Abstract
Telomeres protect chromosome ends from being recognized as double-stranded breaks. Telomeric function is ensured by the shelterin complex in which TRF2 protein is an essential player. The G-rich strand of telomere DNA can fold into G-quadruplex (G4) structure. Small molecules stabilizing G4 structures, named G4 ligands, have been shown to alter telomeric functions in human cells. In this study, we show that a guanine-rich RNA sequence located in the 5′-UTR region of the TRF2 mRNA (hereafter 91TRF2G) is capable of forming a stable quadruplex that causes a 2.8-fold decrease in the translation of a reporter gene in human cells, as compared to a mutant 5′-UTR unable to fold into G4. We also demonstrate that several highly selective G4 ligands, the pyridine dicarboxamide derivative 360A and bisquinolinium compounds Phen-DC(3) and Phen-DC(6), are able to bind the 91TRF2G:RNA sequence and to modulate TRF2 protein translation in vitro. Since the naturally occurring 5′-UTR TRF2:RNA G4 element was used here, which is conserved in several vertebrate orthologs, the present data substantiate a potential translational mechanism mediated by a G4 RNA motif for the downregulation of TRF2 expression.
INTRODUCTION
In vitro, single-stranded G-rich nucleic acid sequences can adopt an unusual conformation called a G4-quadruplex (G4) (1). G4 structures result from the successive stacking of G-quartet formed by four guanines located on a plan and interacting by Hoogsteen hydrogen bonds (2). The formation of G4 structures is strongly dependent on monovalent cations such K+ and Na+ and, hence, physiological buffer conditions favor their formation (3). G4 structures can be formed by one or more nucleic acid strands. Numerous physical analyses have revealed that G4 structures are highly polymorphic and can be subgrouped into various families, such as parallel or antiparallel, according to the orientation of the strands (4–6). In vitro, G4 structures are selectively bound and stabilized by small molecules called G4 ligands (7).
The existence of G4s has been demonstrated in vivo in ciliates using specific antibodies (8), whereas only indirect evidence sustained their existence in mammalian cells [for a review see (9,10)]. Bioinformatic analysis of the human genome indicated that it contains as many as 370 000 sequences possessing the Potential G-Quadruplex-forming Sequences (PQS) (11,12). As expected, most of these sequences are located in repetitive DNA regions, such as telomeres and rDNA. In addition, a statistically significant enrichment of PQS was found in regulatory regions such as gene promoters (13), splice sequences and UTRs regions (14) raising the possibility that G4 structures could play a role in the regulation of gene expression.
In eukaryotes, chromosomes ends are protected from DNA repair systems by a particular nucleoprotein structure, the telomere (15). In humans, the telomere is composed of thousands of G-rich double-stranded TTAGGG repeats (16) and a 3′ single-stranded G-rich extension called the G-tail or G-overhang (17). The telomeric DNA is bound by a telomere-specific six-protein complex called shelterin (18). Shelterin stabilizes a special DNA structure, the t-loop, in which the G-tail invades the duplex telomeric repeats, forming a D-loop structure (18). The t-loop masks chromosome ends and blocks the activation of the DNA damage response at telomeres (19).
TRF2 protein plays an essential role in the shelterin function (18). TRF2 has been shown to promote and stabilize loop formation (20). In conjunction with its partner RAP1, TRF2 also triggers the inhibition of the the non homologous end joining relying on the DNA-dependent protein kinase at telomeres (21). Thus, overexpression of dominant-negative mutants of TRF2 induces telomere uncapping triggering end-to-end chromosome fusions (22) or stochastic deletions of telomeric DNA through a homologous recombination-mediated mechanism (23). TRF2 is overexpressed in several human tumors, such as liver hepatocarcinomas (24), breast carcinomas (25) and lung carcinomas (26), suggesting that TRF2 may play a role in tumorigenesis.
In this study, we describe the biophysical and functional characterization of G-rich sequences present within the TRF2 mRNA. We show that a G-rich sequence located in the 5′-UTR region of the TRF2 mRNA adopts a stable intramolecular G4 RNA structure in vitro, which is able to repress the expression of a reporter gene in vitro and in cells. Mutation of this sequence impairing quadruplex stabilization leads to an increased expression.
Furthermore, using biophysical analyses, we show that the G-quadruplex RNA motif adopted by the G-rich sequence located within the 5′-UTR of TRF2 mRNA is bound by several highly selective G-quadruplex ligands. In vitro studies show that the stabilization of the G4 RNA motif has a significant effect on the expression of a reporter gene. These data suggest that G4 formation in the 5′-UTR from TRF2 represents a new mechanism to control TRF2 expression.
MATERIALS AND METHODS
Oligonucleotides
All oligonucleotides described in Table 1 except +75UTRATGTRF2 and mut+75UTRATGTRF2 (Sigma Aldrich) were purchased from Eurogentec.
Table 1.
Sequence of the oligonucleotides used for this study
| 91TRF2:DNA | GGGAGGGCGGGGAGGG |
| 131TRF2:DNA | GGGAGGAGGCGGGAGTAGCGACGGCAGCGGGCGGG |
| 195TRF2:DNA | GGGCGGGCCCGGCGGGGGCGCCACGAGCCGGGG |
| 199TRF2:DNA | GGGCCCGGCGGGGGCGCCACGAGCCGGGGCTGGGGGG |
| 91TRF2G:RNA | CGGGAGGGCGGGGAGGGC |
| mut91TRF2G:RNA | CGUGAGUGCGCUGAGGGC |
| +75UTRATGTRF2 | GATCCGCCGAGGAAGCGGCCCGGCCGGGAGGGCGGGGAGGGCGCGCGGCGATCGGACACGGAATTCATG |
| mut+75UTRATGTRF2 | GATCCGCCGAGGAAGCGGCCCGGCCGTGAGTGCGCTGAGGGCGCGCGGCGATCGGACACGGAATTCATG |
Circular dichroism measurements
Circular dichroism (CD) spectra were recorded on a JASCO-810 spectropolarimeter using 1-cm path length quartz cuvettes in a reaction volume of 580 µl, as previously described (27). Oligonucleotides 91TRF2G:RNA and mut91TRF2G:RNA (Table 1) were prepared as a 4 µM solution in 10 mM lithium cacodylate pH 7.2, 100 mM NaCl or KCl buffer and annealed by heating to 90°C for 2 min, followed by slow cooling to 20°C. Scans were performed at 20°C over a wavelength range of 235–350 nm with a scanning speed of 500 nm/min, a response time of 1 s, 1 nm data pitch and 1 nm bandwidth.
UV melting assays for G4s
Oligonucleotides 91TRF2G:RNA and mut91TRF2G:RNA (Table 1) were synthesized by Eurogentec (Seraing, Belgium) at the 200 nmol scale and used without further purification. Concentrations were estimated using extinction coefficients provided by the manufacturer. Melting assays were performed on a Uvikon 940 spectrophotometer in a 10 mM lithium cacodylate pH 7.2 buffer (supplemented with either 0.1 M KCl or NaCl, hereafter referred to as potassium and sodium conditions, respectively), as previously described (28–30). Melting experiments were typically performed at a concentration of 4 µM per strand. All transitions were reversible, as shown by superimposable heating and cooling profiles at a fixed rate of 0.2°C/min.
