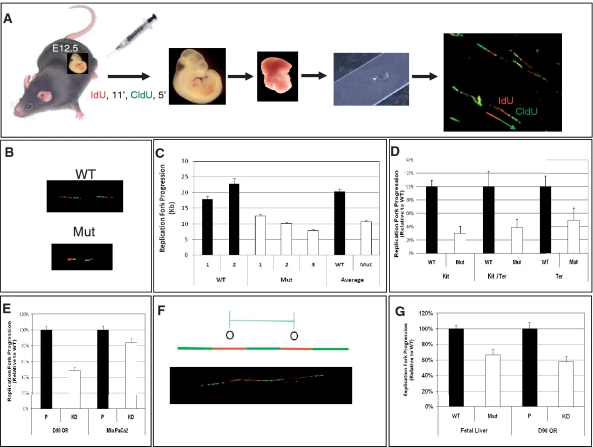
Therefore, we hypothesized that BRG1 might not function as a transcriptional regulator in this context but instead plays a more direct role in the process of DNA replication. To analyze the dynamics of DNA replication, two fluorescently labeled nucleotide analogs were sequentially incorporated into E12.5 embryos in vivo, and DNA fibers were subsequently prepared from dissected FLs and visualized by confocal microscopy (Figure 3A). Adding two analogs sequentially allowed us to determine the directionality of replication and to identify origins of replication as well as continuing and terminating replication forks (33,36). We did not observe a significant difference in the relative ratio of replication intermediates (origin initiations, active replication forks or forks that terminated) between wild-type and mutant samples, suggesting that there is no difference in replication competency (Supplementary Figure 2).
Figure 3.
BRG1 is required for efficient replication fork progression. (A) Schematic of the procedure. Pregnant female mice are injected i.p. with fluorescently labeled IdU (red) followed by CldU (green), E12.5 embryos are removed, FLs are dissected and single-cell suspensions are spotted onto microscope slides. The FL cells are lysed and confocal microscopy is utilized to visualize newly synthesized DNA as pulse-labeled tracks of IdU (red) and CldU (green) incorporation. (B) Representative confocal image of WT and Mut DNA fibers showing decreased track lengths in Muts. (C) Quantification of track lengths from over 200 DNA fibers derived from multiple WT controls (numbered 1 and 2) and Muts (numbered 1, 2 and 3) with average values also presented. The Muts have a significantly smaller track lengths than WT controls (P < 0.0001). (D) Track lengths of Mut relative to WT for flow-sorted cell populations from FLs. (E) Knockdown of BRG1 decreases replication progression rates in D98OR HeLa cells (P < 0.0001) and MiaPaCa2 cells (P < 0.014). Chart shows the relative lengths of replication tracks in parental (P) and knockdown (KD) cells. (F, G) Genetic perturbation of BRG1 results in decreased inter-origin distances. (F) Illustration of the criteria used to measure inter-origin distances. (G) Quantification of inter-origin distances in WT and Mut FL cells or parental (P) and knockdown (KD) D98OR cells. At least 25 inter-origin distances were measured in each sample. Error bars in (C–E) and (G) represent standard errors of the mean.
However, these experiments did demonstrate that replication forks from wild-type samples incorporated significantly more IdU and CldU than mutant samples (Figure 3B). Quantification of track lengths revealed a 20-kb average length for wild-type samples compared to a 10-kb average for mutants (Figure 3C). This method provides an accurate measurement of DNA replication rate: decreased track length of mutant cells is due to reduced incorporation of nucleotide analogs into nascent DNA during the labeling interval (33). These data indicate Brg1 mutants have a 50% reduction in the rate of replication fork progression. Notably, RNAi-mediated knockdown of factors required for replication fork stability such as timeless, claspin and Chk1 also result in a ∼50% reduction of fork progression (36,48).
To rule out the possibility that the altered cellularity of mutant FLs is responsible for their decreased replication fork progression, we repeated our DNA fiber experiments on independent samples that were isolated as specific cell populations using flow sorting. Following IdU and CldU incorporation in vivo, FLs from several WT and mutant E12.5 embryos were pooled and stained with KIT and TER119 to obtain three distinct populations: KIT-positive cells (i.e. hematopoietic stem and progenitor cells), KIT/TER119 double-positive cells (i.e. proerythroblasts and basophilic erythroblasts) and TER119-positive cells (i.e. orthochromatic and polychromatic erythroblasts, and reticulocytes although they lack nuclei and will not incorporate IdU or CldU) as depicted schematically in Figure 1D. In each of these cell types, similar to whole FLs, fork progression rates are decreased ∼50–70% in mutants (Figure 3D). These findings confirm that BRG1 is required for efficient replication fork progression.
To further confirm that BRG1 is required for efficient replication fork progression, we repeated our DNA fiber experiments using an independent experimental system. For these experiments, we performed RNAi to knockdown BRG1 in a HeLa cell subclone (D98OR) and a human pancreatic carcinoma cell line deficient for BRM (MiaPaCa2) (31,49). BRM was also knocked down in the D98OR cells to prevent it from compensating and to resemble the situation in FL cells, which do not express BRM (Figure 2). Western blot analysis demonstrated a robust knockdown (Supplementary Figure 3). Compared to parental controls, the knockdown cells had a ∼50% and ∼20% reduction in replication fork progression in the D98OR and MiaPaCa2, respectively (Figure 3E). The D98OR knockdown phenocopies the FL results.