FRET melting of F21T
Tm of F21T (0.2 µM) was recorded alone, in the presence of 1 µM of G4 ligands, in the presence of 3 µM competitor or in presence of G4 ligands + competitor. The competitors were a double-stranded (ds26:5′-CAATCGGATCGAATTCGATCCGATTG-3′), or 91TRF2G:RNA or Mut91TRF2G:RNA. All experiments were performed in duplicate. The emission of fluorescein is normalized between 0 and 1, and the T1/2 is defined as the temperature for which the normalized emission is 0.5 (31).
Gel electrophoresis
Formation of G4 RNA was confirmed by nondenaturing PAGE. Oligonucleotides were directly visualized by UV shadow (see Supplementary Data). Prior to the incubation, the DNA samples were heated at 90°C for 5 min and slowly cooled (2 h) to room temperature. Samples were incubated at 30 µM strand concentration in Tris–HCl 10 mM pH 7.5 buffer with 100 mM Na+ or K+. Ten percent sucrose was added just before loading. Oligothymidylate markers (dT15, dT21 or dT30) or double-stranded markers (Dx9:5′-d-GCGTATCGG + 5′-d-CCGATACGC; Dx12:5′-d-GCGTGACTTCGG + 5′-d-CCGAAGTCACGC) were also loaded on the gel. One should note that the migration of the dTn oligonucleotides does not necessarily correspond to single strands (32): these oligonucleotides were chosen here to provide an internal migration standard, not to identify intramolecular or higher order structures.
Construction of plasmids
To construct pWUTRF2 and pMUTRF2 plasmids, a synthetic DNA duplex carrying either the +65UTRATGTRF2 or the mut+65UTRATGTRF2 sequence was inserted at the BamHI–EcoRI sites within the pcDNA3 vector (Invitrogen). The resulting plasmids were utilized to clone an EcoRI–XhoI DNA fragment encoding the green fluorescent protein (GFP) obtained by PCR amplification of the GFP sequence from pEGFP-C1 plasmid (Clontech). The insertion of correct sequences was verified by sequencing.
TRF2ΔB construction result from the insertion of a 5′ deleted form of TRF2 cDNA (23) into the BamHI–EcoR1 sites of the pcDNA3 plasmid (Invitrogen).
In vitro transcription and translation
In vitro coupled transcription–translation of pWUTRF2 and pMTRF2 plasmids was carried out using the rabbit reticulocyte lysate-based transcription–translation coupled system (TNT® Quick Coupled Transcription/Translation System, Promega) according to the manufacturer’s instructions. Briefly, a 1 µg plasmid mixture consisting of 0.2 µg of the TRF2ΔB plasmid and either 0.8 µg of pWUTRF2 or pMUTRF2 plasmid was mixed to 12.5 µl of TNT rabbit reticulocyte system, 1 µl of TNT reaction buffer, 0.5 µl of amino acid mixture minus methionine, 1 µl of [35S]-l-methionine at 1175 Ci/mmol (PerkinElmer) and 0.5 µl of T7 RNA polymerase in a total reaction volume of 25 µl. The reaction mixture was incubated at 30°C for 90 min and immediately put at −20°C. To evaluate GFP production, 5 µl of each reaction was diluted 5-fold with 1X Laemmli buffer, heated at 70°C for 10 min and loaded on a 12% SDS–polyacrylamide gel. After separation, [35S]-methionine incorporation was visualized by a Phosphorimager (Typhoon 9210 Amersham). To study the effect of G4 ligands on in vitro gene expression indicated, concentrations of molecules (in a volume of 2.5 µl) were added to the complete reaction mixture before incubation at 30°C.
Cell culture and transfection
The 293T cells were growth at 37°C in a humidified atmosphere containing 5% CO2 in Dulbecco’s modified Eagles medium supplemented with 10% heat-inactivated fetal calf serum and the antibiotics penicillin and streptomycin.
Transfection was carried out in six-well plates using 3 µg of each plasmid and Lipofectamine 2000 (Invitrogen) transfectant reagent according to the manufacturer’s instructions. Twenty-four hours after transfection, cells were recovered by trypsination and the cellular pellet was divided into two equal portions to proceed to western blot and Quantitative real time PCR (RT–qPCR) analyses.
Western blotting
SDS gel electrophoresis was performed in a 12% polyacrylamide gels with a protein content of 50 µg at 150 V in 0.025 M Tris pH 8.3, 0.192 M glycine, 0.1% SDS. Gels were blotted onto Immobilon-P polyvinylidene difluoride membrane (Millipore). The membrane was blocked with 5% dry milk in PBS-T [(1× phosphate buffer saline, 0.1% v/v Tween-20 (Sigma-Aldrich)] for 30 min at room temperature and incubated with the anti-GFP primary antibody (1:1000 in 1× phosphate buffer saline, 0.1% v/v Tween-20) for 1 h at room temperature. A secondary horseradish peroxidase conjugated antibody (1:10 000, 45 min at room temperature) was used and detected using the enhanced chemiluminescence kit (ImmunofaxA, Yelen).
Quantitative real-time PCR assay
Total cellular RNA was isolated using RNeasy Plus Mini Kit (Qiagen). For quantification of mRNA levels, 1 µg of total RNA was used in a 20 µl reverse transcription reaction using the RevertAid H Minus First Strand cDNA synthesis Kit (Fermentas) in the presence of random hexamer. A control reaction, without reverse transcriptase enzyme, was performed in parallel to ensure the absence of plasmid DNA contamination. Real-time PCR was performed with 25 ng of cDNA and 300 nM concentration of both sense and antisense GFP primers (sense 5′-CGACCACATGAAGCAG and antisense 5′-TCCTGGACGTAGCCTTCGG) in a final volume of 25 µl using the SYBR Green TaqMan Universal PCR master mix (Applied Biosystems). Fluorescence was monitored and analyzed in a GeneAmp 7300 detection system instrument (Applied Biosystems). Analysis of 18S ribosomal RNA and GAPDH RNA (Applied Biosystems TaqMan probes) was performed in parallel to normalize for gene expression.
For RNA quantification of in vitro transcription/translation products, 5 µl of this mix was used in 20 µl reverse transcription reaction. Real-time PCR was performed with 5 µl of reverse transcription products.