Many licensed replication origins are not utilized during S phase, but remain dormant and thus the region in which they reside is replicated by neighboring origins (50–52). These dormant origins can be activated when replication is inhibited or slowed and become essential for the completion of replication and cell survival (53–57). The activation of dormant origins can be inferred when the average distance between origins (the inter-origin distance) decreases (56,57). We wondered whether mutant FL cells or D98OR cells lacking BRG1 and BRM have a decreased inter-origin distance as compared to controls since these cells replicate slower. In each case, we found that the cells lacking BRG1 and BRM had a reduced inter-origin distance (∼35% and ∼45%, respectively) (Figure 3F). This decrease in inter-origin distance suggests that cells lacking BRG1 and BRM try to compensate for the reduced rate of replication by initiating more origins and increasing the density of replication forks.
BRG1 co-localizes with replication factors at sites of DNA replication on extended chromatin fibers
We recently analyzed the distribution of ORCs, GINS complex proteins, and PCNA at high resolution by performing immunofluorescence (IF) confocal microscopy on extended chromatin fibers (37). To determine whether BRG1 is involved in DNA replication, we used this same methodology to evaluate the distribution of BRG1 relative to these DNA replication associated proteins. Control experiments demonstrated that two different BRG1 antibodies co-localize on extended chromatin fibers from normal human fibroblasts (NHF1-hTERT cells) (Supplementary Figure 4A). We also demonstrated, as a negative control, a lack of BRG1 staining on extended chromatin fibers prepared from the BRG1-deficient SW13 cancer cell line (Supplementary Figure 4B). The BRG1 antibody used for the rest of this study (Santa Cruz G-7) has been used on western blots by many groups including our own to demonstrate it is absolutely specific (27).
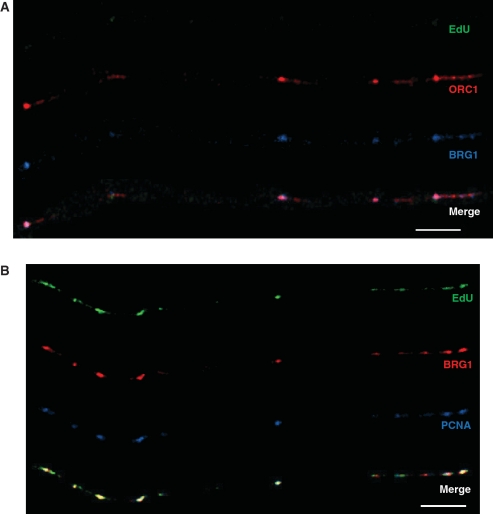
Next, we analyzed extended chromatin fibers from E12.5 FLs and identified significant overlap between BRG1 and ORC1 (Figure 4A) as well as between BRG1 and PCNA at sites of DNA replication (Figure 4B). To detect active sites of DNA replication, the nucleotide analog EdU was injected i.p. into pregnant female mice and incorporated into E12.5 embryos immediately before FLs were dissected and processed. EdU was used for these studies because, unlike IdU and CldU, sites of incorporation can be detected without removing chromatin-bound proteins or denaturing the DNA. Overall, ORC1 co-localized with BRG1 35% the time, while BRG1 co-localized with ORC1 68% of the time (11 fibers, ∼59 Mb). BRG1 co-localized even more strongly with PCNA (Figure 4B), and this co-localization was highly enriched at sites of DNA replication based on the presence of EdU (Figure 4B). Measurement of extended chromatin fibers from mouse FLs revealed that PCNA and BRG1 signal co-localized 84% of the time at sites of replication (14 fibers, ∼70 Mb) compared to only 15% at sites not undergoing replication. It is true that BRG1 is associated with enhancers and promoters in the context of transcription and that some of these elements might coincide with origins of replication. However, loci undergoing DNA replication are not transcribed simultaneously [(58) and references within]. Therefore, BRG1 that co-localizes with the replication protein PCNA at sites of EdU incorporation must be functioning in the context of DNA replication.
Figure 4.
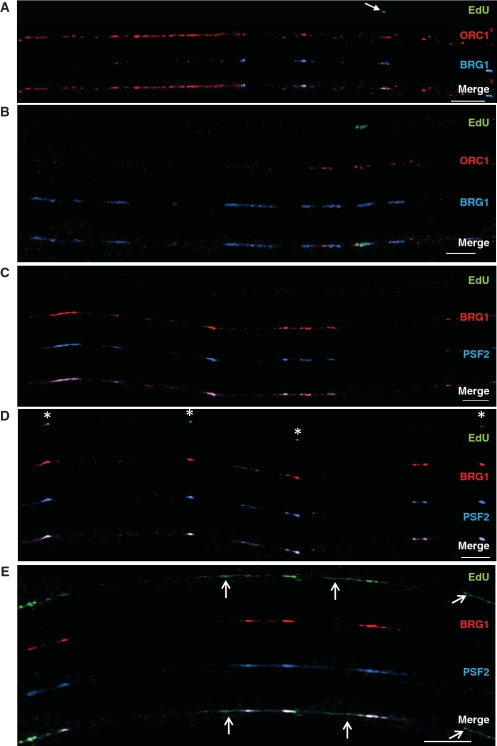
BRG1 co-localizes with ORC1 and PCNA at sites of DNA replication on chromatin fibers from mouse fetal liver cells. (A) ORC1 (red signal) and BRG1 (blue signal) can be seen on this fiber and overlap at several sites. EdU (green signal) is not observed on this fiber, which is consistent with the fact that the vast majority of ORCs are bound to DNA in G1 prior to the onset of DNA replication. (B) BRG1 (red signal) co-localizes with PCNA (blue signal) exclusively at sites of active DNA replication based on EdU incorporation (green signal). Bar ∼25 µm (∼400 kb; bottom right of each panel).