RESULTS
Putative G4 elements within the 5′-part of the human TRF2 mRNA
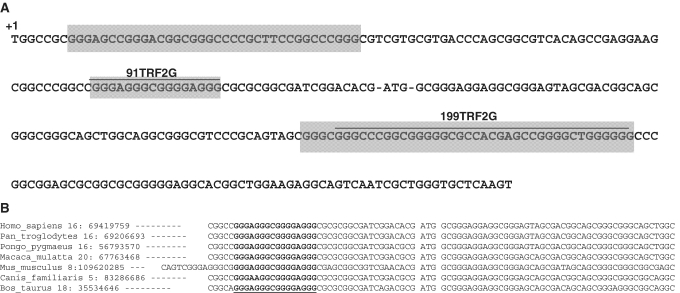
Sequence analysis of the 5′-part of the human TRF2 mRNA shows the presence of several G-rich sequences surrounding the AUG start codon (Figure 1A). Bioinformatic analysis of the TRF2 mRNA using QGRS Mapper program (33) shows the existence of several potential G4 structures within the 5′-part of the TRF2 mRNA. Two sequences present high G-score and are favorable for G4 folding: the first one matching the sequence 5′-GGGAGGGCGGGGAGGG-3′ (91TRF2G) is located within the 5′-UTR, 19 nt upstream of the AUG start codon and the second one matching the sequence 5′-GGGCCCGGCGGGGGCGCCACGAGCCGGGGCUGGGGGG-3′ (199TRF2G) at 66 nt downstream of the translation start site. The DNA sequence alignment of human TRF2 with its orthologs present in six amniota vertebrates shows that the G-rich sequence present in the 5′-UTR of human TRF2 is evolutionarily conserved across species (Figure 1B), indicating a potential regulatory role. Thus we decided to further characterize these sequences, both at the molecular and functional level.
Figure 1.
Sequence analysis of the 5′-part of the TRF2 cDNA. (A) G-rich sequences corresponding to the consensus 5′-G3-Nx-G3-Nx-G3-Nx-G3-3′ are in a gray background. The 191TRF2G and 199TRF2G sequences (G4 structures predicted by QGRS Mapper software) and translation start site are indicated. (B) Conservation of the 91TRF2G motif in the 5′-UTR of TRF2. Nucleotides underlined are runs of guanines able to form G4s.
In a first attempt to determine which G-rich TRF2 sequences may adopt a G4 DNA structure in vitro, we performed a rapid screening by electrophoretic mobility shift assays with several DNA oligonucleotides designed from the PQS (91TRF2G:DNA, 131TRF2G:DNA, 195TRF2G:DNA and 199TRF2G:DNA; see Table 1) and purified recombinant human Topoisomerase IIIα (Topo III), in the presence of increasing concentrations of the 360A pyridine dicarboxamide derivative, a potent and highly specific G4 ligand (Supplementary Figure S1 and Supplementary Data).
We conclude from these experiments that 91TRF2G:DNA sequence is the best candidate in the 5′-UTR of the TRF2 mRNA to adopt a G4 DNA structure in vitro, and we decided to further characterize its biophysical and biological properties.
91TRF2G sequence adopt a G4 RNA structure of extreme stability
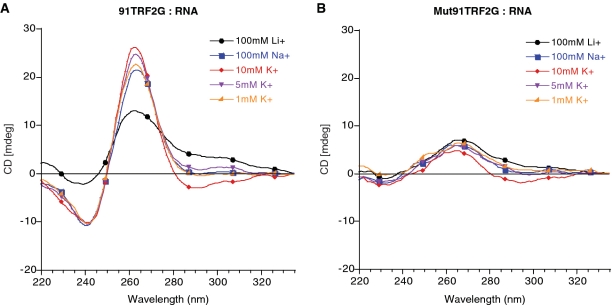
To test whether the 91TRF2G sequence forms a G4 RNA structure, we carried out biophysical analyses on the RNA oligonucleotide 5′-CGGGAGGGCGGGGAGGGC-3′. CD experiments using different ionic conditions (Na+, K+ and Li+) at physiological pH (see ‘Materials and Methods’ section) were performed, and spectra show the characteristic features of parallel G4 structures, with a positive peak at 263 nm and a trough at 240 nm the amplitude of which is higher in Na+ or K+ conditions than in Li+ (Figure 2A) (34). Under the same conditions, thermal difference spectrum analysis of the 91TRF2G:RNA is characterized by a negative minima at 262 and 295 nm, positive maxima at 255 and 270 nm, a shoulder at 245 nm, which are the particular signatures of the G4 structures (data not shown) (29).
Figure 2.
CD spectra of (A) 91TRF2:RNA and (B) Mut91TRF2:RNA sequences. CD spectra were recorded at 20°C on a JASCO-810 using 1-cm path length quartz cuvettes. Oligonucleotides were prepared as a 4 µM and annealed by heating to 90°C for 2 min, followed by slow cooling to 20°C. Buffers containing 10 mM lithium cacodylate at pH 7.2 supplemented with the indicated cation.
The thermal melting of quadruplex nucleic acids can be characterized by an inverse UV transition at 295 nm, and has generally been found to have significant and characteristic cation dependence. The monovalent ion dependence for the stabilization of folded 91TRF2G:RNA as judged by Tm, was in the order K+ > Na+ > Li+ (Table 2), which is characteristic of G4 nucleic acids (3).
Table 2.
Tm-values for 91TRF2G:RNA in the presence of various cations
| Name |
Tma |
||||
|---|---|---|---|---|---|
| 100 mM Li+ | 100 mM Na+ | 10 mM K+ | 5 mM K+ | 1 mM K+ | |
| 91TRF2G:RNAb | 39.5 | 64.0 | >77.0 | 72.0 | 61.8 |
| Mut91TRF2G:RNAb | −c | −c | −c | −c | −c |
For all experiments, the buffer consisted of 10 mM lithium cacodylate (pH 7.2) in the presence of the indicated cation. Tm-values were obtained by measuring the inverted UV transition at 295 nm (30)
aTm in °C, with ±0.5°C precision; determined from the analysis of UV melting profiles at 295 nm.
bOligonucleotides were prepared at 4 µM in 10 mM lithium cacodylate buffer at pH 7.2.
cThis sequence does not form a quadruplex.
At 100 mM KCl, the folded 91TRF2G RNA quadruplex could not be unfolded, even at 95°C, which is indicative of a very stable quadruplex. The 91TRF2G G4 RNA was very stable (Tm = 61.58°C) even in the presence of low-salt concentrations (1 mM KCl). Studies over a 10-fold strand concentration range (from 2 to 20 μM) showed no change in the Tm-value (Table 3), this last result being consistent with the folding of an intramolecular quadruplex.
Table 3.
Tm-values for 91TRF2G:RNA at various concentrations
For all experiments, the buffer consisted of 10 mM lithium cacodylate (pH 7.2) in the presence of 10 mM KCl. Tm-values were obtained by measuring the inverted UV transition at 295 nm (30).
aTm in °C, with ±0.5°C precision; determined from the analysis of UV melting profiles at 295 nm.
bOligonucleotide tested at four strand concentrations (2 µM, 4 µM, 10 µM and 20 µM) in 10 mM lithium cacodylate buffer at pH 7.2 with 10mM KCl and 90 mM LiCl.