To assess BRG1 occupancy in more detail, we used normal human fibroblasts (NHF1-hTERT cells) because of our previous experience visualizing and quantifying replication proteins on extended chromatin fibers from these cells. First, we analyzed BRG1 occupancy in comparison to ORC1 and tracks of recently replicated DNA (Figure 5A and B). To detect active sites of DNA replication, cells were incubated with EdU for 20 or 30 min, as indicated, before being collected. In our previous study, we found that ORC1 is densely distributed on fibers before replication begins (presumably cells in G1 and early S phase) and dissociates from sites after replication is completed (37). GINS complex proteins and PCNA also dissociate from chromatin following replication, and this is thought to prevent re-replication.
Figure 5.
BRG1 co-localizes with ORC1 and PSF2 to different extents on extended chromatin fibers from human fibroblasts. Normal human fibroblasts were labeled with EdU for 20 min (A–D) or 30 min (E). Bars ∼25 µm (∼400 kb; bottom right of each panel). (A) The left side of this chromatin fiber has an abundance of ORC1 (red signal), little BRG1 (blue signal), and no replication tracks. The right side of the fiber has several regions where BRG1 co-localizes with ORC1, including one site that is undergoing DNA replication (green EdU labeling, arrow). (B) This fiber has a higher density of BRG1 than the fiber in (A) where DNA replication activity is marked by green EdU labeling. About half of this fiber has no bound ORC1 and most likely represents a region where ORCs have disassociated from the chromatin after DNA replication occurred (prior to the addition of the EdU label). (C) Both BRG1 (red signal) and the GINS protein PSF2 (blue signal) can be found along this fiber but with no EdU, suggesting that these proteins co-localize on chromatin before DNA replication begins. (D) BRG1, GINS complexes and EdU (green signal, white asterisks) are all present. The small tracks of EdU co-localize with BRG1 and PSF2. (E) Increased incubation time with EdU allows for the visualization of longer tracks of newly replicated DNA. Sites of co-localization of BRG1 and PSF2 can still be found, but there are also regions of EdU that are devoid of these two proteins (arrows). The localization of BRG1, PSF2 and DNA replication tracks on this fiber suggests that as DNA replication in a given replicon is completed, BRG1 and GINS disassociate from chromatin.
The left side of the fiber in Figure 5A shows a high density of ORC1 staining that is typical for sites that have not yet replicated. Alternatively, the right side of the same fiber has a much sparser distribution of ORC1 and at least one active site of replication (arrow). BRG1 overlap with ORC1 is relatively weak at sites that have not replicated but relatively strong where DNA replication is ongoing (the left and right side of the fiber in Figure 5A, respectively). The fiber in Figure 5B also has a sparse distribution of ORC1 that overlaps BRG1 regions, particularly at sites of EdU incorporation. This distribution pattern suggests BRG1 is not involved in the binding of ORCs to origins of replication but has a subsequent function in licensing pre-RCs and/or facilitating the initiation or elongation of replication.
BRG1 was also bound to some sites that likely had already replicated, as indicated by lack of ORC1 binding, suggesting it participates in a process other than DNA replication (Figure 5B and Supplementary Figure 5). This finding is not unexpected considering that BRG1 has a well-established role in transcriptional regulation. To quantify the extent of BRG1-ORC1 co-localization, we analyzed 19 fibers corresponding to ∼93 Mbp of DNA. BRG1 co-localizes with ORC1 62.8% of the time while ORC1 co-localizes with BRG1 55.2% of the time (333 BRG1 sites, 374 ORC1 sites). We also compared the distribution of BRG1 relative to the GINS complex protein PSF2. As we previously reported (37), GINS complex proteins are found on chromatin fibers before DNA replication begins (Figure 5C; absence of EdU staining) and during DNA replication (Figure 5D; asterisks at sites of EdU incorporation). The fibers in Figure 5C–E also show the distribution of BRG1 relative to PSF2. We found that BRG1 overlaps with PSF2 52% of the time while PSF2 overlaps with BRG1 78% of the time (23 fibers, 118 Mb, 376 BRG1 sites, 260 PSF2 sites). This level of co-localization is nearly as high as the overlap between two members of the GINS complex (85% for PSF1 and PSF2) (37). PCNA associates with BRG1 on extended chromatin fibers at levels similar to PSF2 (Figure 6A and B). BRG1 overlaps with PCNA 52% of the time, and PCNA overlaps with BRG1 73% of the time (22 fibers, 132 Mb, 408 BRG1 sites, 289 PCNA sites). These data provide further support that BRG1 in not part of the pre-RC but is instead part of licensed pre-ICs plus elongating replication forks.
Figure 6.
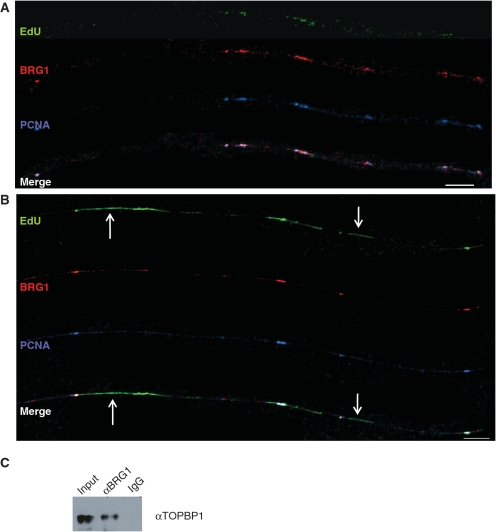
BRG1 co-localizes with PCNA and physically associates with TOPBP1. (A, B) Analysis of BRG1 and PCNA on extended chromatin fibers from human fibroblasts. Normal human fibroblasts were labeled with EdU for 30 min. Bars ∼25 µm (∼400 kb; bottom right of each panel). (A) Both BRG1 (red signal) and PCNA (blue signal) can be seen at sites where replication is beginning (green signal, EdU). (B) Longer tracks of newly replicated DNA can be seen on this fiber. Sites of co-localization of BRG1 and PCNA can still be found, but there are also regions of EdU that are devoid of these two proteins (arrows). (C) BRG1 physically associates with TOPBP1. Shown is a western blot probed with αTOPBP1. Samples include E12.5 FLs (Input) and protein lysates immunoprecipitated with αBRG1 (BRG1) or IgG as a negative control.
The fibers shown in Figure 5A–D were from cells incubated with EdU for 20 min. As mentioned above, when regions complete replication, GINS complex proteins and PCNA disassociate from chromatin. To determine whether this is also the case for BRG1, we increased EdU labeling times to 30 min (Figured 5E and 6A and B), which gave us a wider range of EdU track sizes. In the fiber shown in Figure 5E, regions with EdU staining are devoid of PSF2 and BRG1 (arrows), indicating that the timing of BRG1 disassociation from chromatin in these newly replicated regions is similar to that for PSF2. Likewise, Figure 6A shows that short tracks of EdU (green EdU staining) still have bound BRG1 and PCNA, while regions with longer tracks of newly replicated DNA in Figure 6B (arrows) are devoid of PCNA and BRG1.
The co-localization of BRG1 with replication factors at sites of DNA replication suggests BRG1 is at replication forks. To provide further support for this idea, we performed co-immunoprecipitation (co-IP) assays. These experiments demonstrate that BRG1 physically associates with TOPBP1, which is a component of the replication fork and a sensor of replication stress (Figure 6C). This finding suggests that BRG1 is recruited to DNA replication forks by TOPBP1, and there is evidence that this could be mediated by the BRCT domain of TOPBP1 (38). This result is also compatible with BRG1 mutants having a reduction in replication fork progression since this is known to trigger replication stress (36,48,59–61).
DISCUSSION
To provide additional insight into the mechanism(s) of DNA replication, including its role in development and cancer, it will be necessary to determine the temporal–spatial and functional relationship between chromatin-remodeling factors and the DNA replication machinery. Here, we demonstrate that the BRG1 catalytic subunit of mammalian SWI/SNF-related complexes is physically associated with TOPBP1 and co-localizes with ORCs, GINS complexes and PCNA at sites of DNA replication on extended chromatin fibers. The strength of this approach is that native chromatin is visualized at high resolution (∼16 kb/µm) and the DNA replication status for specific regions of the genome (i.e. not started, in progress or completed) can be determined. As a result, we are able to conclude that BRG1 does not play a role in the binding of ORCs to origins of replication, referred to as site selection, but is instead involved in the firing of origins and the process of replication elongation. The latter conclusion is supported by our observation that the efficiency of replication fork progression in Brg1 mutants and knockdown HeLa cells is reduced to 50% of normal.
Considering that BRG1 breaks DNA–histone contacts to alter the conformation and position of nucleosomes, we propose BRG1 changes nucleosome structure at replication forks to facilitate the removal of H2A–H2B dimers by FACT and/or H3–H4 tetramers by ASF1. This model is supported by several lines of evidence. First, BRG1 and other SWI/SNF subunits are present with FACT in non-canonical WINAC complexes. Second, RSC complexes cooperate with another histone chaperone, NAP1 (nucleosome assembly protein 1), to remove H2A–H2B dimers from nucleosomes in vitro (62). Third, the Brg1 ortholog snf2 exhibits a genetic interaction with asf1 in S. cerevisiae based on double mutants exhibiting a more severe phenotype than either single mutant (63). Moreover, in Drosophila melanogaster,brahma (brm) and two other swi/snf genes (swi3/moira/Baf155 and swi1/osa/Baf250) genetically interact with asf1, and these complexes associate with ASF1 based on co-IP and GST pull-down assays (64). These physical associations, along with our BRG1–TOPBP1 interaction data, provide a plausible explanation for how BRG1 is recruited to sites of replication. ASF1 is known to bind bromodomain-containing proteins, and the bromodomains of BRG1 and BAF180 may reinforce binding to sites with acetylated histones (65,66). Some of our own preliminary data support this notion. On chromatin fibers from normal human fibroblasts during S phase, sites of H3K9 acetylation were found 70% of the time at sites of BRG1 occupancy (10 fibers, 47.5 Mb, 111 H3K9ac sites, 152 BRG1 sites) (Supplementary Figure 6). This correlation was even higher at sites of active replication; 87% of sites with co-localization of H3K9ac and EdU also had BRG1. It is also possible that the bromodomains of BRG1 and BAF180 bind to acetylated lysines on other proteins. Recent proteomic data indicate BRG1 and other SWI/SNF subunits are acetylated and replication-associated protein(s) such as MCMs are frequently acetylated compared to other proteins (67).