Formation of G4 structures is dependent on Hoogsteen bonding established between guanines. Thus, single guanine replacements in the 91TRF2G:RNA sequence should prevent the folding of this sequence in a G4 RNA structure. Mut91TRF2G:RNA sequence 5′ CGUGAGUGCGCUGAGGGC 3′ was designed by replacing four key guanine nucleotides present in the 91TRF2G:RNA sequence at positions G3, G7, G11 and G12. As shown in Figure 2B, CD spectrum analysis of Mut91TRF2G sequence, in the presence of Na+ or K+ ionic concentrations, did not show the main features associated to G4 formation. In addition, native gel electrophoresis revealed that Mut91TRF2G:RNA migrates at its expected molecular weight position, as compared to 91TRF2G:RNA whose migration is lowered, as expected for a G4 structure (Supplementary Figure S2).
Altogether, these data indicate that under near-physiological pH and salt conditions, 91TRF2G:RNA folds into a very stable, intramolecular G4 RNA that presents the characteristic features of a parallel G4. Since CD spectra only provide indirect evidence for G4 topology, additional experiments are necessary to confirm this finding.
91TRF2G sequence inhibits translation of GFP both in vitro and in vivo
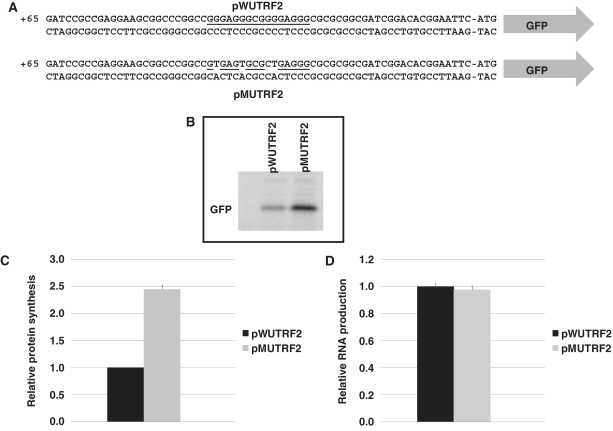
To evaluate the impact of the G4 structure formed by the 91TRF2G sequence on gene expression, we cloned the 62 nt of the 5′-UTR from TRF2, containing either the wild-type 91TRF2G sequence (plasmid pWUTRF2) or its mutated version Mut91TRF2G (plasmid pMUTRF2), upstream of the GFP-coding sequence and immediately downstream of the minimal T7 promoter present on pcDNA3 plasmid (Figure 3A). Using an in vitro coupled transcription–translation assay, based on rabbit reticulocyte lysate system, the GFP expression resulting from these constructions was evaluated by radioactive quantification of [35S]-methionine incorporation and showed a 2.45-fold increase in GFP synthesis for the mutated sequence relative to the wild-type sequence (Figure 3B and C). To further characterize the repressive effect of 91TRF2G sequence on gene expression in vitro, we next proceeded to quantification of RNA molecules produced by the transcription–translation coupled system. As shown in Figure 3D, RT-PCR analysis did not show any significant difference of the in vitro transcription of two constructions. This result indicates that the presence of a G4 sequence within the 5′-UTR of the gene reporter construction does not affect the transcriptional process. Together, these results suggest that the presence of a G4 motif within the 5′-UTR of the TRF2 mRNA inhibits the gene expression at a posttranscriptional level.
Figure 3.
Effect of the 91TRF2G sequence on GFP production in vitro. (A) Schematic representation of the plasmids used to investigate the effect of the 5′-UTR sequence of TRF2 mRNA on expression. The plasmids contain 62 nt of the 5′-UTR TRF2 sequence cloned just upstream of GFP coding sequence. The plasmid pWUTRF2 carries the wild-type 91TRF2G sequence (underlined) which is replaced by the mutant Mut91TRF2G sequence in the pMUTRF2 plasmid (modified nucleotides are not underlined). (B) Expression of the GFP protein in vitro using transcription–translation system. GFP production was visualized by radioactive signal obtained via [35S-]methionine incorporation. (C) Histogram representing the ratio of in vitro GFP production obtained from in vitro transcription and translation of 1 µg of pWUTRF2 or pMUTRF2 plasmids. The ratio of the plasmid pWUTRF2 was set to 1 and the value observed for pMUTRF2 was normalized accordingly. (D) Relative GFP RNA levels for pWUTRF2 and pMUTRF2 constructs determined by real-time PCR assays. Relative Ct for pWUTRF2 was set to 1 and pMUTRF2 Ct was normalized accordingly. The results shown are mean values (± SD) from three independent experiments.
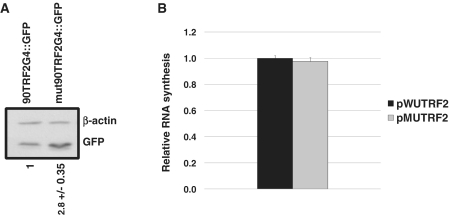
In order to investigate the impact of 91TRF2G G4 structure in a cellular context, 293T cells were transfected with plasmids pWUTRF2 or pMUTRF2. Twenty-four hours after transfection, cells were harvested and protein lysates were prepared. Western blot analysis of GFP expression indicated that 293T cells transfected by pMUTRF2 present a 2.8-fold higher GFP protein level (normalized to actin), as compared to the signal obtained using pWUTRF2 (Figure 4A). As RNA quadruplexes in genes promoters have been reported to modulate transcription levels of several genes (35–37), and to exclude any variation due to the transfection efficiency, mRNA levels arising from transcription of pWUTRF2 and pMUTRF2 plasmids were quantified using RT–qPCR assays. Relative quantification of GFP mRNA molecules produced by pWUTRF2 or pWUTRF2 showed no significant difference of the transcriptional expression between the two constructions (Figure 4B). This result together with our in vitro data showing the formation of a highly stable parallel G4 RNA structure under physiological conditions for 91TRF2G:RNA strongly suggest that the repression of GFP expression observed in vivo is due to the ability of 91TRF2G:RNA G4 structure to reduce translation in human cells.
Figure 4.
Effect of the 91TRF2G motif on translation in human cells. (A) Western blot analysis of GFP expression in 293T cells transfected with 3 µg of pWUTRF2 or pMUTRF2 plasmid. Twenty-four hours after transfection, cells were harvested and lysates were prepared. (B) Relative GFP mRNA levels for pWUTRF2 and pMUTRF2 constructs determined by real-time PCR assays. GFP mRNA levels were normalized to GAPDH and β-actin mRNAs. Relative Ct for pWUTRF2 was set to 1 and pMUTRF2 Ct was normalized accordingly. The results shown are mean values (±SD) from three independent experiments.
91TRF2G:RNA G4 structure is bound by G4 ligands in vitro
The interaction of the 91TRF2G:RNA sequence with G4 ligands has been investigated by a competition FRET assay using a real-time PCR apparatus and the doubly labelled F21T oligonucleotide which mimics the human telomeric single-strand overhang, as previously described by Mergny and coworkers (31). To this end, melting of F21T is performed in the presence of three different, highly selective, G4 ligands: the 360A molecule (38) and two bisquinolinium compounds Phen-DC(3) and Phen-DC(6) (39) (see structures in Supplementary Figure S3), and various competitors: a 26bp duplex (ds26), the 91TRF2G:RNA sequence and its mutant counterpart, the Mut91TRF2G.