More experiments will be required to determine precisely how BRG1 alters nucleosome conformation and/or position to promote octamer disassembly at the replication fork. BRG1 can slide or evict nucleosomes, which is referred to as cis- and trans-displacement, respectively (23). BRG1 can also create a small loop of DNA that protrudes away from the octamer to make the nucleosome more accessible without moving it; this might be an intermediate step for cis- and trans-displacement, but other ATPase chromatin remodelers such as SNF2H apparently cannot perform this function (23). In addition, we and others have demonstrated recently that BRG1 can form chromatin loops between enhancers and promoters to facilitate transcription (29,68,69). Chromatin loops also have been shown to occur at replication foci during S phase so BRG1 might be involved in this process as an additional possibility.
The data presented here demonstrate BRG1 plays a novel role in a fundamental biological process, DNA replication, which is compatible with its known function in embryonic development and cancer prevention. For example, Brg1 null homozygotes die at the blastocyst stage just prior to implantation (25). Although it might have been assumed that this early embryonic lethality is due to deregulated transcription, it is difficult or impossible to ascertain the relative contribution of defective transcription and DNA replication. In this regard, it is notable that null mutations of many replication-associated genes such as Psf1, which encodes a GINS complex member, also cause early embryonic lethality (70). The small size and poor accessibility of the early mouse embryo make it relatively intractable for molecular studies, but BRG1 is also dosage sensitive and functions as a haploinsufficient tumor suppressor. We previously identified many copy-number gains (i.e. duplications and amplifications) and losses (i.e. deletions) in DNA from Brg1 null heterozygous mammary tumors (47). We did not propose that BRG1 maintains genomic stability because there was not a plausible mechanism and genomic instability could have been a secondary effect since it is a general feature of tumors. Considering the role of BRG1 in replication and the strong links between DNA replication and repair, however, it seems likely that BRG1 is, in fact, crucial for the process of maintaining genome stability. Additional evidence supports this view. For example, BRG1 also physically interacts with BRCA1 (71,72). A direct role for BRG1 in replication/repair is further supported by a recent study that used the Cre/loxP system to generate Brg1 null homozygous fibroblasts (73). Following adenoviral delivery of Cre, Brg1 mutant cells exhibited decreased BrdU incorporation, delayed cell-cycle progression and compromised genome integrity manifest by micronuclei and aneuploidy. Because these are primary, non-immortalized/transformed cells as opposed to cancer cells, it is clear that BRG1 plays a direct role in maintenance of genomic stability. All of these findings are consistent with recent reports that BRG1 and SWI/SNF-related complexes are involved in both double-strand break repair and nucleotide excision repair (74–77). Therefore, when considering the function of BRG1 in vivo, in both development and cancer prevention, one should consider the DNA replication/repair-based mechanism described here in addition to transcriptional regulation of downstream target genes such as those in the RB pathway.
FUNDING
The American Institute for Cancer Research and National Institutes of Health CA125237 (to S.J.B.); National Institutes of Health CA138841 to B.E.W.; and National Institutes of Health CA084493 to D.G.K. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
ACKNOWLEDGEMENTS
S.M.C. and P.D.C. contributed equally to this study. We thank Dr. Bruna Brylawski for performing tissue culture as well as Dr. Bob Bagnell and the UNC Microscopy Service Laboratory for providing advice on confocal microscopy. Anti-BRG1 (J1) was a generous gift of Drs. W. Wang and J. Crabtree. We would also like to acknowledge Dr. Bill Kaufmann and Dr. Nicole Francis for comments on this manuscript.
REFERENCES
- 1.Lebofsky R, Heilig R, Sonnleitner M, Weissenbach J, Bensimon A. DNA replication origin interference increases the spacing between initiation events in human cells. Mol. Biol. Cell. 2006;17:5337–5345. doi: 10.1091/mbc.E06-04-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Annu. Rev. Genet. 2007;41:237–280. doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DePamphilis ML. Cell cycle dependent regulation of the origin recognition complex. Cell Cycle. 2005;4:70–79. doi: 10.4161/cc.4.1.1333. [DOI] [PubMed] [Google Scholar]
- 4.DePamphilis ML, Blow JJ, Ghosh S, Saha T, Noguchi K, Vassilev A. Regulating the licensing of DNA replication origins in metazoa. Curr. Opin. Cell. Biol. 2006;18:231–239. doi: 10.1016/j.ceb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Berezney R, Dubey DD, Huberman JA. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma. 2000;108:471–484. doi: 10.1007/s004120050399. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura H, Morita T, Sato C. Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp. Cell. Res. 1986;165:291–297. doi: 10.1016/0014-4827(86)90583-5. [DOI] [PubMed] [Google Scholar]
- 7.Goren A, Tabib A, Hecht M, Cedar H. DNA replication timing of the human beta-globin domain is controlled by histone modification at the origin. Genes Dev. 2008;22:1319–1324. doi: 10.1101/gad.468308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M. Histone acetylation regulates the time of replication origin firing. Mol. Cell. 2002;10:1223–1233. doi: 10.1016/s1097-2765(02)00702-5. [DOI] [PubMed] [Google Scholar]
- 9.Gilbert DM. Replication timing and transcriptional control: beyond cause and effect. Curr. Opin. Cell. Biol. 2002;14:377–383. doi: 10.1016/s0955-0674(02)00326-5. [DOI] [PubMed] [Google Scholar]
- 10.Cohen SM, Brylawski BP, Cordeiro-Stone M, Kaufman DG. Same origins of DNA replication function on the active and inactive human X chromosomes. J. Cell. Biochem. 2003;88:923–931. doi: 10.1002/jcb.10429. [DOI] [PubMed] [Google Scholar]
- 11.Gomez M, Brockdorff N. Heterochromatin on the inactive X chromosome delays replication timing without affecting origin usage. Proc. Natl Acad. Sci. USA. 2004;101:6923–6928. doi: 10.1073/pnas.0401854101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiratani I, Takebayashi S, Lu J, Gilbert DM. Replication timing and transcriptional control: beyond cause and effect–part II. Curr. Opin. Genet. Dev. 2009;19:142–149. doi: 10.1016/j.gde.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barton MC, Crowe AJ. Chromatin alteration, transcription and replication: What's the opening line to the story? Oncogene. 2001;20:3094–3099. doi: 10.1038/sj.onc.1204334. [DOI] [PubMed] [Google Scholar]
- 14.Groth A. Replicating chromatin: a tale of histones. Biochem. Cell. Biol. 2009;87:51–63. doi: 10.1139/O08-102. [DOI] [PubMed] [Google Scholar]
- 15.Tan BC, Chien CT, Hirose S, Lee SC. Functional cooperation between FACT and MCM helicase facilitates initiation of chromatin DNA replication. EMBO J. 2006;25:3975–3985. doi: 10.1038/sj.emboj.7601271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mousson F, Ochsenbein F, Mann C. The histone chaperone Asf1 at the crossroads of chromatin and DNA checkpoint pathways. Chromosoma. 2007;116:79–93. doi: 10.1007/s00412-006-0087-z. [DOI] [PubMed] [Google Scholar]
- 17.Annunziato AT. Split decision: what happens to nucleosomes during DNA replication? J. Biol. Chem. 2005;280:12065–12068. doi: 10.1074/jbc.R400039200. [DOI] [PubMed] [Google Scholar]
- 18.Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science. 328:94–98. doi: 10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]
- 19.de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat. Rev. Genet. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- 20.Mohrmann L, Verrijzer CP. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta. 2005;1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Sudarsanam P, Winston F. The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet. 2000;16:345–351. doi: 10.1016/s0168-9525(00)02060-6. [DOI] [PubMed] [Google Scholar]
- 22.Tsukiyama T. The in vivo functions of ATP-dependent chromatin-remodelling factors. Nat. Rev. Mol. Cell. Biol. 2002;3:422–429. doi: 10.1038/nrm828. [DOI] [PubMed] [Google Scholar]
- 23.Racki LR, Narlikar GJ. ATP-dependent chromatin remodeling enzymes: two heads are not better, just different. Curr. Opin. Genet. Dev. 2008;18:137–144. doi: 10.1016/j.gde.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flanagan JF, Peterson CL. A role for the yeast SWI/SNF complex in DNA replication. Nucleic Acids Res. 1999;27:2022–2028. doi: 10.1093/nar/27.9.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 26.Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc. Natl Acad. Sci. USA. 2008;105:6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bultman SJ, Gebuhr TC, Magnuson T. A Brg1 mutation that uncouples ATPase activity from chromatin remodeling reveals an essential role for SWI/SNF-related complexes in beta-globin expression and erythroid development. Genes Dev. 2005;19:2849–2861. doi: 10.1101/gad.1364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim SI, Bultman SJ, Jing H, Blobel GA, Bresnick EH. Dissecting molecular steps in chromatin domain activation during hematopoietic differentiation. Mol. Cell. Biol. 2007;27:4551–4565. doi: 10.1128/MCB.00235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim SI, Bultman SJ, Kiefer CM, Dean A, Bresnick EH. BRG1 requirement for long-range interaction of a locus control region with a downstream promoter. Proc. Natl Acad. Sci. USA. 2009;106:2259–2264. doi: 10.1073/pnas.0806420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim SI, Bresnick EH, Bultman SJ. BRG1 directly regulates nucleosome structure and chromatin looping of the {alpha} globin locus to activate transcription. Nucleic Acids Res. 2009;37:6019–6027. doi: 10.1093/nar/gkp677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosson GB, Bartlett C, Reed W, Weissman BE. BRG1 loss in MiaPaCa2 cells induces an altered cellular morphology and disruption in the organization of the actin cytoskeleton. J. Cell. Physiol. 2005;205:286–294. doi: 10.1002/jcp.20397. [DOI] [PubMed] [Google Scholar]
- 32.Link KA, Burd CJ, Williams E, Marshall T, Rosson G, Henry E, Weissman B, Knudsen KE. BAF57 governs androgen receptor action and androgen-dependent proliferation through SWI/SNF. Mol. Cell. Biol. 2005;25:2200–2215. doi: 10.1128/MCB.25.6.2200-2215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chastain PD, II, Heffernan TP, Nevis KR, Lin L, Kaufmann WK, Kaufman DG, Cordeiro-Stone M. Checkpoint regulation of replication dynamics in UV-irradiated human cells. Cell Cycle. 2006;5:2160–2167. doi: 10.4161/cc.5.18.3236. [DOI] [PubMed] [Google Scholar]