As shown in Table 4, in the presence of sodium or potassium, the three G4 ligands display a good selectivity for the quadruplex over the control duplex, since stabilization of F21T is only moderately affected by a 15 M excess of the ds26 DNA duplex. In contrast, when the 91TRF2G:RNA sequence was added, the competition pattern shows a strong decrease in ΔT1/2 for all compounds. In the presence of sodium or potassium, 91TRF2G:RNA sequence provokes a complete loss of the stabilization of the F21T by bisquinolinium compounds. For the pyridine dicarboxamide derivate, the 360A molecule, competition analyses show a differential binding to 91TRF2G:RNA sequence dependent on ionic conditions. While in the presence of sodium, 91TRF2G:RNA sequence induces a complete displacement of 360A from F21T oligonucleotide; in the presence of potassium, the G4 ligand presents a lower affinity for the RNA sequence as compared to F21T sequence although a significant decrease in ΔT1/2 is observed (ΔT1/2 decreases of 14°C). In contrast, the mutant sequence, Mut91TRF2G:RNA, did not compete with the stabilization of F21T by G4 ligands (Table 4). Together, our results demonstrate that the 91TRF2G:RNA sequence is able to bind G4 ligands and that the binding to these molecules is associated with its capacity to adopt a G4 structure.
Table 4.
FRET melting of F21T
| ΔT1/2 (°C) | No compound | 1 µM Phen-DC(3) | 1 µM Phen-DC(6) | 1 µM 360A |
|---|---|---|---|---|
| 100 mM NaCl | ||||
| No competitor | 0.1 | 27.2 | >29.7 | 25.4 |
| Ds26 | 0.0 | 24.0 | >28.1 | 24.0 |
| 91TRF2G:RNA | 0.6 | 0.1 | 0.0 | 1.0 |
| Mut91TRF2G:RNA | −0.5 | 24.8 | >20.6 | 24.1 |
| 10 mM KCl + 90 mM LiCl | ||||
| No competitor | 0.0 | 19.2 | >31.6 | 26.5 |
| ds26 | −0.2 | 18.3 | >31.3 | 26.5 |
| 91TRF2G:RNA | −0.5 | 0.6 | −0.5 | 12.0 |
| Mut91TRF2G:RNA | −1.4 | 17.6 | >22.4 | 25.5 |
The values of melting were determined in presence of G4 ligands and some competitors. Values represent means from three independent experiments. Compounds were tested at 1 µM. Prior to the Tm FRET, the oligonucleotides were preformed in 10 mM lithium cacodylate buffer at pH 7.2 with 100 mM NaCl or 10 mM KCl + 90 mM LiCl.
91TRF2G:RNA G4 structure modulates the expression of a gene reporter in vitro
To further demonstrate that the repressive effect of 5′-UTR 91TRF2G sequence was due to the formation of a G4 structure, we studied the effect of G4 ligands on GFP synthesis in vitro.
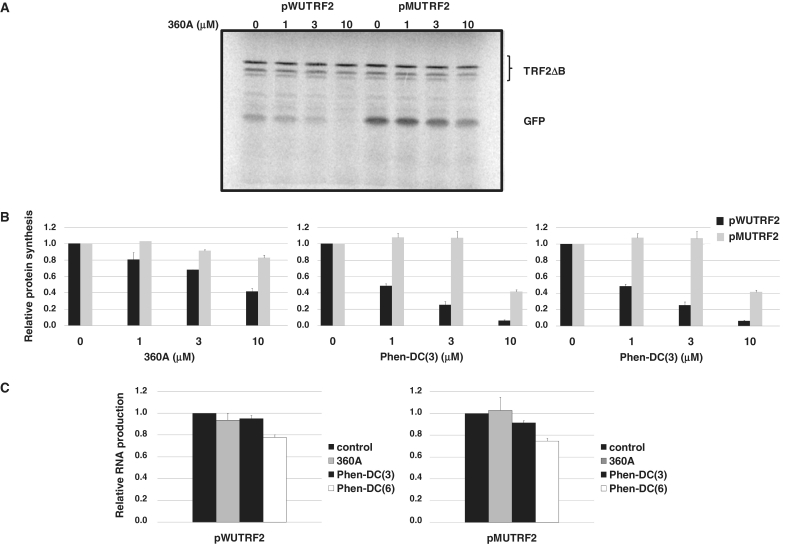
As shown in Figure 5B, all of the three G4 ligands used in this assay have a more pronounced effect, concentration dependent, on GFP expression from the wild-type (pWUTRF2) than from the mutant construction (pMUTRF2).
Figure 5.
Effect of G4 ligands on GFP expression in vitro. The pWUTRF2 or pMUTRF2 plasmid (0.8 µg) andcontrol plasmid TRF2ΔB (0.2 µg), lacking both 5′-UTR and G-rich sequences located just downstream of initiation codon, were transcribed and translated in vitro in the presence of various G4 ligands at the indicated concentrations. (A) Gel analysis of GFP expression from pWUTRF2 and pMUTRF2 plasmids in the presence of increasing concentrations of the 360A. (B) Quantification of the effect on the GFP production by G4 ligands. GFP synthesis was quantified by [35S]-methionine incorporation and was normalized to TRF2ΔB protein production to discard any nonspecific effect of G4 ligands on global protein production. The ratio of the GFP in the absence of G4 ligands was set to 1 and the values observed in the presence of different G4 ligands concentrations were normalized accordingly. (C) Relative GFP RNA levels for pWUTRF2 (left) and pMUTRF2 (right) constructs determined by real-time PCR assays. G4 ligands were used at 10 µM. The results shown are mean values (±SD) from three independent experiments.
GFP expression from the wild-type construction (pWUTRF2 containing the 91TRF2G motif), is decreased by 15 and 60% using 1 and 10 µM 360A, respectively (Figure 5A and B, left). In contrast, up to 10 µM 360A causes a very limited reduction of GFP protein synthesis of the mutant construction (∼10% at 10 µM; Figure 5A and B, left).
The bisquinolinium compound Phen-DC(3) exhibits a more marked inhibition effect. At 3 µM concentration, Phen-DC(3) inhibits GFP expression from pWUTRF2 by four-fold, while at the same concentration this molecule did not show any significant effect on pMUTRF2 construction. Although at 10 µM concentration, Phen-DC(3) showed a marked effect on GFP expression of from the mutant construction (∼60%), GFP expression from the wild-type construction was inhibited by 10-fold (Figure 5B, central).
When compared to the two others molecules, Phen-DC(6) compound shows a more pronounced nonspecific effects. At 3 µM, the GFP expression for the mutant reaches 40% inhibition and this value was of 50% at 10 µM. Nevertheless, at the same concentrations, the inhibition of GFP synthesis from the wild-type construction reaches 75 and 90%, respectively (Figure 5B, right).