- 34.Frum RA, Chastain PD, II, Qu P, Cohen SM, Kaufman DG. DNA replication in early S phase pauses near newly activated origins. Cell Cycle. 2008;7:1440–1448. doi: 10.4161/cc.7.10.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCall CM, Miliani de Marval PL, Chastain PD, II, Jackson SC, He YJ, Kotake Y, Cook JG, Xiong Y. HIV-1 Vpr-binding protein VprBP, a WD40 protein associated with the DDB1-CUL4 E3 ubiquitin ligase, is essential for DNA replication and embryonic development. Mol. Cell. Biol. 2008;28:5621–5633. doi: 10.1128/MCB.00232-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Unsal-Kacmaz K, Chastain PD, Qu PP, Minoo P, Cordeiro-Stone M, Sancar A, Kaufmann WK. The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Mol. Cell. Biol. 2007;27:3131–3142. doi: 10.1128/MCB.02190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen SM, Chastain PD, II, Cordeiro-Stone M, Kaufman DG. DNA replication and the GINS complex: localization on extended chromatin fibers. Epigenetics Chromatin. 2009;2:6. doi: 10.1186/1756-8935-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu K, Luo Y, Lin FT, Lin WC. TopBP1 recruits Brg1/Brm to repress E2F1-induced apoptosis, a novel pRb-independent and E2F1-specific control for cell survival. Genes Dev. 2004;18:673–686. doi: 10.1101/gad.1180204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Betz BL, Strobeck MW, Reisman DN, Knudsen ES, Weissman BE. Re-expression of hSNF5/INI1/BAF47 in pediatric tumor cells leads to G1 arrest associated with induction of p16ink4a and activation of RB. Oncogene. 2002;21:5193–5203. doi: 10.1038/sj.onc.1205706. [DOI] [PubMed] [Google Scholar]
- 40.Dunaief JL, Strober BE, Guha S, Khavari PA, Alin K, Luban J, Begemann M, Crabtree GR, Goff SP. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 41.Hendricks KB, Shanahan F, Lees E. Role for BRG1 in cell cycle control and tumor suppression. Mol. Cell. Biol. 2004;24:362–376. doi: 10.1128/MCB.24.1.362-376.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang H, Cui K, Zhao K. BRG1 controls the activity of the retinoblastoma protein via regulation of p21CIP1/WAF1/SDI. Mol. Cell. Biol. 2004;24:1188–1199. doi: 10.1128/MCB.24.3.1188-1199.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol. Cell. Biol. 2008;28:3457–3464. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts CW, Orkin SH. The SWI/SNF complex–chromatin and cancer. Nat. Rev. Cancer. 2004;4:133–142. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- 45.Strobeck MW, DeCristofaro MF, Banine F, Weissman BE, Sherman LS, Knudsen ES. The BRG-1 subunit of the SWI/SNF complex regulates CD44 expression. J. Biol. Chem. 2001;276:9273–9278. doi: 10.1074/jbc.M009747200. [DOI] [PubMed] [Google Scholar]
- 46.Weissman B, Knudsen KE. Hijacking the chromatin remodeling machinery: impact of SWI/SNF perturbations in cancer. Cancer Res. 2009;69:8223–8230. doi: 10.1158/0008-5472.CAN-09-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bultman SJ, Herschkowitz JI, Godfrey V, Gebuhr TC, Yaniv M, Perou CM, Magnuson T. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene. 2008;27:460–468. doi: 10.1038/sj.onc.1210664. [DOI] [PubMed] [Google Scholar]
- 48.Petermann E, Helleday T, Caldecott KW. Claspin promotes normal replication fork rates in human cells. Mol. Biol. Cell. 2008;19:2373–2378. doi: 10.1091/mbc.E07-10-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weissman B, Stanbridge EJ. Characterization of ouabain resistant, hypoxanthine phosphoribosyl transferase deficient human cells and their usefulness as a general method for the production of human cell hybrids. Cytogenet. Cell Genet. 1980;28:227–239. doi: 10.1159/000131536. [DOI] [PubMed] [Google Scholar]
- 50.Taylor JH. Increase in DNA replication sites in cells held at the beginning of S phase. Chromosoma. 1977;62:291–300. doi: 10.1007/BF00327029. [DOI] [PubMed] [Google Scholar]
- 51.Santocanale C, Sharma K, Diffley JF. Activation of dormant origins of DNA replication in budding yeast. Genes Dev. 1999;13:2360–2364. doi: 10.1101/gad.13.18.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gilbert DM. Replication origin plasticity, Taylor-made: inhibition vs recruitment of origins under conditions of replication stress. Chromosoma. 2007;116:341–347. doi: 10.1007/s00412-007-0105-9. [DOI] [PubMed] [Google Scholar]
- 53.Woodward AM, Gohler T, Luciani MG, Oehlmann M, Ge X, Gartner A, Jackson DA, Blow JJ. Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J. Cell. Biol. 2006;173:673–683. doi: 10.1083/jcb.200602108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ge XQ, Jackson DA, Blow JJ. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 2007;21:3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ibarra A, Schwob E, Mendez J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc. Natl Acad. Sci. USA. 2008;105:8956–8961. doi: 10.1073/pnas.0803978105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blow JJ, Ge XQ. A model for DNA replication showing how dormant origins safeguard against replication fork failure. EMBO Rep. 2009;10:406–412. doi: 10.1038/embor.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rao VA, Conti C, Guirouilh-Barbat J, Nakamura A, Miao ZH, Davies SL, Sacca B, Hickson ID, Bensimon A, Pommier Y. Endogenous gamma-H2AX-ATM-Chk2 checkpoint activation in Bloom's syndrome helicase deficient cells is related to DNA replication arrested forks. Mol. Cancer Res. 2007;5:713–724. doi: 10.1158/1541-7786.MCR-07-0028. [DOI] [PubMed] [Google Scholar]
- 58.Malyavantham KS, Bhattacharya S, Alonso WD, Acharya R, Berezney R. Spatio-temporal dynamics of replication and transcription sites in the mammalian cell nucleus. Chromosoma. 2008;117:553–567. doi: 10.1007/s00412-008-0172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maya-Mendoza A, Petermann E, Gillespie DA, Caldecott KW, Jackson DA. Chk1 regulates the density of active replication origins during the vertebrate S phase. EMBO J. 2007;26:2719–2731. doi: 10.1038/sj.emboj.7601714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petermann E, Caldecott KW. Evidence that the ATR/Chk1 pathway maintains normal replication fork progression during unperturbed S phase. Cell Cycle. 2006;5:2203–2209. doi: 10.4161/cc.5.19.3256. [DOI] [PubMed] [Google Scholar]
- 61.Petermann E, Maya-Mendoza A, Zachos G, Gillespie DA, Jackson DA, Caldecott KW. Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol. Cell. Biol. 2006;26:3319–3326. doi: 10.1128/MCB.26.8.3319-3326.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lorch Y, Maier-Davis B, Kornberg RD. Chromatin remodeling by nucleosome disassembly in vitro. Proc. Natl Acad. Sci. USA. 2006;103:3090–3093. doi: 10.1073/pnas.0511050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Korber P, Barbaric S, Luckenbach T, Schmid A, Schermer UJ, Blaschke D, Horz W. The histone chaperone Asf1 increases the rate of histone eviction at the yeast PHO5 and PHO8 promoters. J. Biol. Chem. 2006;281:5539–5545. doi: 10.1074/jbc.M513340200. [DOI] [PubMed] [Google Scholar]
- 64.Moshkin YM, Armstrong JA, Maeda RK, Tamkun JW, Verrijzer P, Kennison JA, Karch F. Histone chaperone ASF1 cooperates with the Brahma chromatin-remodelling machinery. Genes Dev. 2002;16:2621–2626. doi: 10.1101/gad.231202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen W, Xu C, Huang W, Zhang J, Carlson JE, Tu X, Wu J, Shi Y. Solution structure of human Brg1 bromodomain and its specific binding to acetylated histone tails. Biochemistry. 2007;46:2100–2110. doi: 10.1021/bi0611208. [DOI] [PubMed] [Google Scholar]
- 66.Singh M, Popowicz GM, Krajewski M, Holak TA. Structural ramification for acetyl-lysine recognition by the bromodomain of human BRG1 protein, a central ATPase of the SWI/SNF remodeling complex. ChemBiochem. 2007;8:1308–1316. doi: 10.1002/cbic.200600562. [DOI] [PubMed] [Google Scholar]
- 67.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 68.Ni Z, Abou El Hassan M, Xu Z, Yu T, Bremner R. The chromatin-remodeling enzyme BRG1 coordinates CIITA induction through many interdependent distal enhancers. Nat. Immunol. 2008;9:785–793. doi: 10.1038/ni.1619. [DOI] [PubMed] [Google Scholar]
- 69.Kim SI, Bresnick EH, Bultman SJ. BRG1 directly regulates nucleosome structure and chromatin looping of the alpha globin locus to activate transcription. Nucleic Acids Res. 2009;37:6019–6027. doi: 10.1093/nar/gkp677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ueno M, Itoh M, Kong L, Sugihara K, Asano M, Takakura N. PSF1 is essential for early embryogenesis in mice. Mol. Cell. Biol. 2005;25:10528–10532. doi: 10.1128/MCB.25.23.10528-10532.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bochar DA, Wang L, Beniya H, Kinev A, Xue Y, Lane WS, Wang W, Kashanchi F, Shiekhattar R. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell. 2000;102:257–265. doi: 10.1016/s0092-8674(00)00030-1. [DOI] [PubMed] [Google Scholar]
- 72.Hill DA, de la Serna IL, Veal TM, Imbalzano AN. BRCA1 interacts with dominant negative SWI/SNF enzymes without affecting homologous recombination or radiation-induced gene activation of p21 or Mdm2. J. Cell. Biochem. 2004;91:987–998. doi: 10.1002/jcb.20003. [DOI] [PubMed] [Google Scholar]
- 73.Bourgo RJ, Siddiqui H, Fox S, Solomon D, Sansam CG, Yaniv M, Muchardt C, Metzger D, Chambon P, Roberts CW, et al. SWI/SNF deficiency results in aberrant chromatin organization, mitotic failure, and diminished proliferative capacity. Mol. Biol. Cell. 2009;20:3192–3199. doi: 10.1091/mbc.E08-12-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chai B, Huang J, Cairns BR, Laurent BC. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 2005;19:1656–1661. doi: 10.1101/gad.1273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hara R, Sancar A. The SWI/SNF chromatin-remodeling factor stimulates repair by human excision nuclease in the mononucleosome core particle. Mol. Cell. Biol. 2002;22:6779–6787. doi: 10.1128/MCB.22.19.6779-6787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Park JH, Park EJ, Lee HS, Kim SJ, Hur SK, Imbalzano AN, Kwon J. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. EMBO J. 2006;25:3986–3997. doi: 10.1038/sj.emboj.7601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao Q, Wang QE, Ray A, Wani G, Han C, Milum K, Wani AA. Modulation of nucleotide excision repair by mammalian SWI/SNF chromatin-remodeling complex. J. Biol. Chem. 2009;284:30424–30432. doi: 10.1074/jbc.M109.044982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.