To investigate if the inhibition of the GFP synthesis induced by G4 ligands was due to a posttranscriptional effect, we next quantified the RNA molecules. As shown in Figure 5C, 10 µM 360A did not show any significant effect on RNA production from either pWUTRF2 plasmid or its mutant counterpart pMUTRF2 (Figure 5C, left and right). For Phen-DC(3), a limited 10% inhibition effect was observed only on RNA production from pMUTRF2 plasmid. In contrast, Phen-DC(6) shows a significant inhibition effect on RNA production from both pWUTRF2 and pMUTRF2 plasmids. At 10 µM, Phen-DC(6) provokes a 25% inhibition of the RNA production for both constructions (Figure 5C) which cannot account for the profound and preferential inhibition of GFP synthesis observed from the pWUTRF2 plasmid under these conditions (Figure 5B, right). Together, these results indicate that the inhibition of the protein production induced by the G4 ligands used in this report is mainly due to their ability to interfere with a posttranscriptional process.
DISCUSSION
In the past years, several works have reported the presence of stable G4 structures in RNA molecules. For instance RNA G4-forming sequences have been found in the HIV-genomic RNA, hTERT, FMR1, FGF-2, NRAS, Zic-1, IGF-II, ESR1 and MT3-MMP mRNAs (40–48). In this report, we show that a G4-forming sequence, located 19 nt upstream of the initiation codon of the TRF2 protein, the 91TRF2G sequence, represses protein synthesis in vitro and is able to reduce the translation rate of a reporter gene in human cells.
Biophysical studies of the 91TRF2G:RNA sequence using CD, UV melting and FRET analysis suggest that the G4 RNA structure is extremely stable and could be formed under physiological conditions. Remarkably, this sequence may adopt the features of a stable G4 (typical G4 CD spectrum and ΔTm >20°C) for K+ concentrations as low as 1 mM. Native gel electrophoresis, as well as the independence of the Tm-values over a 10-fold strand concentration range, is consistent with the formation of an intramolecular quadruplex rather than a dimer. The formation of G4 structures is entirely dependent on Hoogsteen hydrogen bonding established between guanine nucleotides, and the guanine modifications introduced in the mut91TRF2G:RNA sequence completely abolish the formation of the RNA quadruplex, as evidenced by CD and UV melting analysis. Interestingly, the 91TRF2G:RNA sequence is characterized by a ‘Type I spectra’ with a trough at 240 nm and a positive peak at 263 nm, which are the main features of parallel G4 structures (34). Although the structural characterization of this quadruplex remains to be determined, the parallel folding of a RNA quadruplex is highly probable due to the more favored C3′-endo conformation of the ribose ring (49), although a combination of C2′-endo and C3′-endo has also been observed (50).
Stable secondary RNA structures located in the 5′-UTRs of mRNA have been shown to regulate translation (51). For example, in bacteria, a RNA G4 motif located directly upstream of the Shine–Dalgarno sequence showed a very strong inhibition of the translation of a reporter gene (52). In human cells, the RNA G4 motif located in the 5′-UTR region of Zic-1 and MT3-MMP mRNAs have been shown to repress translation (40,46). Furthermore, using in vitro and functional assays, Halder and coworkers (53) have shown that natural and synthetic G4 motifs introduced into the 5′-UTR region of a mRNA reporter system act as universal translational suppressors in mammalian cell lines.
In our study, using a GFP reporter system, we show that the 91TRF2:RNA G4-forming sequence, located in the 5′-UTR region of the TRF2 mRNA, causes a 2.4-fold decrease of the expression of a reporter gene in vitro and a 2.8-fold decrease in the rate of expression in human cells (Figures 3 and 4). Interestingly, our results indicate that the 91TRF2G quadruplex motif does not affect the GFP mRNA accumulation both in vitro and in cellulo, as compared to its mutated counterpart (Figures 3 and 4). Taken together, these observations show that the G4 motif formed by the 91TRF2G sequence located within the 5′-UTR region of the TRF2 mRNA downregulates gene expression by a translational suppressor mechanism.
In a previous work, Kumari and colleagues (54) demonstrated that translation modulation by an RNA G4-forming sequence is dependent on its position in the 5′-UTR of the message, with the inhibition of translation occurring only when the G4-forming-sequence is rather proximal to the 5′-cap. In our constructions, the G4 motif adopted by the 91TRF2G:RNA sequence is located at 92 bases downstream of the putative transcriptional start site of T7 polymerase or at 125 bases downstream of the putative transcriptional start site of CMV promoter. Since in the normal context, this sequence is located closer to the 5′-end of the TRF2 mRNA (90 bases), we can reasonably suppose that its repressive effect on translation may be at least as efficient as what we observe with the reporter constructions.
The search for specific G4 ligands was initially directed to target DNA telomere repeats and lead to the synthesis of compounds that present marked effects on telomere replication and capping (55). Interestingly, these compounds did not present potent selectivity towards other G4 structures (56), and several reports indicated that these ligands should not be considered as simple telomere ‘binders’ (57) and may act in extratelomeric regions (58,59), or at the mRNA level (60) to modulate gene promoter expression (61) and rDNA biosynthesis (62).
The 360A molecule, a pyridine dicarboxamide derivate, Phen-DC(3) and Phen-DC(6), two bisquinolinium compounds, are three G4 ligands among the most potent ones reported so far (63). In this study, we demonstrated that these G4 ligands bind to the 91TRF2G:RNA sequence and that this binding is dependent on the ability of this sequence to adopt a G4 structure. Thus, as evaluated by competition FRET experiments, these ligands exhibit a clear affinity for the 91TRF2G:RNA sequence, while they are not able to bind the mutant counterpart, which did not show the main features associated to G4 formation (Figure 2).
Using an in vitro coupled transcription–translation system, we demonstrated that the binding of G4 ligands to the 91TRF2G:RNA was correlated with their ability to inhibit the protein synthesis in vitro. Thus, even if at 10 µM G4 ligands exhibit an inhibitory effect on the protein synthesis from the control plasmid pMUTF2 (∼20% inhibition for 360A and 60% inhibition for bisquinolinium compounds): the inhibition effect on the expression of the GFP protein from the construction containing the G4-forming sequence was ∼60% for the 360A and reached 90% for bisquinolinium compounds. In our assay, bisquinolinium compounds are more active than the 360A molecule. While, at 1 µM 360A molecule has a limited inhibitory effect on the expression of GFP protein from pWUTRF2 plasmid, Phen-DC(3) and Phen-DC(6) inhibit the protein synthesis by two-fold. Interestingly, competition FRET studies show that, in the presence of potassium, the binding of 360A to the 91TRF2G:RNA was weaker than the binding of bisquinolinium compounds to the same sequence (Table 4). When compared to two others molecules used in this study, Phen-DC(3) exhibits a more selective effect. At 3 µM, Phen-DC(3) molecule inhibits GFP expression from pWUTRF2 by four-fold, whereas it does not affect the expression of the GFP protein from the control plasmid. For comparison, the commonly used G4 ligand TMPyP4, show no RNA G4 specificity (64), and in a recent publication Bugaut and coworkers (64) identified a pyridine-2,6-bis-quinolino-dicarboxamide derivate, RR82 that exhibits an two-fold inhibitory effect on NRAS 5′-UTR G4-forming sequence at 10 µM concentration.
More interestingly, we demonstrated that G4 ligands used in this report do not affect the RNA production and/or stability, as evaluated by RT-PCR analyses (Figure 5C). Although we cannot exclude that in a natural chromatin context, the folding of the 91TRF2G sequence in a G4 structure could also affect the transcription of the endogenous TRF2 mRNA; all reports so far that demonstrate the impact of G4 DNA structures on transcriptional processes concern G4-forming sequences located in the promoter regions (13).
Taken together, these observations are consistent with G4 ligands inhibiting protein synthesis via interactions that stabilize the 91TRF2G:RNA G4 motif. Furthermore, our study demonstrated the proof-of-concept for G4 ligand-mediated regulation of translation by targeting natural G4 RNA motifs.
Mice overexpressing TRF2 in the skin develop both spontaneous and carcinogenic-induced epithelial tumors (65). Conversely, downregulation of TRF2 was found to reduce the tumorigenic potential of grafted tumors in mice (66). Thus, these studies suggest that the level of TRF2 protein in human cells may play an important role on cell fate and cancer development. Several reports show that TRF2 is overexpressed in human tumors (24–26). Although most of these works have evaluated TRF2 expression at the mRNA level, Nijjar and coworkers (25) have shown that the upregulation of the TRF2 protein in tumor cells, as compared to normal cells results from differences in posttranscriptional regulation, since their mRNA levels remained comparable. According to the data reported here, it may be interesting to check if G4-forming sequences in TRF2 mRNA could be implicated in part of the changes in TRF2 protein level associated with cancer.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Institut National Contre le Cancer (TELINCA program) and Ligue Nationale Contre le Cancer (‘équipes labellisées’, to P.C., D.G., B.S., J-.F.R. and A.G., in part). Funding for open access charge: CNRS - Délégation Midi-Pyrénées.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
P.C. is a scientist from INSERM, France.
REFERENCES
- 1.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc. Natl Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sen D, Gilbert W. Guanine quartet structures. Meth. Enzymol. 1992;211:191–199. doi: 10.1016/0076-6879(92)11012-8. [DOI] [PubMed] [Google Scholar]
- 3.Sen D, Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 4.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Webba da Silva M. Geometric formalism for DNA quadruplex folding. Chemistry. 2007;13:9738–9745. doi: 10.1002/chem.200701255. [DOI] [PubMed] [Google Scholar]
- 7.Monchaud D, Teulade-Fichou MP. A hitchhiker’s guide to G-quadruplex ligands. Org. Biomol. Chem. 2008;6:627–636. doi: 10.1039/b714772b. [DOI] [PubMed] [Google Scholar]
- 8.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Johnson JE, Smith JS, Kozak ML, Johnson FB. In vivo veritas: using yeast to probe the biological functions of G-quadruplexes. Biochimie. 2008;90:1250–1263. doi: 10.1016/j.biochi.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008;36:1321–1333. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oganesian L, Karlseder J. Telomeric armor: the layers of end protection. J. Cell. Sci. 2009;122:4013–4025. doi: 10.1242/jcs.050567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl Acad. Sci. USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 19.de Lange T. T-loops and the origin of telomeres. Nat. Rev. Mol. Cell Biol. 2004;5:323–329. doi: 10.1038/nrm1359. [DOI] [PubMed] [Google Scholar]
- 20.Stansel RM, de Lange T, Griffith JD. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 2001;20:5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bombarde O, Boby C, Gomez D, Frit P, Giraud-Panis MJ, Gilson E, Salles B, Calsou P. TRF2/RAP1 and DNA-PK mediate a double protection against joining at telomeric ends. EMBO J. 2010;29:1573–1584. doi: 10.1038/emboj.2010.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 23.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 24.Oh BK, Kim YJ, Park C, Park YN. Up-regulation of telomere-binding proteins, TRF1, TRF2, and TIN2 is related to telomere shortening during human multistep hepatocarcinogenesis. Am. J. Pathol. 2005;166:73–80. doi: 10.1016/S0002-9440(10)62233-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nijjar T, Bassett E, Garbe J, Takenaka Y, Stampfer MR, Gilley D, Yaswen P. Accumulation and altered localization of telomere-associated protein TRF2 in immortally transformed and tumor-derived human breast cells. Oncogene. 2005;24:3369–3376. doi: 10.1038/sj.onc.1208482. [DOI] [PubMed] [Google Scholar]
- 26.Hsu CP, Ko JL, Shai SE, Lee LW. Modulation of telomere shelterin by TRF1 [corrected] and TRF2 interacts with telomerase to maintain the telomere length in non-small cell lung cancer. Lung Cancer. 2007;58:310–316. doi: 10.1016/j.lungcan.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 27.Guedin A, Lacroix L, Mergny JL. Thermal melting studies of ligand DNA interactions. Methods Mol. Biol. 2010;613:25–35. doi: 10.1007/978-1-60327-418-0_2. [DOI] [PubMed] [Google Scholar]
- 28.Mergny JL, Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- 29.Mergny JL, Li J, Lacroix L, Amrane S, Chaires JB. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005;33:e138. doi: 10.1093/nar/gni134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mergny JL, Phan AT, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 31.De Cian A, Guittat L, Kaiser M, Sacca B, Amrane S, Bourdoncle A, Alberti P, Teulade-Fichou MP, Lacroix L, Mergny JL. Fluorescence-based melting assays for studying quadruplex ligands. Methods. 2007;42:183–195. doi: 10.1016/j.ymeth.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 32.Kejnovska I, Kypr J, Vorlickova M. Oligo(dT) is not a correct native PAGE marker for single-stranded DNA. Biochem. Biophys. Res. Commun. 2007;353:776–779. doi: 10.1016/j.bbrc.2006.12.093. [DOI] [PubMed] [Google Scholar]
- 33.Kikin O, D’Antonio L, Bagga PS. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balagurumoorthy P, Brahmachari SK, Mohanty D, Bansal M, Sasisekharan V. Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res. 1992;20:4061–4067. doi: 10.1093/nar/20.15.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palumbo SL, Memmott RM, Uribe DJ, Krotova-Khan Y, Hurley LH, Ebbinghaus SW. A novel G-quadruplex-forming GGA repeat region in the c-myb promoter is a critical regulator of promoter activity. Nucleic Acids Res. 2008;36:1755–1769. doi: 10.1093/nar/gkm1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Granotier C, Pennarun G, Riou L, Hoffschir F, Gauthier LR, De Cian A, Gomez D, Mandine E, Riou JF, Mergny JL, et al. Preferential binding of a G-quadruplex ligand to human chromosome ends. Nucleic Acids Res. 2005;33:4182–4190. doi: 10.1093/nar/gki722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Cian A, Delemos E, Mergny JL, Teulade-Fichou MP, Monchaud D. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 2007;129:1856–1857. doi: 10.1021/ja067352b. [DOI] [PubMed] [Google Scholar]
- 40.Arora A, Dutkiewicz M, Scaria V, Hariharan M, Maiti S, Kurreck J. Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA. 2008;14:1290–1296. doi: 10.1261/rna.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balkwill GD, Derecka K, Garner TP, Hodgman C, Flint AP, Searle MS. Repression of translation of human estrogen receptor by G-quadruplex formation. Biochemistry. 2009;48:11487–11495. doi: 10.1021/bi901420k. [DOI] [PubMed] [Google Scholar]
- 42.Bonnal S, Schaeffer C, Creancier L, Clamens S, Moine H, Prats AC, Vagner S. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J. Biol. Chem. 2003;278:39330–39336. doi: 10.1074/jbc.M305580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christiansen J, Kofod M, Nielsen FC. A guanosine quadruplex and two stable hairpins flank a major cleavage site in insulin-like growth factor II mRNA. Nucleic Acids Res. 1994;22:5709–5716. doi: 10.1093/nar/22.25.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gomez D, Aouali N, Londono-Vallejo A, Lacroix L, Megnin-Chanet F, Lemarteleur T, Douarre C, Shin-ya K, Mailliet P, Trentesaux C, et al. Resistance to the short term antiproliferative activity of the G-quadruplex ligand 12459 is associated with telomerase overexpression and telomere capping alteration. J. Biol. Chem. 2003;278:50554–50562. doi: 10.1074/jbc.M308440200. [DOI] [PubMed] [Google Scholar]
- 45.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morris MJ, Basu S. An unusually stable G-quadruplex within the 5′-UTR of the MT3 matrix metalloproteinase mRNA represses translation in eukaryotic cells. Biochemistry. 2009;48:5313–5319. doi: 10.1021/bi900498z. [DOI] [PubMed] [Google Scholar]
- 47.Schaeffer C, Bardoni B, Mandel JL, Ehresmann B, Ehresmann C, Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sundquist WI, Heaphy S. Evidence for interstrand quadruplex formation in the dimerization of human immunodeficiency virus 1 genomic RNA. Proc. Natl Acad. Sci. USA. 1993;90:3393–3397. doi: 10.1073/pnas.90.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang CF, Shafer RH. Engineering the quadruplex fold: nucleoside conformation determines both folding topology and molecularity in guanine quadruplexes. J. Am. Chem. Soc. 2006;128:5966–5973. doi: 10.1021/ja0603958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng J, Xiong Y, Sundaralingam M. X-ray analysis of an RNA tetraplex (UGGGGU)(4) with divalent Sr(2+) ions at subatomic resolution (0.61 A) Proc. Natl Acad. Sci. USA. 2001;98:13665–13670. doi: 10.1073/pnas.241374798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pelletier J, Sonenberg N. The involvement of mRNA secondary structure in protein synthesis. Biochem. Cell Biol. 1987;65:576–581. doi: 10.1139/o87-074. [DOI] [PubMed] [Google Scholar]
- 52.Wieland M, Hartig JS. RNA quadruplex-based modulation of gene expression. Chem. Biol. 2007;14:757–763. doi: 10.1016/j.chembiol.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 53.Halder K, Wieland M, Hartig JS. Predictable suppression of gene expression by 5′-UTR-based RNA quadruplexes. Nucleic Acids Res. 2009;37:6811–6817. doi: 10.1093/nar/gkp696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumari S, Bugaut A, Balasubramanian S. Position and stability are determining factors for translation repression by an RNA G-quadruplex-forming sequence within the 5′ UTR of the NRAS proto-oncogene. Biochemistry. 2008;47:12664–12669. doi: 10.1021/bi8010797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neidle S. Human telomeric G-quadruplex: the current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 2010;277:1118–1125. doi: 10.1111/j.1742-4658.2009.07463.x. [DOI] [PubMed] [Google Scholar]
- 56.Lemarteleur T, Gomez D, Paterski R, Mandine E, Mailliet P, Riou JF. Stabilization of the c-myc gene promoter quadruplex by specific ligands’ inhibitors of telomerase. Biochem. Biophys. Res. Commun. 2004;323:802–808. doi: 10.1016/j.bbrc.2004.08.150. [DOI] [PubMed] [Google Scholar]
- 57.De Cian A, Cristofari G, Reichenbach P, De Lemos E, Monchaud D, Teulade-Fichou MP, Shin-Ya K, Lacroix L, Lingner J, Mergny JL. Reevaluation of telomerase inhibition by quadruplex ligands and their mechanisms of action. Proc. Natl Acad. Sci. USA. 2007;104:17347–17352. doi: 10.1073/pnas.0707365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pennarun G, Granotier C, Gauthier LR, Gomez D, Hoffschir F, Mandine E, Riou JF, Mergny JL, Mailliet P, Boussin FD. Apoptosis related to telomere instability and cell cycle alterations in human glioma cells treated by new highly selective G-quadruplex ligands. Oncogene. 2005;24:2917–2928. doi: 10.1038/sj.onc.1208468. [DOI] [PubMed] [Google Scholar]
- 59.Gomez D, Wenner T, Brassart B, Douarre C, O’Donohue MF, El Khoury V, Shin-Ya K, Morjani H, Trentesaux C, Riou JF. Telomestatin-induced telomere uncapping is modulated by POT1 through G-overhang extension in HT1080 human tumor cells. J. Biol. Chem. 2006;281:38721–38729. doi: 10.1074/jbc.M605828200. [DOI] [PubMed] [Google Scholar]
- 60.Gomez D, Lemarteleur T, Lacroix L, Mailliet P, Mergny JL, Riou JF. Telomerase downregulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Res. 2004;32:371–379. doi: 10.1093/nar/gkh181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Drygin D, Siddiqui-Jain A, O’Brien S, Schwaebe M, Lin A, Bliesath J, Ho CB, Proffitt C, Trent K, Whitten JP, et al. Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Cancer Res. 2009;69:7653–7661. doi: 10.1158/0008-5472.CAN-09-1304. [DOI] [PubMed] [Google Scholar]
- 63.Monchaud D, Allain C, Bertrand H, Smargiasso N, Rosu F, Gabelica V, De Cian A, Mergny JL, Teulade-Fichou MP. Ligands playing musical chairs with G-quadruplex DNA: a rapid and simple displacement assay for identifying selective G-quadruplex binders. Biochimie. 2008;90:1207–1223. doi: 10.1016/j.biochi.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 64.Bugaut A, Rodriguez R, Kumari S, Hsu ST, Balasubramanian S. Small molecule-mediated inhibition of translation by targeting a native RNA G-quadruplex. Org. Biomol. Chem. 2010;8:2771–2776. doi: 10.1039/c002418j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Munoz P, Blanco R, Flores JM, Blasco MA. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat. Genet. 2005;37:1063–1071. doi: 10.1038/ng1633. [DOI] [PubMed] [Google Scholar]
- 66.Biroccio A, Rizzo A, Elli R, Koering CE, Belleville A, Benassi B, Leonetti C, Stevens MF, D’Incalci M, Zupi G, et al. TRF2 inhibition triggers apoptosis and reduces tumourigenicity of human melanoma cells. Eur. J. Cancer. 2006;42:1881–1888. doi: 10.1016/j.ejca.2006.03.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.